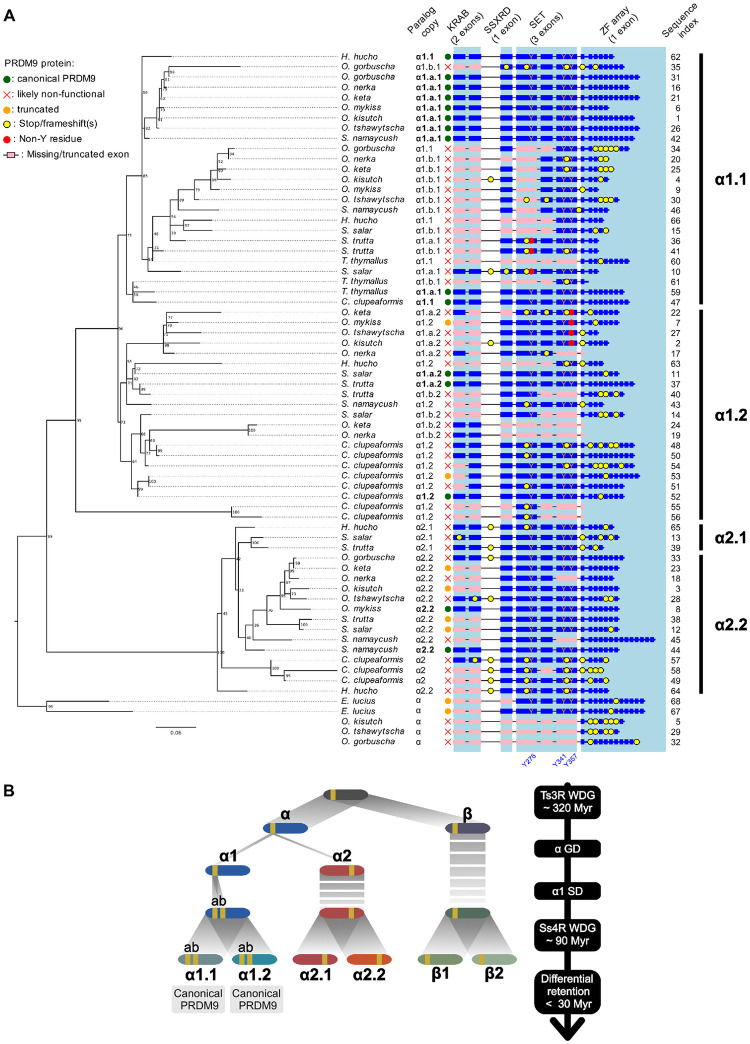
Fig 1. Prdm9 duplication history in salmonids.
(A) Phylogenetic tree of Prdm9α paralogs in 12 salmonids and northern pike (Esox lucius) as outgroup species. Prdm9β is shown in S1 Fig. The phylogenetic tree was computed on the concatenated 6 exons of the 3 canonical PRDM9 domains KRAB, SSXRD, and SET, with 1,000 bootstrap replicates (values shown). The columns, from left to right, indicate the (i) species name; (ii) annotated paralog copy (in bold: full-length copy without pseudogenization); (iii) Prdm9 copy status. Prdm9α clusters into 2 main groups (α1 and α2) that are divided in 2 subgroups (α1.1/α1.2 and α2.1/α2.2). The scale bar is in unit of substitution per site. The right panel shows the coding potential of each paralog, and indicates the presence of frame-shifting mutations or stop codons, and of substitutions in the catalytic tyrosines of the SET domain (Y276, Y341, and Y357). Canonical (full length) Prdm9 proteins contain 4 key domains: KRAB (encoded by 2 exons), SSXRD (encoded by 1 exon), SET (encoded by 3 exons), and the ZF array (encoded by 1 exon). Complete exons are shown in blue. Missing or truncated exons are shown in pink. Other regions of the protein (upstream of the KRAB domain, and between KRAB and SSXRD) are encoded by additional exons (not shown here), that are not conserved between α1 and α2 clades. Paralogs were classified as “canonical PRDM9” if they contained all exons encoding the 4 key domains, without any frameshift/non-sense mutation (at least up to the first ZF) [NB: some sequences contain frameshifts or non-sense mutations in the ZF array. This leads to a shortened ZF array, but does not necessarily impair the function of PRDM9]. Paralogs were classified as “likely non-functional” if they contained frameshifts or non-sense mutations, or if they missed at least 1 SET exon. Other cases were classified as “truncated.” The 3 last α copies, belonging to O. kisutch, O. tshawytscha, and O. gorbuscha, have lost the 3 domains KRAB, SSXRD, and SET, but have kept their ZF exons, and were therefore added below the phylogenetic tree. The last column indicates the sequence indexes referring to the S1 Table with additional information on the corresponding copy. (B) Consensus history of Prdm9 duplication events in salmonids. After the teleost-specific WGD (Ts3R WDG), the chromosomes of the common ancestor of teleosts were duplicated. Two ohnolog chromosomes arose from the one carrying the ancestral Prdm9 locus: one carrying the Prdm9α copy and the other the Prdm9β copy. GD of the α paralog (referred to as α1) led to the appearance of a new α copy (α2) on another chromosome. The α1 copy (becoming α1.a) then underwent an SD, generating a α1.b copy in tandem on the same chromosome. By this time, the β paralog had lost the KRAB and SSXRD domains. Lastly, the 4 copies were duplicated during the salmonids-specific Ss4R WGD, with the newly formed paralogs (annotated α1.a.2, α1.b.2, α2.2, β2) on ohnolog chromosomes. One full-length copy was retained in each species. The Salmo genus (S. trutta and S. salar) retained the α1.2 copy, whereas all other salmonids retained the α1.1 copy. A second full-length PRDM9 was also retained in C. clupeaformis (α1.2), O. mykiss (α2.2), and S. namaycush (α2.2). Ohnolog chromosomes are represented with similar color shades (i.e., blue, red, and green) and Prdm9 locus in yellow. This global picture of the duplication events in the salmonid history does not show other independent lineage-specific duplication events and losses. The data and codes underlying this figure can be found in https://doi.org/10.5281/zenodo.11083953. GD, gene duplication; WGD, whole genome duplication; ZF, zinc finger.