Abstract
A photoreactive analogue of spermine, N1-azidobenzamidino (ABA)-spermine, was covalently attached after irradiation to Escherichia coli 30S ribosomal subunits or naked 16S rRNA. By means of RNase H digestion and primer extension, the cross-linking sites of ABA-spermine in naked 16S rRNA were characterised and compared with those identified in 30S subunits. The 5′ domain, the internal and terminal loops of helix H24, as well as the upper part of helix H44 in naked 16S rRNA, were found to be preferable binding sites for polyamines. Association of 16S rRNA with ribosomal proteins facilitated its interaction with photoprobe, except for 530 stem–loop nt, whose modification by ABA-spermine was abolished. Association of 30S with 50S subunits, poly(U) and AcPhe-tRNA (complex C) further altered the susceptibility of ABA-spermine cross-linking to 16S rRNA. Complex C, modified in its 30S subunit by ABA-spermine, reacted with puromycin similarly to non-photolabelled complex. On the contrary, poly(U)-programmed 70S ribosomes reconstituted from photolabelled 30S subunits and untreated 50S subunits bound AcPhe-tRNA more efficiently than untreated ribosomes, but were less able to recognise and reject near cognate aminoacyl-tRNA. The above can be interpreted in terms of conformational changes in 16S rRNA, induced by the incorporation of ABA-spermine.
INTRODUCTION
The bacterial 30S subunit is a large ribonucleoprotein complex composed of a 1542-nt RNA (16S rRNA) and 21 proteins, with a great wealth of available functional and structural information (1–3). The 16S rRNA is subdivided into three major structural domains, and one minor domain (4): the 5′ domain (residues ∼26–557), the central domain (residues ∼558–912), the 3′ major domain (residues ∼926–1391) and the 3′ minor domain (residues ∼1392–1542). Physical proximity of proteins and 16S rRNA within the 30S subunit has been measured by chemical protection methods, cross-linking techniques, computational methods and recently by crystallographic analysis (2,3,5 and references therein). The small subunit decodes the genetic information, governs mRNA and tRNA translocation, and controls the fidelity of codon–anticodon interactions. However, none of these functional properties can be detected in free 16S rRNA because proteins and ions are required for crucial adjustments of the 16S rRNA functional conformation (6).
In the course of structural and functional studies, there has been a significant effort to understand how ribosomal activity is influenced by the ionic environment. Thus, much has been learned about the role of divalent and monovalent ions for the formation and maintenance of active 30S subunits (7–11). Furthermore, metal-promoted cleavage assays have been used to map metal ion binding sites within the eubacterial 70S and human 80S ribosomes (12–15). Nevertheless, polyamines, such as spermine and spermidine, are also essential for optimal translational accuracy and efficiency (16,17). Despite the fact that much has been accomplished in the elucidation of polyamine action on tRNA positioning to ribosome and regulation of the translation process at several levels (18,19), there is still relatively little information available concerning the contacts between polyamines and ribosome. As a consequence, the molecular basis of polyamine action on ribosomal functions remains elusive. In a recent report, we have presented evidence that spermine at 300 µM inhibits the puromycin reaction by acting at the elbow and the 3′ strand of the acceptor stem of the donor substrate (20). On the contrary, the stimulatory effect observed at low concentrations of spermine seems to be related with polyamine binding to RNA and protein constituents of the ribosome (21).
Under physiological pH conditions, spermine carries a net positive charge +4. Interactions of spermine with RNA are less well understood than with DNA, and crystallographic studies are almost exclusively limited to tRNA (19). Progress in identifying binding sites for polyamines in the ribosome has been made using cross-linking methods with homo-bifunctional reagents (22–24). However, these methods have failed to cross-link spermine to rRNA, probably because the reagents used are amine-reactive cross-linkers and their reactivity towards RNA is low. Furthermore, the use of such symmetrical reagents may cause side effects by inducing protein–protein and intra-RNA cross-linking. An alternative approach, which we have successfully applied for the mapping of spermine binding sites in AcPhe-tRNA bound at the P-site of Escherichia coli poly(U)-programmed ribosomes (20) or in ribosomal proteins (21), is the use of a photoreactive analogue of spermine that has an arylazido group attached to one of the terminal amino groups of the molecule. This photoprobe, N1-azidobenzamidino (ABA)-spermine, retains almost all biochemical properties of the parent compound, spermine (20,21). On the other hand, photolabelled ribosomal complexes with ABA-spermine exhibit similar functionality to that obtained with untreated ribosomal complexes reacting in solution (reversible interaction) with spermine (21). Thus, it is reasonable to believe that ABA-spermine binds specifically to the ribosome. In addition, this ribosome-bound ligand can easily react through irradiation with a variety of adjacent groups in both proteins and rRNA. By linking to a nucleoside of 16S rRNA, the photoprobe is expected to act as a barrier for reverse transcriptase. The site of cross-linking can, therefore, be localised exactly from the size of the reverse transcription product. The objective of this study is to gain a comprehensive view of the 16S rRNA nucleotide residues involved in polyamine binding and to correlate these sites with conformational states and functional properties of the 30S ribosomal subunit.
MATERIALS AND METHODS
Materials
Puromycin dihydrochloride, spermine tetrahydrochloride, 4-aminobenzonitrile, dimethyl sulfate (DMS), DMS stop-solution, tRNAPhe and heterogeneous tRNA from E.coli were obtained from Sigma-Aldrich. AMV reverse transcriptase and RNase H were from Roche Diagnostics GmbH and Promega, respectively. dNTPs and ddNTPs were obtained from Boehringer Mannheim. All radio-chemicals mentioned were from Amersham. Cellulose nitrate filters (type HA, 24 mm diameter, 0.45 µm pore size) were from Millipore. ABA-spermine was synthesised from methyl 4-azidobenzoimidate and spermine and purified by chromatography on a sulfopropyl–Sephadex column (25). Reduced ABA-spermine (rABA-spermine) was prepared by treatment of ABA-spermine with dithiothreitol according to Staros et al. (26).
Biochemical preparations
Salt-washed (0.5 M NH4Cl) and polyamine-depleted ribosomes from E.coli B cells, partially purified translation factors, S100-enzymes, crude Ac[3H]Phe-tRNA charged with 15.8 pmol of [3H]Phe per A260 unit, 70S ribosomes and native 50S and 30S ribosomal subunits were prepared as described previously (21,27). 16S rRNA was isolated from 30S ribosomal subunits by phenol extraction and ethanol precipitation and it was further purified by electrophoresis on 8% polyacrylamide/7 M urea gels. Before their use, 16S rRNA or ribosomal subunits were activated in buffer 50 mM HEPES–KOH pH 7.2, 20 mM magnesium acetate, 100 mM NH4Cl, by heating for 20 min at 42°C. Tubes were cooled to 0°C on ice and the concentration of Mg2+ ions was normalised to 6 mM. Initiation ternary ribosomal complex, i.e. the Ac[3H]Phe-tRNA·poly(U)·ribosome complex (complex C), was prepared at 6 mM Mg2+ in the presence or in the absence of translation factors, and purified through adsorption on cellulose nitrate filters by established procedures (27). In some experiments, 30S ribosomal subunits labelled with ABA-spermine were incubated for 30 min with 2 molar equivalents of native 50S subunits in buffer containing 50 mM HEPES–KOH pH 7.2, 15 mM MgCl2, 100 mM NH4Cl and 6 mM 2-mercaptoethanol at 37°C. The resulting mixture, cooled to 0°C on ice, was normalised at a concentration of Mg2+ ions equal to 6 mM, and then used for complex C formation.
