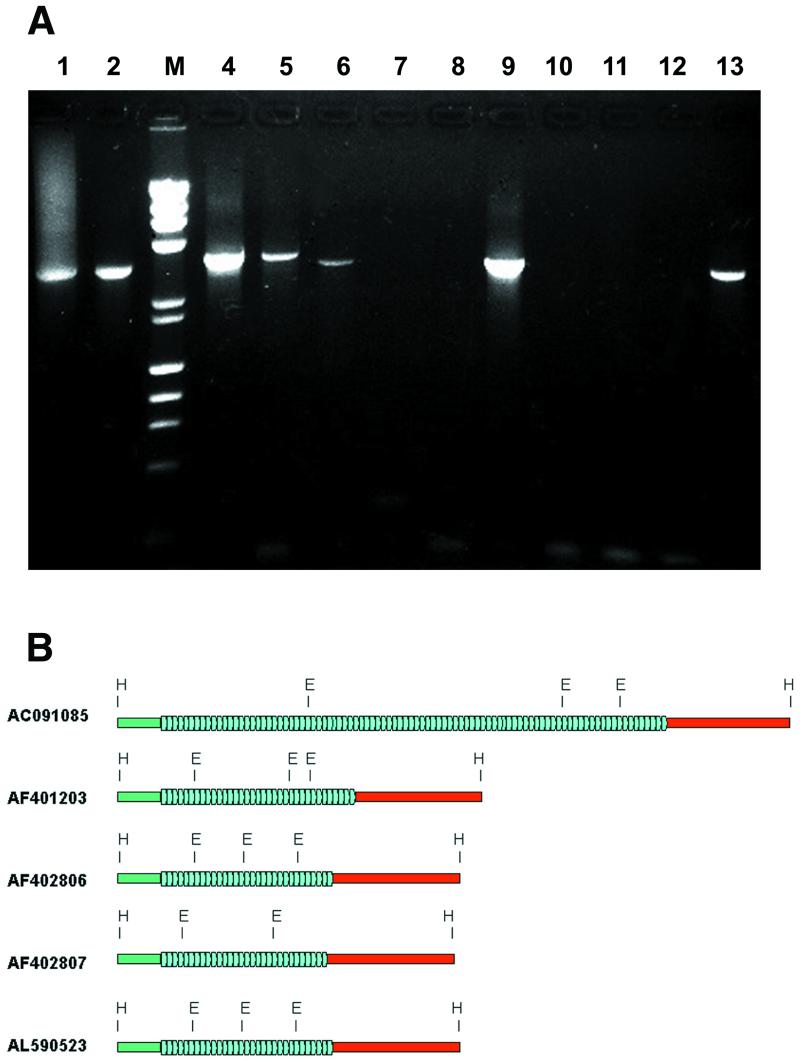
Figure 3.
(A) PCR amplification of the chAB4–YNF1 junction region. Amplification was done using primers JCT F (chAB4 side) and JCT R (YNF1 side) on human genomic DNA (lane 1), YAC 857_b_10 (lane 2), and PAC clones 635H14, 635J14, 654D10, 654O8, 655I18, 659M14, 662K13, 663D9, 668N10 and 670G14 (lanes 4–13). Extension step during amplification was 10 min. M, size marker lane, mixture of λ HindIII + ΦX174 HaeIII digests. (B) Structure of five independent chAB4–NF1 family junctions. Flanking sequences on the chAB4 side (green bar) and the NF1-related side (red bar), and the tandemly repeated array of degenerate 48 bp satellite units (blue arrowheads) are indicated. The organization of junctions from AC091085 sequence from chromosome 17 (from GenBank), PAC 635H14 (AF401203, this work), PAC 659M14 (AF402806, this work), chromosome 22 YAC 857_b_10 (AF402807, this work), and from the chromosome 22-derived GenBank entry AL590523 are compared. In all cases, these connecting sequences reside in a HindIII restriction fragment. Sequences on either side of the satellite array are highly conserved: 360 bp (green bars) on the chAB4 side was found to be 96% identical in all five junctions. Similarly, the NF1-like regions (red bars) displayed 94% sequence homology. However, substantial heterogeneity was observed in the number of satellite repeats. On the one hand, the copy-number of 48 bp repeats is variable (blue arrowheads); on the other hand, the repeat units show sequence divergence. Polymorphic EcoRI sites in the 48 bp satellite are indicated.