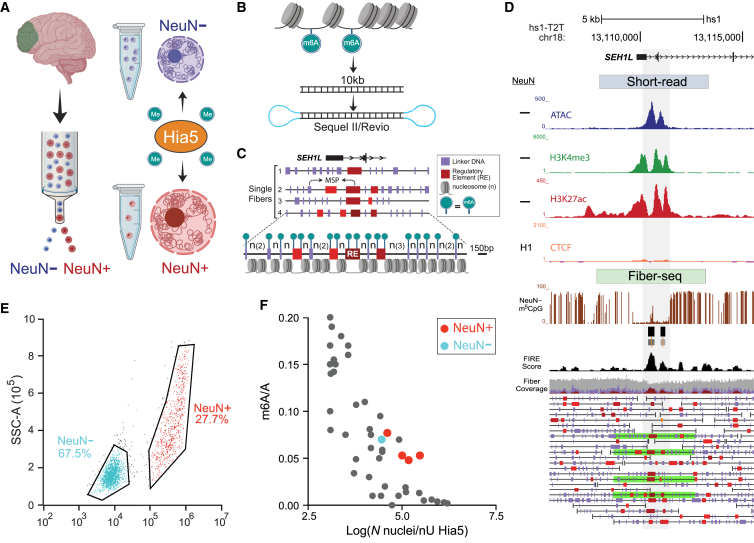
Figure 1.
Single chromatin fiber long-read epigenomic profiling by cell type in human brain
(A–D) Work flow. (A) Fluorescence-activated cell sorting (FACS) of NeuN immunostained neuronal and non-neuronal nuclei from the region of interest (PFC) and in situ incubation of the intact nuclei with adenine methyltransferase Hia5 for m6A tagging of extranucleosomal genomic DNA, followed by (B) amplification-free PacBio (Sequel II or Revio) long-read sequencing of ∼10-kb single DNA molecules derived directly from the brain nuclei. (C and D) Early step computational analyses include annotation to the telomere-to-telomere T2T-CHM13v2.0/hs1 genome, mapping of Hia5 methylation-sensitive patches (MSPs)/nucleosome-depleted regions, and calling of linker DNA (purple) and regulatory elements (red, brown), such as promoters and enhancers, at single-fiber resolution. (C) Example fiber visualizations were taken from the NeuN− Fiber-seq reference set, showing the promoter region of the chr18 SEH1L gene locus. Portions of fibers shown (fiber 1–4) are marked by green bars in (D). Fiber 4 in (C) is shown at enlarged scale, highlighting Fiber-seq-generated nucleosomal position map at single-nucleosome resolution. (D) hs1-T2T genome browser shots for PFC non-neuronal chromatin profiles at the SEH1L locus, including (top to bottom) “peak” landscapes for (blue) ATAC, (green) H3K4me3, (red) H3K27ac-seq, and (orange) CTCF ENCODE (Enc.) neural cell line (CTCF short-read-based peak landscapes). Fiber-seq long-read sequencing: (brown) m5CpG tracks, (black) Fiber-seq FIRE scores of MSPs. NeuN− single DNA molecules = single chromatin fibers as indicated; green-shaded single DNA molecules/chromatin fibers are highlighted as representative fiber examples shown in (C), including the fourth fiber at 150 bp resolution. Purple vertical rectangles mark linker DNA. Red/brown MSPs scoring as regulatory elements (P, promoter, and other REs, regulatory elements) on fiber (FIRE false discovery rate [FDR], orange, 0.10 ≥ q > 0.05; bright red, 0.05 ≥ q > 0.01; dark red-brown, q ≤ 0.01).
(E) Representative FACS showing efficient separation of non-neuronal (NeuN−) from neuronal (NeuN+) nuclei.
(F) Genome-scale proportion of methyladenines (m6A/total A; y axis) in 46 Fiber-seq-processed samples of human brain nuclei. Each sample/aliquot was sequenced at low depth (see text), and the m6A/A was computed as an average across all DNA molecules of a sample. The x axis marks the number of nuclei/nU Hia5 enzyme for each sample of nuclei (log scale, x axis). Blue dot, NeuN− reference sample sequenced with 11,143,795 reads/median of 30-fold genomic coverage; red dots, NeuN+ samples (4 aliquots × 2 brains) picked for sequencing to a total of 4,749,556 reads/median of 20-fold genomic coverage.