Photo-affinity labelling, chemical modification and mapping of cross-linking sites of ABA-spermine in 16S rRNA
Naked 16S rRNA, 30S ribosomal subunits or complex C were photolabelled with ABA-[14C]spermine as described previously (20,21). After the irradiation step, 100 µM dithiothreitol was added to the incubation mixture, to scavenge any unreacted photoprobe. The photolabelled product was purified by gel-filtration on Sephadex G50 column, by micro-dialysis in buffer A (50 mM HEPES–KOH pH 7.2, 6 mM magnesium acetate, 100 mM NH4Cl) and by ethanol precipitation. Extraction of the final product with 0.6 N perchloric acid and analysis of the extract by HPLC (28) ensured that the photolabelled material was discharged from non-covalently bound photoproducts. Aliquots of labelled or non-labelled complex C (100 pmol) in 300 µl buffer A were modified with 1 µl of DMS for 2 h at 4°C. The DMS reactions were stopped by adding 75 µl of DMS stop-solution, mixed and incubated for 10 min on ice, and then ethanol precipitated. Pellets were resuspended in 0.3 M sodium acetate, extracted with phenol and chloroform, and precipitated twice with ethanol.
To make an approximate localisation of the photoreactive probe cross-linking sites in 16S rRNA, aliquots of rRNA isolated from photolabelled 30S ribosomal subunits or complex C preparations were first hybridised with selected pairs of 11-deoxynucleotides, complementary to 16S rRNA at positions 200 nt apart, digested with RNase H, and applied to a 5% polyacrylamide/7 M urea gel for analysis (29). For the amplification of the radioactive signal, gels were treated with 2,5-diphenyloxazole in dimethyl sulfoxide according to Bonner and Laskey (30), dried and subjected to autoradiography. Precise identification of the cross-linking sites was made by primer extension analysis according to the primer hybridisation procedure of Stern et al. (31) and the extension and gel electrophoresis protocols of Stahl et al. (32). The stops in cDNA synthesis were visualised on a gel autoradiogram and characterised by reference to dideoxy sequencing reactions on 16S rRNA that were run in parallel. Intensities of bands were scored by visual inspection in combination with image analysis. Only those bands reproduced at least three times were taken into account. Control experiments were performed, in which primer extension was performed on unmodified ribosomes, or in the simultaneous presence of a 250-fold excess of spermine in the incubation mixture, or by replacing ABA-spermine with rABA-spermine.
Peptide bond formation assay
The peptidyltransferase (PTase) activity of partially or totally photolabelled complex C was titrated by the puromycin reaction performed at 25°C in the presence of 6 mM Mg2+ and 100 mM NH4+. Under these conditions, the reaction between complex C and excess puromycin (S) follows pseudo-first-order kinetics (28), and the relationship
k′obs = (kmax[S])/(Ks + [S]) 1
holds, where k′obs is the first-order rate constant that can be calculated from the slope of semi-logarithmic time plots, Ks is the dissociation constant of the encounter complex between puromycin and complex C, and kmax represents the catalytic rate constant of PTase which is a function of photoprobe concentration (28). The values of kmax and Ks were estimated from double-reciprocal plots of equation 1.
Binding of Ac[3H]Phe-tRNA to the ribosomal A-site
Poly(U)-programmed 70S ribosomes (62.2 pmol) at the initiation state Pi with only one tRNAPhe bound to the ribosomal P-site were prepared in the presence of rABA-spermine (step 1), according to Dabrowski and Nierhaus (33). Following incubation at 37°C for 10 min to fill all P-sites, the mixture was cooled to 25°C, and 2 A260 U of Ac[3H]Phe-tRNA were added. The binding was carried out at 25°C within specified time intervals (step 2). The reaction was stopped by diluting aliquots (25 µl) of the incubation mixture in 2 ml ice-cold buffer A containing 6 mM 2-mercaptoethanol and rABA-spermine at the proper concentration, and by adsorption and wash of the diluted mixture on cellulose nitrate filter disks. The level of Ac[3H]Phe-tRNA binding to the ribosomal A-site was calculated by measuring the trapped radioactivity on the filters in a scintillation spectrometer. Whenever required, pre-labelled 70S ribosomes in their 30S ribosomal subunit with ABA-spermine were used, instead of untreated 70S ribosomes. The resulting incubation mixture was then treated as already described, with the exception that no rABA-spermine was included in the binding and wash buffer.
Binding of Ac[3H]Phe-tRNA to the ribosomal P-site
The binding mixture (25 µl) contained 0.4 mM GTP, 8 µg poly(U), 10.4 pmol ribosomes (untreated or labelled in their 30S subunit with ABA-spermine), 5.3 pmol Ac[3H]Phe-tRNA, and 6 mM 2-mercaptoethanol in buffer A. When untreated ribosomes were used, rABA-spermine at final concentrations ranging from 0 to 300 µM was also added to the above mixture. The time course of the reaction was monitored up to 30 min at 25°C. The level of total Ac[3H]Phe-tRNA binding was measured by nitrocellulose filtration. The P-site bound Ac[3H]Phe-tRNA was titrated by puromycin (2 mM, 2 min at 25°C).
Translation of poly(U) templates by labelled ribosomes
ABA-spermine was incubated in the dark with ribosomes for 10 min at 25°C, in buffer A containing 0.5 mM GTP, 640 µg/ml poly(U), 64 A260 U/ml ribosomes and 27 A260 U/ml AcPhe-tRNA. The mixture was cooled in an ice bath and irradiated at 300 nm for 20 min (20). After the irradiation step, dithiothreitol was added at a final concentration of 100 µM to scavenge any unreacted photoprobe. The binding mixture was purified by gel filtration over a Sephadex G-50 column, dialyzed in buffer A containing 6 mM 2-mercaptoethanol, and then transferred to an equal volume of charging mix containing 50 mM HEPES–KOH pH 7.2, 6 mM magnesium acetate, 100 mM NH4Cl, 10 mM K2HPO4, 6 mM 2-mercaptoethanol, 2 mM ATP, 0.5 mM GTP, 80 mM phosphocreatine, 80 µg/ml creatine kinase, 280 A260 U/ml crude tRNA from E.coli, 2.8 mg/ml (protein) S100-enzymes, and 6 nmol/ml of l-[3H]phenylalanine (33.6 Ci/mmol) and 4.6 nmol/ml of l-leucine (for the incorporation of phenylalanine experiments) or 6 nmol/ml of l-[3H]leucine (33.6 Ci/mmol) and 4.4 nmol/ml of l-phenylalanine (for the mis-incorporation of leucine experiments). Before receiving the binding mixture, the charging mix was pre-warmed for 5 min at 25°C. The combined mixture was incubated at 25°C for 45 min. The level of l-[3H]phenylalanine and l-[3H]leucine incorporation per A260 U of ribosomes as well as the error frequency were calculated according to Dresios et al. (34).
