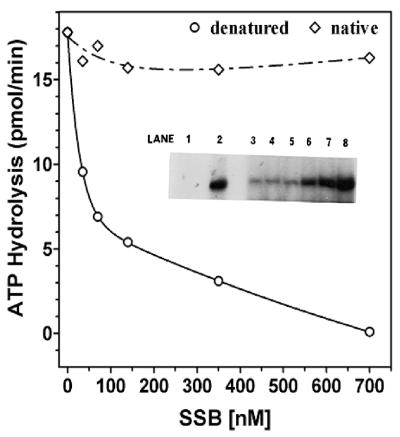
Figure 6.
Influence of DNA structure on SSB inhibition of ssDNA stimulated ATPase. Standard ATPase assays were carried out for a period of 10 min in the presence of 200 pmol (as nucleotide) of poly(dT):oligo(dA)35–(dC)6. The reaction mixtures contained 3.33 nM DnaB protein and varying amounts of SSB as indicated. Heat-denatured DNA co-factor (circles); native DNA co-factor (diamonds). Inset, influence of E.coli SSB on DNA unwinding of poly(dT):oligo(dA)35–(dC)6. A standard helicase assay was carried out in the presence of increasing amounts of E.coli SSB using 16.7 nM DnaB, and poly(dT):oligo(dA)35–(dC)6 as a substrate. Increased DNA unwinding corresponds to increased amounts of liberated [α-32P]oligo(dA)35–(dC)6. Lane 1, native substrate; lane 2, heat-denatured substrate; lane 3, no SSB; lane 4, 35 nM SSB; lane 5, 70 nM SSB; lane 6, 140 nM SSB; lane 7, 0.35 µM SSB; lane 8, 0.7 µM SSB.