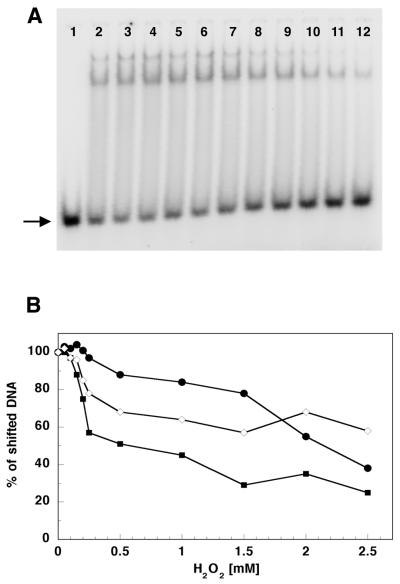
Figure 4.
Hydrogen peroxide affects the ability of p140, p55 and p140/p55 complex to bind to dsDNA. (A) An example of an autoradiogram of a gel mobility shift experiment showing p140 with no treatment (lane 2) or treatment with 0.05, 0.1, 0.15, 0.2, 0.25, 0.50, 1.0, 1.5, 2.0 and 2.5 mM H2O2 (lanes 3–12, respectively). Lane 1 contains the oligonucleotide substrate without addition of protein. Oligonucleotide substrate preparation and reaction conditions are described in Materials and Methods. Products were separated on a 15% polyacrylamide non-denaturating gel and quantitated on a Molecular Dynamics PhosphorImager as described in Materials and Methods. Two shifted products were observed corresponding to either one or two p140 molecules bound per oligonulceotide substrate. (B) DNA binding efficiency of p140 (filled squares), p55 (closed circles) and p140/p55 complex (open diamonds) after preincubation at increasing H2O2 concentrations. The arrow indicates the unshifted dsDNA substrate.