RESULTS
Localization of ABA-spermine cross-linking sites in 16S rRNA
Under irradiation at 300 nm, the arylazido group of ABA-spermine is converted to nitrene, a highly reactive species that is able to react with a variety of adjacent groups of the target molecule (25). Accordingly, ABA-spermine is photo-incorporated into both the RNA and protein fractions of 30S subunits. We have previously found that >75% of incorporated photoprobe is preferentially distributed to 16S rRNA (21). Evidence for target site-specific photo-incorporation was sought by performing control experiments, in which a 250-fold excess of spermine was added in the incubation mixture during photolabelling. As shown in Figures 1–3, the radioactive fragments resulted by RNase H digestion and the authentic stops of reverse transcriptase are essentially abolished under these conditions. Furthermore, experiments performed in the absence of ABA-spermine or in the presence of rABA-spermine were used to distinguish artifacts originated from breakages induced upon incubation of the target molecule in the modification solution, or from natural stops in reverse transcription.
Figure 1.
Analysis of ABA-spermine–16S rRNA cross-links by RNase H digestion. 16S rRNA, isolated from irradiated complex C (minus translation factors) in the presence of (A) 50 µM or (B) 300 µM ABA-[14C]spermine, was incubated with cDNA probes, and then digested with RNase H. The products were resolved on 5% polyacrylamide/7 M urea gels, and the radioactive fragments were visualised by autoradiography. Lanes 1 and 2, undigested ABA-[14C]spermine–16S rRNA samples from complex C irradiated in the absence and in the presence of excess spermine, respectively. Lanes 3–9, ABA-[14C]spermine–16S rRNA samples hybridised (lane 3) with cDNA 363–373, (lane 4) with cDNAs 363–373 and 560–570, (lane 5) with cDNAs 560–570 and 654–664, (lane 6) with cDNAs 654–664 and 839–851, (lane 7) with cDNAs 839–851 and 1042–1053, (lane 8) with cDNAs 1042–1053 and 1381–1391, (lane 9) with cDNA 1381–1391, and then digested with RNase H. Numbers indicated to the left of the gel correspond to the sizes (nt) of RNA markers.
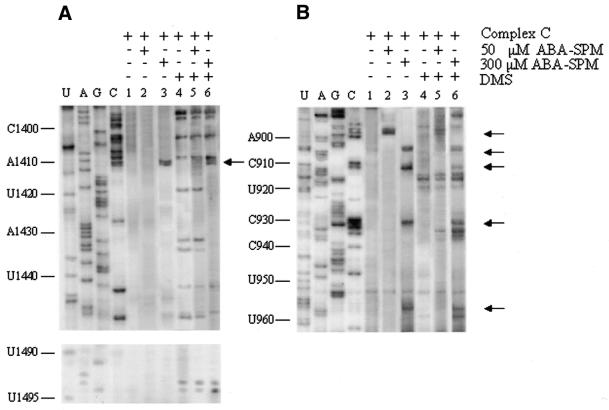
Figure 3.
Gel electrophoresis fractionation of products resulting from ABA-spermine cross-linking to 30S ribosomal subunits or complex C in the presence or absence of translation factors, as monitored by primer extension analysis. The primer for the reverse transcriptase reaction was complementary to 16S rRNA positions 838–854. The stops of reverse transcriptase reaction are indicated on the right margin, while the positions at 10-nt intervals are shown in the left margin. Photolabelling of 30S subunits was carried out with 50 µM (lanes 3 and 5) or with 300 µM ABA-spermine (lane 7), in the absence (lanes 3 and 7) or in the presence (lane 5) of translation factors. Lanes 1 and 2 correspond to 30S subunits irradiated without photoprobe (controls), in the absence and in the presence of translation factors, respectively. Lanes 4 and 6 correspond to the same samples as in lanes 3 and 5, but photolabelled in the simultaneous presence of excess spermine. Lane 8 corresponds to complex C photolabelled with 300 µM ABA-spermine in the absence of translation factors. 30S subs, 30S subunits; Transl. factors, translation factors; SPM, spermine.
The ABA-spermine–16S rRNA cross-links were first analysed using electrophoretic and autoradiographic analysis of the products of RNase H cleavage of labelled 16S rRNA, hybridised with several pairs of deoxyoligonucleotides complementary to the rRNA. The results obtained with naked 16S rRNA, 30S ribosomal subunits or complex C, are summarised in Table 1. Also, representative autoradiograms are shown in Figure 1. Naked 16S rRNA appears to be less susceptible to cross-linking than 16S rRNA in 30S ribosomal subunits or in complex C. Interestingly, the region encompassing positions 1048–1386 seems to escape cross-linking with ABA-spermine (Fig. 1, lanes 8, and Table 1), whether the target rRNA molecule is naked or associated with ribosomal proteins.
Table 1. Summary of ABA-[14C]spermine cross-linking results, as determined by RNase H digestiona.
| Regions of 16S rRNA labelled by ABA-[14C]spermine | Translation factors during photolabelling | Target moleculeb | |||||
|---|---|---|---|---|---|---|---|
| Naked 16S rRNA | 30S ribosomal subunit | Complex C | |||||
| A | B | A | B | A | B | ||
| 1–368 | (–) | – | + | + | + | + | + |
| (+) | n.d. | n.d. | + | n.d. | + | n.d. | |
| 369–565 | (–) | + | + | + | + | + | + |
| (+) | n.d. | n.d. | + | n.d. | + | n.d. | |
| 566–659 | (–) | – | – | + | + | + | + |
| (+) | n.d. | n.d. | + | n.d. | + | n.d. | |
| 660–844 | (–) | + | + | + | + | + | + |
| (+) | n.d. | n.d. | + | n.d. | + | n.d. | |
| 845–1047 | (–) | – | – | + | + | + | + |
| (+) | n.d. | n.d. | + | n.d. | + | n.d. | |
| 1048–1208 | (–) | – | – | – | – | – | – |
| (+) | n.d. | n.d. | – | n.d. | – | n.d. | |
| 1209–1302 | (–) | – | – | – | – | – | – |
| (+) | n.d. | n.d. | – | n.d. | – | n.d. | |
| 1303–1386 | (–) | – | – | – | – | – | – |
| (+) | n.d. | n.d. | – | n.d. | – | n.d. | |
| 1387–1542 | (–) | – | + | + | + | – | + |
| (+) | n.d. | n.d. | + | n.d. | – | n.d. | |
aABA-[14C]spermine–16S rRNA cross-links were analysed by RNase H digestion, as described in the legend to Figure 1. The target molecule was irradiated in the presence (+) or in the absence (–) of translation factors. The concentration of photoprobe was (A) 50 or (B) 300 µM.
b–, absence of cross-links in the corresponding RNase H fragment; +, presence of cross-links; n.d., not determined.
