Figure 2.
Hia5 m6A tagging of accessible chromatin and internucleosomal DNA in neurons and non-neurons at single-molecule resolution
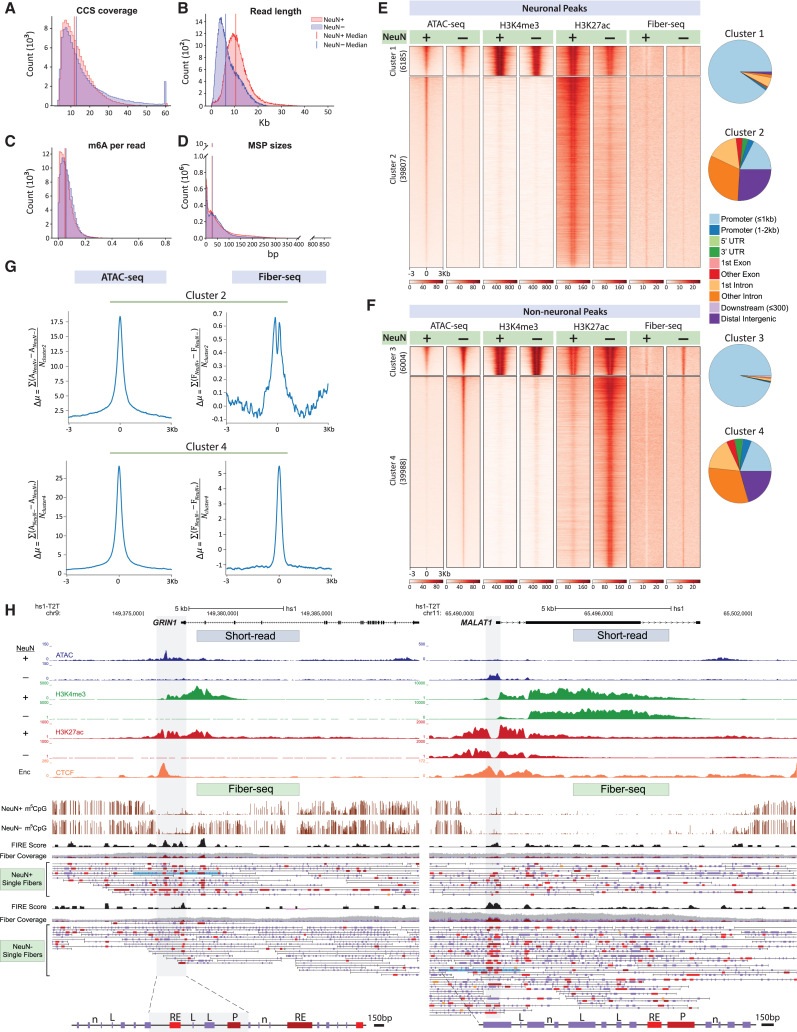
(A–D) Quality controls for PFC NeuN+ (red) and NeuN− (blue) Fiber-seq, including (A) proportional representation of DNA molecules by circular consensus sequence (CCS) coverage, (B) read length, (C) m6A/A per read, and (D) genome-wide counts of (Hia5) methylation sensitive patches (MSPs) (y axis) by base pair length (x axis).
(E and F) Heatmap for 6-kb sequences centered on 45,992 ATAC-seq peaks each for (E) NeuN+ and (F) NeuN−, split by k-means (k = 2) into promoter prominent (NeuN+) ATAC cluster 1 and (NeuN−) cluster 3 and enhancer prominent (NeuN+) ATAC cluster 2 and (NeuN−) cluster 4, as indicated. Numbers in parentheses indicate number of peaks by cluster. Pie charts display the genomic annotation of each cluster. Cell-type-specific alignments (NeuN+, NeuN−) are shown for (left to right) ATAC-seq, H3K4me3 and H3K27ac ChIP-seq, and m6A signal in the (by cell type) corresponding Fiber-seq libraries (see text). Notice strong cell-type-specific NeuN+ versus NeuN− differential regulation for enhancer-dominated clusters 2 and 4 compared to promoter-dominated clusters 1 and 3.
(G) Signal difference by cell type (NeuN+ versus NeuN−) for each of the 39,807 neuronal ATAC-seq peaks from cluster 2 (E) and for each of the 39,988 non-neuronal ATAC-seq peaks from cluster 4 (F), using the formula shown on the y axis of each plot. For each set of enhancer peaks, the difference in ATAC counts and percentage accessibility was computed for every peak at each position and then divided by the total number of peaks in the cluster to determine the average difference in ATAC-seq counts and Fiber-seq percentage accessibility at each position relative to the center of the ATAC-seq peaks. For NeuN+ enhancers, NeuN− signal was subtracted from NeuN+ signal, while for NeuN− enhancers, NeuN+ signal was subtracted from NeuN− signal. The exact equation used to calculate the per-position difference in ATAC-seq counts and Fiber-seq percentage accessibility is noted in the y axis label of each plot. The ATAC-seq counts for NeuN+ and NeuN− are denoted as ANeuN+ and ANeuN−, respectively, while for Fiber-seq percentage accessibility they are referred to as FNeuN+ and FNeuN−. The NeuN+ and NeuN− enhancer ATAC peaks are denoted as Ncluster2 and Ncluster4.
(H) Representative browser shots, ∼15 kb wide, comparing hs1-T2T genome annotated neuronal and non-neuronal chromatin from PFC (top) short-read epigenomic landscapes with (bottom) Fiber-seq signals for (left) GRIN1 NMDA receptor gene locus (chromosome 9q34.3) and (right) long non-coding RNA MALAT1 locus (chromosome 11q13.1) (right). Top to bottom, as indicated, GRIN1 gene map, NeuN+- and, separately, NeuN−- specific “peak” landscapes for (blue) ATAC, (green) H3K4m3, (red) H3K27ac-seq, and (orange) H1 neural cell line (ENCODE) CCCTC-binding factor (CTCF) peak landscape. Fiber-seq long-read sequencing: (brown) m5CpG tracks by cell type, (black) NeuN+ Fiber-seq FIRE scores of methylation-sensitive patches (MSPs); NeuN+ and NeuN− single DNA molecules are as indicated; blue-shaded single DNA molecules/chromatin fibers are highlighted as representative examples and shown at the bottom again at a higher (150 bp) resolution, with purple-colored vertical rectangles marking linker DNA (examples marked by “L” on fiber), with single-nucleosome (“n” on fiber) resolution, and red/brown-colored MSPs scoring as regulatory elements (P, promoter, and RE, regulatory element on fiber) by FIRE analyses (orange, FDR 0.10 ≥ q > 0.05; bright red, FDR 0.05 ≥ q > 0.01; dark red-brown FDR q ≤ 0.01). Notice prominence of FIRE nucleosome-depleted regions (NDRs) for GRIN1 in neuronal and for MALAT1 in non-neuronal fibers.