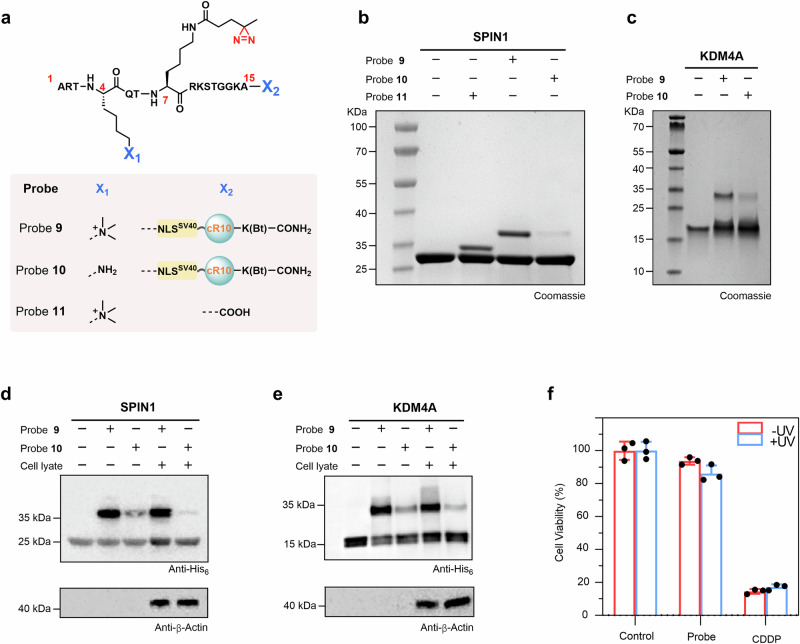
Fig. 3. Nucleus-targeted histone-tail-based photoaffinity probe to capture readers of H3K4me3 in vitro.
a Chemical structure of probes 9 to 11. b, c SDS-PAGE analysis of the labeling of SPIN1 and KDM4A by probe 9. Probes 10 and 11 were used for comparison for SPIN1 (b), and probe 10 was used for comparison for KDM4A (c). Gel images shown in (b, c) are representative of independent biological replicates (n = 2). d, e Cell lysate profiling using probe 9 with probe 10 as a comparison. Labeled proteins were analyzed by immunoblotting using an anti-His6 antibody. Immunoblotting images shown in (d, e) are representative of independent biological replicates (n = 2). f Viability of HeLa cells treated with 60 μM probe 9. Data were plotted as mean ± SEM of three independent biological replicates.