The precise position of the cross-linking sites was determined by primer extension analysis, making use of the fact that reverse transcriptase pauses or stops one position before a modified nucleoside by ABA-spermine. The modified nucleoside is, therefore, determined exactly from the size of the reverse transcription product. The entire 16S rRNA, except its extreme 3′ end (nucleotides 1497–1542), was analysed in this way. The modified nucleosides by ABA-spermine are summarised in Table 2. Also, representative autoradiograms are given in Figures 2–4. Reverse transcriptase stops almost always at a single nucleoside. Only 10 exceptions were recorded, in which two adjacent nucleosides were found as stops, the one closer to the 5′ terminus probably constituting the cross-linking site. In agreement with the results obtained by the RNase H experiments, ABA-spermine cross-links to most of the 16S rRNA structure, except region 1034–1399. The cross-linking sites are dispersed throughout the rRNA, in both helices and loops of the secondary structure, but are detected more frequently (65%) in single-stranded regions.
Table 2. Summary of ABA-spermine cross-linking results, as monitored by primer extension analysis.
| Target molecule | Translation factors during photolabelling | ABA-spermine concentration (µM) | Cross-linking positions |
|---|---|---|---|
| Naked 16S rRNA | – | 50 | A414, C470, G791 |
| – | 300 | A77, G128, G187, A315, A414, C470, A496, G505, G521, G778, G791, C1400, C1411 | |
| 30S ribosomal subunit | – | 50 | G113, G147, C307, C422, U464, C470, U594, C732, A759, G791, C896, A1433, A1441 |
| + | 50 | G147, C307, C422, U464, C470, U594, C732, G760, C896, A1433, A1441 | |
| – | 300 | A80, G82, G128, A131, G147, A179, U208, G241, A250, C267, C307, G399, C422, G462, C470, C477, U594, G670, A759, A787, U789, A792, C795, U904, U911, C931, U956, U982, C999, C1001, C1011, G1033, C1400, C1411, A1433, A1441 | |
| Complex C | – | 50 | G113, G147, C307, C422, U464, C470, U594, A759, C896 |
| + | 50 | G147, C307, C422, U464, C470, U594, A759, C896 | |
| – | 300 | A80, G82, G128, A131, G147, A179, U208, G241, C267, C307, C422, G462, C470, C477, U594, A759, U904, U911, C931, U956, U982, C999, C1001, C1011, G1033, C1411 |
Figure 2.
Gel electrophoresis fractionation of products resulting from ABA-spermine cross-linking to naked 16S rRNA or 30S subunits, as monitored by primer extension analysis. Reverse transcription was done with a primer complementary to 16S rRNA positions 561–577. The stops of reverse transcriptase reaction are listed on the right margin. Positions at 10-nt intervals, read from sequencing products (lanes U, A, G and C), are shown in the left margin. Naked 16S rRNA (lanes 3–5) or 30S subunits (lanes 6–8) were labelled with 50 µM (lanes 3 and 6) or 300 µM (lanes 4 and 7) ABA-spermine in the absence of translation factors, as described in Materials and Methods. Lanes 1 and 2 are the respective controls (untreated samples) for 16S rRNA and 30S subunits. Lanes 5 and 8 correspond to 16S rRNA and 30S subunits, respectively, photolabelled with 50 µM ABA-spermine in the presence of excess spermine. 30S subs, 30S subunits; SPM, spermine.
Figure 4.
Gel electrophoresis fractionation of products resulting from ABA-spermine cross-linking to complex C, as monitored by primer extension analysis. The primers for reverse transcription were complementary to 16S rRNA positions (A) 1505–1527 and (B) 1047–1063. The stops of reverse transcriptase reaction are listed in the right margin, while the positions at 10-nt intervals, read from the sequencing products (lanes U, A, G and C) are indicated in the left margin. Photolabelling of complex C was performed at 50 µM (lane 2) or at 300 µM (lane 3) in the absence of translation factors. Lane 1, untreated complex C (control); lane 4, modified complex C by DMS; lanes 5 and 6, complex C labelled in the absence of translation factors with 50 or 300 µM ABA-spermine, respectively, and then modified by DMS. SPM, spermine.
An important feature of Table 2 is that the pattern of ABA-spermine cross-linking depends on whether 16S rRNA is photolabelled naked or in complex with ribosomal proteins. In addition, the photoprobe concentration as well as the presence of translation factors during photolabelling influences both the number and the intensity of the autoradiography bands. Thus, photolabelling of naked 16S rRNA with 50 µM ABA-spermine causes three double stops or pauses in reverse transcription, one of them presented in Figure 2, suggesting probe photo-incorporation into A414, U470 and G791. Probing with 300 µM ABA-spermine results in enrichment of the cross-linking pattern by additional sites (A77, G128, G187, A315, A496, G505, G521, G778, C1400 and C1411). It is obvious that the 5′ domain, the internal and terminal loops of helix H24, as well as the upper region of helix H44 [helices numbered according to Brimacombe (35)], are preferable polyamine binding regions in naked 16S rRNA. Assembly of 30S ribosomal subunit facilitates 16S rRNA interaction with photoprobe, except for nucleosides localised at the 530 stem–loop whose modification by ABA-spermine is abolished (Fig. 2). Namely, increased interaction of ABA-spermine with helices H6a, H8, H12, H16, H17, H20, H21, H27 and H44 is observed at 50 µM, while at higher concentrations of photoprobe additional sites in helices H4, H6, H7, H10, H11, H22, H24, H27, H28, H31, H33, H33a,b and H44 become accessible to cross-linking (Table 2). With the exception of nucleosides localised at the loop-790 whose susceptibility to ABA-spermine disappears (Fig. 3), the cross-linking pattern is not altered noticeably in the presence of translation factors. Also, no detectable changes in the cross-linking pattern are observed when complex C is labelled with 50 µM ABA-spermine, except in two cases: interaction of photoprobe with nucleosides A1433 and A1441 in the lower part of helix H44 is abolished (Table 2) and susceptibility of C732 and loop-790 to ABA-spermine is decreased (Fig. 3). Additional changes are detected when complex C is labelled with 300 µM ABA-spermine (Figs 3 and 4). Namely, nucleosides A250, G399, G670 and C1400 in complex C are protected, while the reactivity at positions A80, G82, U911 and C1411 increases.
Chemical probing with DMS of labelled and non-labelled complex C by ABA-spermine indicated that spermine cross-linking alters the 16S rRNA reactivity towards DMS. Complex C labelled by 50 µM ABA-spermine exhibits seven positions (A81, C188, A250, A432, A441, A915 and A935) of increased reactivity towards DMS, and five positions (A130, A1394, A1433, A1447 and A1556) of decreased reactivity, compared with unlabelled complex C. Photolabelling at 300 µM ABA-spermine alters the reactivity of additional sites. Thus, the reactivity of nucleosides A119, A129, A149, C194, A382, A412, A414, C422, A430, A448, A665, A889, A1408 and A1493 is enhanced, while the reactivity at positions A72, C316, A498, A510, C526, A546, A814, A816, A1004, A1014, A1016, A1035, A1418, A1431 and A1468 is reduced. In spite of perturbations caused by ABA-spermine cross-linking, the sedimentation profile of complex C is not significantly altered as revealed in a previous study (21).
