Figure 3.
Fiber-seq FIRE peak-based analysis of nucleosome-depleted regions (NDRs)
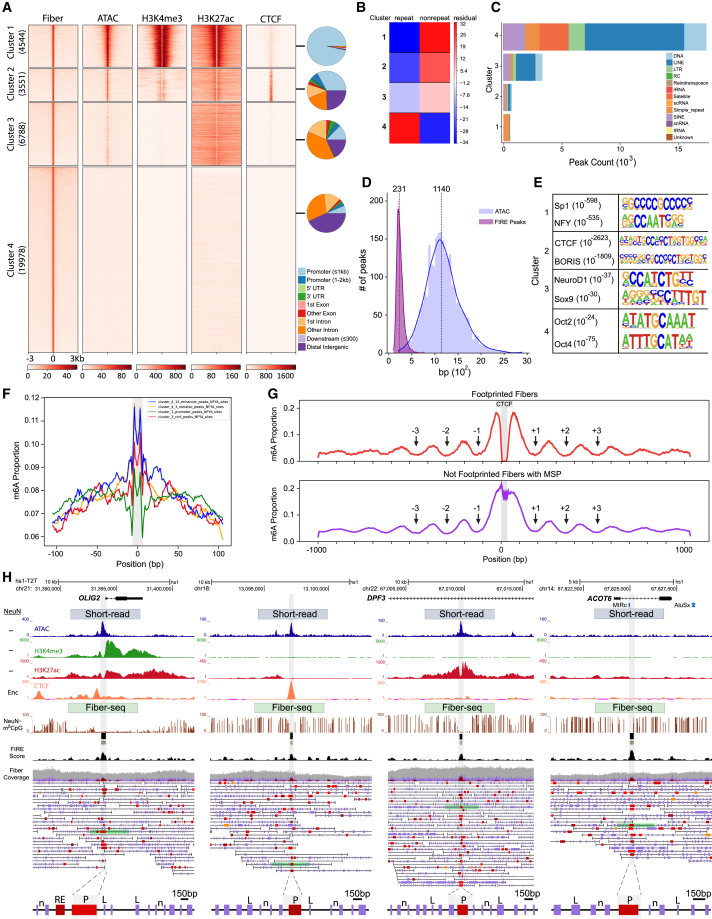
(A) Fiber-seq FIRE-defined peak heatmap of PFC NeuN− nuclei population, with k-means Fiber-seq clusters 1–4 and their alignments by corresponding sequences from PFC NeuN− open chromatin ATAC-seq, transcriptional histone marks H3K4me3 and H3K27ac, and Encode H1 neural cell line CTCF ChIP-seq conventional short-read sequencing.
(B) Cluster-specific enrichment scores for repeat and non-repeat sequences; note that repeat enrichment is specific for cluster 4 (chi-squared, df = 3, p = 2.2 × 10−16).
(C) Total peak count by type of DNA repeat and Fiber-seq cluster, as indicated. Notice that retroelements, including LINEs, SINEs, and LTR-associated fiber peaks (NDRs), are overwhelmingly locating to fiber cluster 4 with only very minimal contributions by clusters 1–3.
(D) PFC NeuN− Fiber-seq and ATAC-seq peak size distribution profiles; notice several-fold sharper peak size in former compared to latter.
(E) Top scoring HOMER regulatory motifs to each for the four Fiber-seq peak clusters, 1–4 (p, Kolmogorov-Smirnov).
(F) Fine-grained regulatory motif footprint for NFY TF with fiber cluster-specific binding scores as indicated (∗∗∗p < 1e−16, Kruskal-Wallis); note sharp footprints centered in NDRs.
(G) Plots depicting the average proportion of adenines that are methylated in fibers centered on n =1,855 specific CTCF binding sites (see text), with fibers with (top) MSP harboring a CTCF footprint and (bottom) MSP with no footprint. Arrows point to local minima adjacent to MSP, indicating strong phasing with well-positioned nucleosomes and cis actuation of flanking nucleosomal array. See also, as additional control, Figure S6F for fibers with a CTCF motif but no MSP.
(H) Representative hs1-T2T genome browser shots of cluster-specific genomic loci, with single-fiber resolution by cluster. (Left to right) Cluster 1 OLIG2, cluster 2 intergenic, cluster 3 DPF3, and cluster 4 ATOC6 gene locus. (Top to bottom) PFC NeuN−-specific “peak” landscapes for (blue) ATAC, (green) H3K4me3, (red) H3K27ac-seq, and (orange) CTCF ENCODE neural cell line CTCF short-read-based peak landscapes. Fiber-seq long-read sequencing: (brown) m5CpG tracks by cell type, (black) NeuN+ Fiber-seq FIRE scores of MSPs; NeuN+ and NeuN− single DNA molecules are as indicated; green-shaded single DNA molecules/chromatin fibers are highlighted as representative examples and shown at the bottom again at a higher (150 bp) resolution, with purple vertical rectangles marking linker DNA (examples marked by “L” on fiber), and single-nucleosome (“n” on fiber) resolution. Red/brown-colored MSPs scoring as regulatory elements (P, promoter, and other RE, regulatory element, on fiber) (FIRE FDR, orange, 0.10 ≥ q > 0.05; bright red, 0.05 ≥ q > 0.01; dark red-brown, q ≤ 0.01).