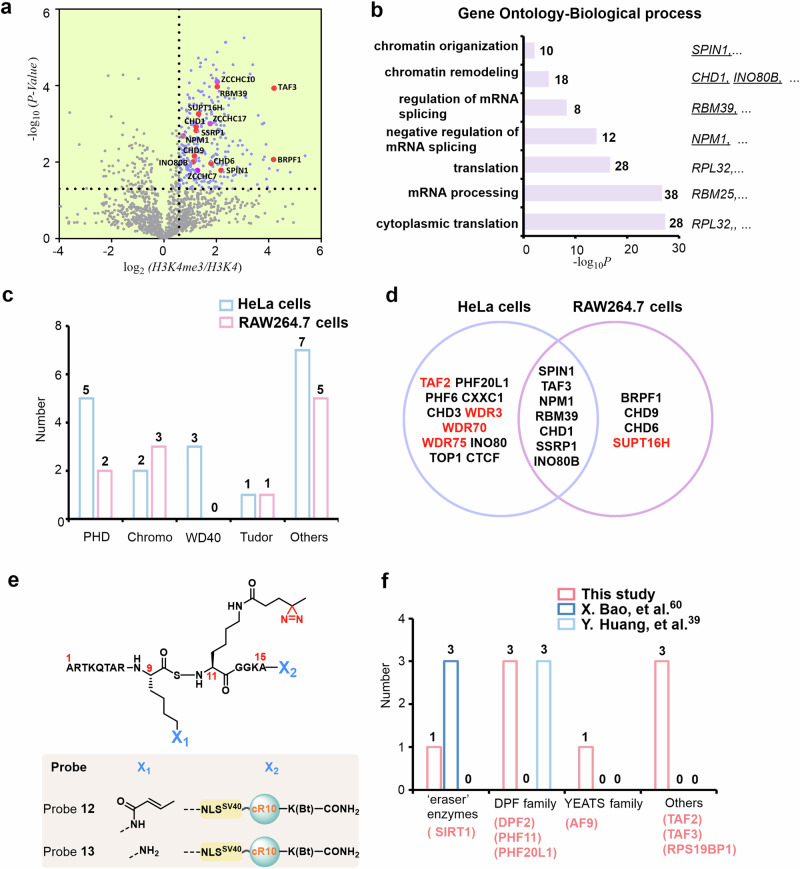
Fig. 5. Determining the interactome of H3K4me3 and H3K9cr in RAW264.7 cells.
a Volcano plots of mass spectrometry results. Hits significantly enriched by probe 9 (p < 0.05, >1.5-fold-change) are colored blue. Some established H3K4me3 reader proteins are highlighted and labeled in red. A two-sided test was used. b GO analysis (biological process) of significantly enriched hits (p < 0.05, >1.5-fold-change) in Fig. 5a. The number of proteins in each GO term is shown. c Number of identified H3K4me3 reader proteins containing PHD, chromo, WD40, and Tudor domains by probe 9 in HeLa and RAW264.7 cells. d Overlap of identified H3K4me3 reader proteins using probe 9 in HeLa and RAW264.7 cells. e Chemical structure of probe 12 and probe 13. f Number of identified H3K9cr reader proteins containing eraser enzymes, DPF, YEATS, and other families in the present study and two previous studies. Red-colored words indicate H3K9cr reader proteins enriched in this study. Source data are provided as a Source Data file.