Effect of 30S subunit interaction with ABA-spermine on ribosomal functions
30S subunits are implicated in important ribosomal functions, such as formation of the initiation ribosomal complex, binding of the aminoacyl-tRNA substrates and translation accuracy (1). Therefore, we attempted to test the ability of modified 30S subunits by ABA-spermine to accomplish these functions.
Effect on the binding of Ac[3H]Phe-tRNA to the P- and A-site of poly(U)-programmed ribosomes. Native or modified 30S subunits by ABA-spermine were incubated with native 50S subunits in a molar ratio of 1:2, and the formation of 70S ribosome was monitored by sucrose gradient sedimentation. These tests provided strong evidence that photolabelled 30S subunits are competent for association with 50S subunits (data not shown). In addition, the peak of 70S ribosomes obtained co-sediments with that of native 70S ribosomes.
In order to test whether ABA-spermine photoincorporation would interfere with AcPhe-tRNA binding in a poly(U)-dependent manner, two approaches were applied. In the first case, the ability of poly(U)-programmed 70S ribosomes to bind Ac[3H]Phe-tRNA was investigated in a binding mixture containing rABA-spermine at final concentrations ranging from 0 to 300 µM, i.e. upon conditions allowing reversible interaction between ribosomes and photoprobe. In the second approach, poly(U)-programmed 70S ribosomes were reconstituted from photolabelled 30S subunits and native 50S subunits, and assayed for their efficiency to bind Ac[3H]Phe-tRNA. In the first series of experiments, Ac[3H]Phe-tRNA binding to P-site is improved by rABA-spermine from 0.04 up to a maximum of 0.34 Ac[3H]Phe-tRNA molecules per 70S ribosome, while binding to the A-site is elevated from 0.06 to 0.14 (Table 3). In the second series of experiments, 70S ribosomes reconstituted from photolabelled 30S subunits and native 50S subunits exhibit higher efficiency for Ac[3H]Phe-tRNA binding to both sites, compared with untreated ribosomes. However, the upper limit of binding stimulation in this case is ∼70% of that observed upon conditions promoting reversible interaction of photoprobe (Table 3).
Table 3. Effect of 30S subunit interaction with ABA-spermine on AcPhe-tRNA binding to the ribosomal P- and A-sites.
| rABA-spermine or ABA-spermine concentration (µM) | P-site bound AcPhe-tRNA per 70S | A-site bound AcPhe-tRNA per 70S | |
|---|---|---|---|
| Ribosomes reacting with rABA-spermine in solutiona | 0 | 0.037 | 0.061 |
| 50 | 0.127 | 0.080 | |
| 300 | 0.343 | 0.140 | |
| Ribosomes labelled in 30S subunit by ABA-spermineb | 50 | 0.115 | 0.074 |
| 300 | 0.250 | 0.093 |
aThe binding mixture (25 µl) contained 50 mM HEPES–KOH pH 7.2, 6 mM magnesium acetate, 100 mM NH4Cl, 0.4 mM GTP, 8 µg poly(U), 5.3 pmol Ac[3H]Phe-tRNA, 6 mM 2-mercaptoethanol, rABA-spermine at final concentrations ranging from 0 to 300 µM, and 10.4 pmol 70S ribosomes pre-filled (A-site binding) or not pre-filled (total binding) in their P-site by tRNAPhe. The time course of the reaction was monitored up to 30 min at 25°C. The values of bound Ac[3H]Phe-tRNA measured by nitrocellulose filtration correspond to the maximal level of binding curves. The P-site bound Ac[3H]Phe-tRNA was calculated from the total binding by titration with puromycin (2 mM, 10 min at 25°C).
bThe composition of the binding mixture was the same as above, with the exception that no rABA-spermine was added and that 70S ribosomes were constructed from 30S subunits photolabelled with 50 or 300 µM ABA-spermine and from native 50S subunits.
Effect on translational accuracy. To investigate the effect of ABA-spermine cross-linking on the mis-incorporation of the near-cognate amino acid l-leucine, an in vitro system for poly(Phe) synthesis was utilised, using poly(U) as template. The accuracy of translation was determined by the error frequency (34). We found that at 6 mM Mg2+ the error frequency of native ribosomes is 0.0015, i.e. 1.5 errors every 103 codons. Photolabelling of 30S subunits with 50 µM ABA-spermine causes a 2.7-fold decrease in the ability of the resulting complex C to accurately translate poly(U) templates. Photolabelling at 300 µM ABA-spermine further reduces the accuracy of translation by a factor of 3.6.
Effect on peptide bond formation. Pre-labelling of complex C in its 30S subunit does not alter the extent of peptide bond formation when translation factors are present (Table 4). In the absence of translation factors, the extent is highly improved by cross-linking of ABA-spermine either to whole complex C or to 30S subunit. With regards to the kinetic phase of peptide bond formation, previous studies have indicated that photo-incorporation of ABA-spermine into complex C can stimulate or inhibit the PTase activity in a manner dependent on the experimental conditions (21). To further investigate this effect, the PTase activity of complex C photolabelled in its 30S subunit under inhibitory (300 µM ABA-spermine, no translation factors) or stimulatory conditions (50 µM ABA-spermine, plus translation factors), was tested by the puromycin-reaction assay. In both cases, cross-linking of ABA-spermine to 30S subunits had no effect on the catalytic properties of the reconstituted complex C (Table 4).
Table 4. Extent and kinetic parameters of AcPhe-puromycin synthesis carried out with complex C totally labelled or labelled in its 30S subunit by ABA-sperminea.
| Complex C species | Translation factors | Photoprobe concentration (µM) | Extent of AcPhe-puromycin synthesis (%) | kmax (min–1) |
|---|---|---|---|---|
| Unlabelled | – | 0 | 38 | 1.54 ± 0.05 |
| + | 0 | 80 | 2.20 ± 0.07 | |
| Totally labelled | – | 300 | 78 | 1.30 ± 0.03 |
| + | 50 | 80 | 3.40 ± 0.05 | |
| Labelled in 30S subunit | – | 300 | 73 | 1.50 ± 0.03 |
| + | 50 | 78 | 2.25 ± 0.07 |
aComplex C unlabelled, totally labelled or labelled in its 30S subunit by ABA-spermine, reacted with puromycin in buffer containing 6 mM Mg2+ and 100 mM NH4+. The kmax and Ks values were obtained from the corresponding double reciprocal plots of equation 1. The Ks value was invariable and equal to 665 ± 30 µM.
