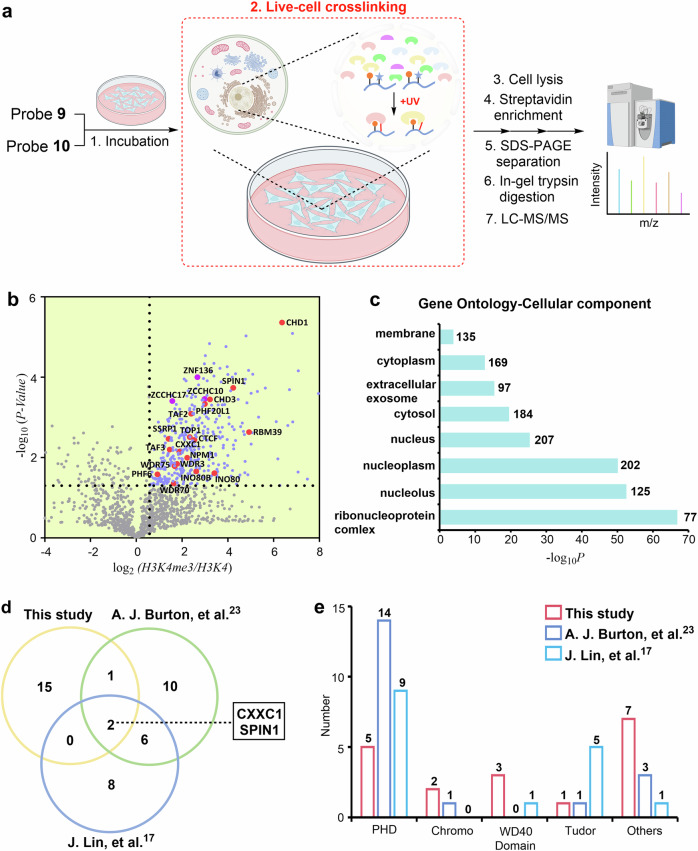
Fig. 4. Determining the in situ interactome of H3K4me3 in HeLa cells.
a Schematic for the LFQ proteomics workflow to determine the interactome of H3K4me3 by probe 9. Probe 10 was used for comparison. Some elements of Fig. 4(a) were created in BioRender. Chu, G. (2024) https://BioRender.com/u07k630. b Volcano plots of the quantitative mass spectrometry results. Significantly enriched hits (p < 0.05, >1.5-fold-change) are colored blue. Some established H3K4me3 reader proteins are highlighted and labeled in red. A two-sided test was used. c GO analysis (cellular components) of significantly enriched hits (p < 0.05, >1.5-fold-change) in Fig. 4b. The number of proteins in each GO term is shown. d Venn diagram comparison of identified H3K4me3 reader proteins in the present study and two previous studies, respectively. e Number of identified H3K4me3 reader proteins containing PHD, Chromo, WD40, and Tudor domains in the present study and two previous studies. Source data are provided as a Source Data file.