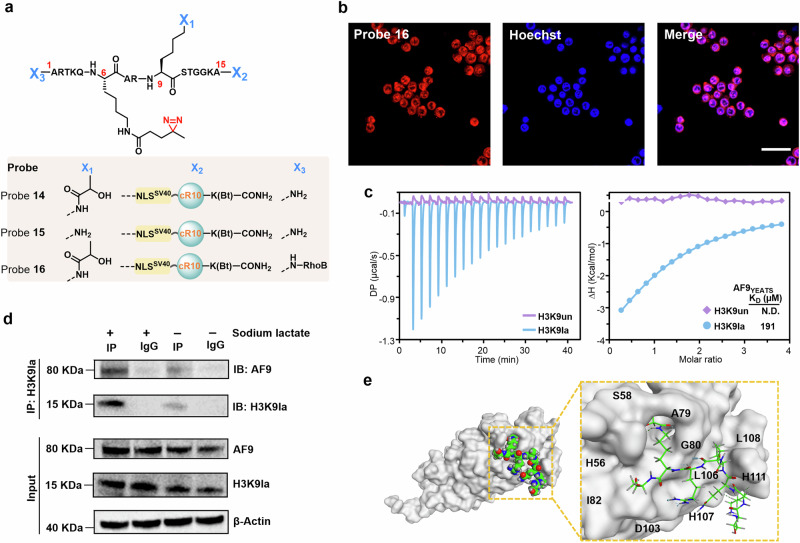
Fig. 6. Determining the in situ interactome of H3K9la.
a Chemical structure of probes 14, 15, and 16. b Confocal microscopy images of RAW264.7 cells treated with probe 14 for 1 h at 37 °C. Probe 14 was visualized using TER fluorescence (red channel), and Hoechst 33258 was utilized for nuclear staining (blue channel). Scale bars: 20 μm. Confocal microscopy images shown in (b) are representative of independent biological replicates (n = 3). c ITC fitting curves of AF9YEATS with H31-10K9la (blue) peptide or H31-10 (red) peptide. d Co-IP assay was conducted to detect the interaction of H3K9la and AF9 in RAW264.7 cells with or without sodium lactate treatment. Immunoblotting images shown in (d) are representative of independent biological replicates (n = 3). e Structural modeling of AF9YEATS and H31-10K9la peptide. The AF9YEATS structure was displayed on a gray surface, the ligand is shown in sphere and sticks, the carbon atoms are green, the oxygen atoms are red, and the nitrogen atoms are blue. Source data are provided as a Source Data file.