DISCUSSION
Our ignorance of the molecular basis of polyamine action on protein synthesis originates from the lack of detailed knowledge of the polyamine binding sites on ribosomes. Recently, we introduced the application of ABA-spermine for polyamine probing of ribosome (20,21). The use of this analogue has several advantages over other approaches. Most important of them is the capability of ABA-spermine to display biological activity similar to that of the parent compound, and to react through irradiation not only with proteins, but also with various groups in rRNA. Also, cross-linking of ABA-spermine to ribosomes can be carried out under mild experimental conditions, a fact that ensures the probing of active ribosome structure. The arylazido tether (ABA-) of the photoprobe is ∼9 Å and, therefore, the cross-links observed to 16S rRNA must lie approximately within one nucleoside from the terminal amino group of spermine, either on the same strand or possibly on the opposite strand.
Our results of the 16S rRNA photoaffinity labelling with ABA-spermine have led to a map of polyamine cross-linking sites in the small ribosomal subunit of E.coli. A summary of ABA-spermine cross-linking sites in a refined secondary structure model of 16S rRNA is given in Figure 5. The number of cross-linking sites found in this work is slightly higher than the number of binding sites calculated by Hill-plot analysis in a previous study (21). However, it should be mentioned that some of the mapped sites are related with adjacent nucleosides in the primary, secondary or tertiary structure of 16S rRNA, and may represent alternative orientations of the arylazido tether of photoprobe attached to the same binding site.
Figure 5.
Summary of ABA-spermine cross-linking sites in 16S rRNA of E.coli. Location of ABA-spermine cross-linking sites in the secondary structure model (cited at http://www.rna.icmb.utexas.edu) is marked with coloured arrows (see colour code). The arrows, discontinuous or solid, indicate sites labelled by 50 or 300 µM ABA-spermine, respectively. Long arrows, strong cross-links; medium arrows, intermediate cross-links; short arrows, weak cross-links. Nucleosides not analysed are shown in red. With the exception of G113 and nucleosides localised at the loop-790, the cross-linking pattern is not altered noticeably in the presence of translation factors and, for the sake of simplicity, is not presented in this diagram.
The cross-linking sites of ABA-spermine are distributed throughout the 16S rRNA, with the sole exception of a large region in the 3′-major domain. Nevertheless, the 3′-major domain is the target of hydrolytic cleavage by divalent cations (13–15). On the other hand, there are regions in 16S rRNA accessible to ABA-spermine, but insensitive to metal ion-catalysed hydrolysis. This finding is consistent with previous observations suggesting that structural and functional changes of ribosomes caused by polyamines may be different from those induced by divalent metal ions (7,8,36). Nevertheless, some regions of ABA-spermine photoincorporation in the 5′ domain, with the most pronounced situated at positions G147, U208, G241 and A250, are in close proximity with parts of 16S rRNA that are accessible to cleaving divalent cations.
Although a clear preference for the modification of single-stranded rRNA regions is evident, it is unsafe to make assumptions about motifs for polyamine binding pockets. In double-stranded regions of 16S rRNA, G and C are the most frequently modified nucleosides by ABA-spermine. G·U and G·A wobble pairs or bulged bases also show high susceptibility to the photoprobe. It is important to bear in mind that ion-binding motifs in RNA have been related with tandem G·U pairs or with G·C pairs followed by a sheared G·A pair (37,38). How the localisation of spermine binding sites in 16S rRNA may explain the effect of polyamines on ribosomal functions is discussed below.
The 5′ domain
In naked 16S rRNA, the 5′-terminal domain shows the highest susceptibility to ABA-spermine, compared with other regions of 16S rRNA. Reverse transcriptase pauses are localised in helix H6, in the lower part of 5′ domain, and in the bulged and terminal loops of helix H18. Helix H6 constitutes the target of ABA-spermine upon any experimental condition. It is relevant that the spur stem–loop of H6 appears to mimic the anticodon stem–loop of P-site bound tRNA (11). This structural resemblance may explain the tendency of ABA-spermine to bind helix H6, given that the anticodon stem of AcPhe-tRNA bound at the ribosomal P-site is one of the preferable polyamine binding sites (20). The presence of polyamine binding sites in the lower part of 5′-domain is most likely related with structural requirements. This region associated with protein S20 appears to serve as an anchor regulating the orientation of the main features in the inter-subunit interface and the decoding region (39). The identification of ABA-spermine cross-links in this region suggests that, apart from the role of ribosomal proteins in the correct folding of rRNA, polyamines may be also important for the attainment of the local tertiary structure. Most of the ribosomal proteins bound into this region are also labelled by ABA-spermine (21).
In the upper part of 5′ domain, three cross-linking sites are located within or in close proximity to helix H18. It is known that the bulged and terminal loops of helix H18 form a pseudoknot structure involved in the interaction of streptomycin with ribosomes (40). Probably, polyamines may contribute to the stabilisation of this pseudoknot. Since similar cross-linking is not observed in 30S subunits, it seems that binding of ribosomal proteins hinders the attachment of polyamines, while assuring by itself the establishment of the functional tertiary structure. Nevertheless, the general rule is that 16S rRNA in 30S subunits is much more accessible to ABA-spermine than naked 16S rRNA. Moreover, various C and A nucleosides in native 30S subunits which are reactive towards DMS show an enhanced reactivity upon photoincorporation of ABA-spermine. This implies that the 5′ domain tends to adopt an apparent overall ‘loosening’ of its structure upon polyamine binding.
The cross-linking pattern observed in complex C is a close approximation to that found in 30S subunits. However, there are two striking examples of abolished cross-linking, concerning nucleosides A250 and G399. Nucleoside A250 may be protected by the association of 30S and 50S ribosomal subunits, while G399 may be protected by P-site bound tRNA or poly(U), as suggested previously (40–42).
The central domain
In naked 16S RNA, there is only one strong cross-linking site at position G791 and a second weaker site at position G778. In 30S subunits labelled in the absence of translation factors, the former cross-linking site still occurs among a long stretch of adjacent cross-links observed within the loop-790. Additional sites appear around the upper and lower three-helix junctions of H22 as well as in helix H27. When photolabelling of 30S subunits was performed in the presence of translation factors, the intensity of bands corresponding to cross-linking sites within loop-790 was substantially reduced, while in complex C the bands were fully eliminated (Fig. 3). This may be due to shielding effects caused by ribosome-bound ligands (translation factors, P-site bound tRNA, etc.) or by subunit association. It is known that the loop-790 (nucleotides 786–796) constitutes part of the B2b inter-subunit bridge in prokaryotic ribosomes (43), protruding into the subunit interface and implicated in several important ribosomal functions, such as subunit association (4), binding of translation factors (44–46), binding of tRNA (41,47) and antibiotic binding (46,48).
The cross-linking of ABA-spermine into helices H20, H21 and H22 is probably related to structural requirements. For instance, U594 is located in close proximity to positions G654 and A655 which may play a direct role in the formation of a putative Mg2+ or polyamine binding site involved in the stabilisation of the lower three-helix (H20, H21 and H22) junction and in the binding of protein S15 (8). Another cross-linking site, G670, is located near the upper three-helix (H22, H23a and H23) junction, which also requires a specific organisation by polyvalent cations for efficient binding of ribosomal proteins S6, S15 and S18 (49). With regard to A759, this is a known hyper-reactive nucleoside (41). Therefore, the specificity of its cross-linking with ABA-spermine has to be regarded with caution.
Helix H27 is the third region in the central domain of 16S rRNA containing strong cross-linking sites of ABA-spermine. H27 is essential for several ribosomal functions, such as subunit association, tRNA binding, drug binding and assembly of ribosomal proteins S4, S5 and S12 (1,40,48,50). It has been proposed that H27 shifts between two alternating base-paired arrangements during translation: an ‘error-prone’ conformation with nucleosides 910–912 paired to 885–887, and a ‘restrictive’ conformation with nucleosides 910–912 paired to 888–890 (51). The ‘error-prone’ conformation has a greater affinity for tRNA than wild-type ribosomes, able to accept even non-cognate tRNAs at the A-site. In contrast, the ‘restrictive’ conformation has reduced capability for tRNA binding to the A-site and an elevated rate of frameshifting. It has been revealed by crystallographic studies that H27 is pointing towards the surface of 30S subunit, packing against the minor groove of the proximal end of H44, just below the decoding site where the A-site tRNA binds (11). Due to the location of H27 on the ribosomal surface, the access of ABA-spermine to this region is relatively easy. Our results show that the cross-linking pattern concerning H27 neither depends on the presence of translation factors nor does change upon subunit association. However, remarkable differences are visible when the concentration of photoprobe increases from 50 to 300 µM. At 50 µM, the cross-linking is positioned at nucleoside C896. At 300 µM, the cross-linking shifts from the above position to U904 and U911. Concomitantly, A889 becomes sensitive to DMS modification. These changes could be accommodated by a mechanism in which preferential attachment of polyamines to U911 stabilises the G·U wobble base pairing and tips the equilibrium between the ‘restrictive’ and ‘error-prone’ conformations to the right. This may significantly affect the topography of the decoding region in 30S subunits and explain the changes observed in AcPhe-tRNA binding (Table 3) and fidelity of decoding poly(U). However, prevailing evidence suggests that polyamines and Mg2+ at the concentrations used would be expected to enhance the fidelity of decoding (16,17,19). Therefore, it seems more probable that photocross-linking of spermine prohibits changes between translational states thus leading to an error-prone phenotype.
Chemical probing of photolabelled complex C with DMS indicates both decreased and increased reactivity at various nucleosides in the central domain, compared with the wild-type reactivity pattern. In other words, ‘loosening’ of the structure in one location appears to be compensated for by a tightening of folding elsewhere.
The 3′-major domain
In naked 16S rRNA, the 3′-major domain is insensitive to cross-linking, at any photoprobe concentration. Cross-linking sites are observed only in 30S subunits and complex C, labelled by 300 µM ABA-spermine. Nevertheless, from a structural and functional point of view, the importance of the rare labelled positions is beyond doubt. G931 is located in between nucleosides G926 and C936, which are implicated in P-, A- and E-site tRNA binding (1,29) and mRNA binding (11). Another cross-linking site, U956, is situated within region 954–963 implicated in protein S9 and tRNA binding to the ribosome (29,52). Remarkable is the labelling of U982, which along with U956 is placed into the loop-980, one of the multiple RNA elements creating the binding pocket of Thx protein in Thermus thermophilus (53). The protein Thx is a small peptide with high content in basic amino acid residues (54). It is believed that Thx fits into this binding pocket, and its positive charge stabilises the organisation of the rRNA elements at the top of the head. Protein Thx is not conserved in E.coli but binding pockets for cations are (53), indicating that polyamine attachment may give extra stability to this region. Consistently, the 3′-major domain achieves a tighter structure upon ABA-spermine photoincorporation, as indicated by DMS-protection experiments.
The 3′-minor domain
Because of the presence of methylated nucleosides at positions 1516, 1518 and 1519 (Fig. 5), which block the progress of reverse transcriptase, only the penultimate stem (H44) has been examined. No cross-linking sites are observed in naked 16S rRNA upon photolabelling at 50 µM of ABA-spermine. By increasing the concentration of photoprobe to 300 µM, two cross-links appear, localised at positions 1400 and 1411. The former site has been identified as one of the rRNA residues making intermolecular contacts between rRNA and P-site bound tRNA (11,43,47,55). It has also been found to participate in the rRNA environment of translation factors IF-3 and EF-G (45,46), indicating that this site possesses a strategic role in fundamental functions of the E.coli ribosome. The cross-linking at C1400 is preserved in 30S ribosomal subunits, but it is abolished in complex harbouring AcPhe-tRNA at the P-site. Given that photoincorporation of ABA-spermine into 30S subunits improves the efficiency of ribosomes for AcPhe-tRNA binding, it is reasonable to assume that attachment of polyamines at C1400 has a beneficial effect on this function. Nevertheless, the extent of binding observed with ‘hybrid’ 70S ribosomes (photolabelled 30S subunits associated with native 50S subunits), is 70% of that obtained with native 70S ribosomes interacting reversibly with photoprobe in solution (Table 3). Under the latter conditions, however, AcPhe-tRNA is also targeted by the spermine analogue, a fact further improving the AcPhe-tRNA efficiency for binding to the ribosome (20).
The second cross-linking site, seen at position C1411 of naked 16S rRNA, continues to exist in 30S subunit as well as in complex C. This site is located in close proximity to the heart of the decoding centre, which organises mRNA and tRNA translocation and controls fidelity in codon–anticodon interaction. Noah and Wollenzien (10) have demonstrated that the decoding region conformation depends on Mg2+ ions. It has also been found by crystallographic analysis that several Mg2+ ions coordinate with nucleosides positioned into or near the decoding region (11,56). Therefore, it is reasonable to suggest that photoprobe bound to C1411, along with other polyamine molecules bound adjacently or within the decoding region (H27, H34), may control changes in the conformation of essential bases. Given that both AcPhe-tRNA binding to the A-site and error frequency increase upon binding of polyamine to these sites, it could be assumed that the induced structural changes may be correlated with a flipping out of A1492 and A1493 bases from their stacked positions inside the internal loop of the A-site, as previously proposed for the paromomycin mechanism of action (56). Consistent with this hypothesis is the observation that bases A1408 and A1493, involved in a non-canonical base pair (57), become more reactive toward DMS upon binding of photoprobe to C1411.
Two other cross-linking sites found in 30S subunits, but not in naked 16S rRNA, concern nucleosides A1433 and A1441. The former nucleoside is included in the internal loop 1431–1434/1467–1469 of H44. Several sites in this loop have been implicated in translation termination (58). Nucleoside A1441 has been characterised as the most reactive adenosine in the whole 3′-minor domain (59). Hence, the specificity of cross-linking to this site is questionable. Both cross-linking sites become completely shielded in complex C. Analogous reduction of A1441 reactivity against Pb2+-induced cleavage or protection of both sites from base-specific probes upon subunit association have been reported previously (12,40).
A general conclusion emerging from our results is that the polyamine binding sites are related with ribosomal regions implicated in several important ribosomal functions, including subunit association, binding of ribosome ligands (mRNA, tRNA, translation factors, antibiotics) and recognition of cognate tRNA. However, binding of polyamines to the small subunit seems to have no effect on the catalytic properties of peptidyltransferase. This is consistent with the localisation of the catalytic centre on the large ribosomal subunit. To date, there have been no reports on the cross-linking sites of spermine with 23S rRNA, and work in our laboratory has been currently orientated to this subject.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs D. Synetos and A. Kazantzi for critical reading of the manuscript. This work was supported by a grant (99ED-605) from the General Secretariat of Research and Technology, Ministry of Development of Greece and the European Social Fund.
REFERENCES
- 1.Green R. and Noller,H.F. (1997) Ribosomes and translation. Annu. Rev. Biochem., 66, 679–716. [DOI] [PubMed] [Google Scholar]
- 2.Moore P.B. (2001) The ribosome at atomic resolution. Biochemistry, 40, 3243–3250. [DOI] [PubMed] [Google Scholar]
- 3.Ramakrishnan V. and Moore,P.B. (2001) Atomic structures at last: the ribosome in 2000. Curr. Opin. Struct. Biol., 11, 144–154. [DOI] [PubMed] [Google Scholar]
- 4.Noller H.F. (1984) Structure of ribosomal RNA. Annu. Rev. Biochem., 53, 119–162. [DOI] [PubMed] [Google Scholar]
- 5.Fink D.L., Chen,R.O., Noller,H.F. and Altman,R.B. (1996) Computational methods for defining the allowed conformational space of 16S rRNA based on chemical footprinting data. RNA, 2, 851–866. [PMC free article] [PubMed] [Google Scholar]
- 6.Burma D.P., Tewari,D.S. and Srivastava,A.K. (1985) Ribosomal activity of the 16S·23S RNA complex. Arch. Biochem. Biophys., 239, 427–435. [DOI] [PubMed] [Google Scholar]
- 7.Michelinaki M., Spanos,A., Coutsogeorgopoulos,C. and Kalpaxis,D.L. (1997) New aspects on the kinetics of activation of ribosomal peptidyltransferase-catalyzed peptide bond formation by monovalent ions and spermine. Biochim. Biophys. Acta, 1342, 182–190. [DOI] [PubMed] [Google Scholar]
- 8.Batey R.T. and Williamson,J.R. (1998) Effects of polyvalent cations on the folding of an rRNA three-way junction and binding of ribosomal protein S15. RNA, 4, 984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orr J.W., Hagerman,P.J. and Williamson,J.R. (1998) Protein and Mg2+-induced conformational changes in the S15 binding site of 16S ribosomal RNA. J. Mol. Biol., 275, 453–464. [DOI] [PubMed] [Google Scholar]
- 10.Noah J.W. and Wollenzien,P. (1998) Dependence of the 16S rRNA decoding region structure on Mg2+, subunit association, and temperature. Biochemistry, 37, 15442–15448. [DOI] [PubMed] [Google Scholar]
- 11.Carter A.P., Clemons,W.M., Brodersen,D.E., Morgan-Warren,R.J., Wimberly,B.T. and Ramakrishnan,V. (2000) Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature, 407, 340–348. [DOI] [PubMed] [Google Scholar]
- 12.Gornicki P., Baudin,F., Romby,P., Wiewiorowski,M., Kryzosiak,W., Ebel,J.P., Ehresmann,C. and Ehresmann,B. (1989) Use of lead (II) to probe the structure of large RNA’s. Conformation of the 3′ terminal domain of E. coli 16S rRNA and its involvement in building the tRNA binding sites. J. Biochem. Struct. Dyn., 6, 971–984. [DOI] [PubMed] [Google Scholar]
- 13.Winter D., Polacek,N., Halama,I., Streicher,B. and Barta,A. (1997) Lead-catalyzed cleavage of ribosomal RNAs. Nucleic Acids Res., 25, 1817–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polacek N. and Barta,A. (1998) Metal ion probing of rRNAs: evidence for evolutionarily conserved divalent cation binding pockets. RNA, 4, 1282–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorner S. and Barta,A. (1999) Probing ribosome structure by europium-induced RNA cleavage. Biol. Chem., 380, 243–251. [DOI] [PubMed] [Google Scholar]
- 16.Jelenc P.C. and Kurland,C.G. (1979) Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc. Natl Acad. Sci. USA, 76, 3174–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartetzko A. and Nierhaus,K.H. (1988) Mg2+/NH4+/polyamine system for polyuridine-dependent polyphenylalanine synthesis with near in vivo characteristics. Methods Enzymol., 164, 650–658. [DOI] [PubMed] [Google Scholar]
- 18.Agrawal R.K., Penezek,P., Grassucci,R.A., Burkhardt,N., Nierhaus,K.H. and Frank,J. (1999) Effect of buffer conditions on the position of tRNA on the 70S ribosomes as visualized by cryoelectron microscopy. J. Biol. Chem., 274, 8723–8729. [DOI] [PubMed] [Google Scholar]
- 19.Cohen S.S. (1998) A Guide to the Polyamines. Oxford University Press, New York.
- 20.Amarantos I. and Kalpaxis,D.L. (2000) Photoaffinity polyamines: interactions with AcPhe-tRNA free in solution or bound at the P-site of Escherichia coli ribosomes. Nucleic Acids Res., 28, 3733–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amarantos I., Xaplanteri,M.A., Choli-Papadopoulou,T. and Kalpaxis,D.L. (2001) Effects of two photoreactive spermine analogues on peptide bond formation and their application for labeling proteins in Escherichia coli functional ribosomal complexes. Biochemistry, 40, 7641–7650. [DOI] [PubMed] [Google Scholar]
- 22.Stevens L. and Pascoe,G. (1972) The location of spermine in bacterial ribosomes as indicated by 1,5-difluoro-2,4-dinitrobenzene and by ethidium bromide. Biochem. J., 128, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernabeu C., Vazques,D. and Ballesta,J.P.G. (1978) Proteins associated with rRNA in the Escherichia coli ribosome. Biochim. Biophys. Acta, 518, 290–297. [DOI] [PubMed] [Google Scholar]
- 24.Kakegawa J., Sato,E., Hirose,S. and Igarashi,K. (1986) Polyamine binding sites on Escherichia coli ribosomes. Arch. Biochem. Biophys., 251, 413–420. [DOI] [PubMed] [Google Scholar]
- 25.Clark E., Swank,R.A., Morgan,J.E., Basu,H. and Matthews,H.R. (1991) Two new photoaffinity polyamines appear to alter the helical twist of DNA in nucleosome core particles. Biochemistry, 30, 4009–4020. [DOI] [PubMed] [Google Scholar]
- 26.Staros J.V., Bayley,H., Standring,D.N. and Knowles,J.R. (1978) Reduction of aryl azides by thiols: implications for the use of photoaffinity reagents. Biochem. Biophys. Res. Commun., 80, 568–572. [DOI] [PubMed] [Google Scholar]
- 27.Synetos D. and Coutsogeorgopoulos,C. (1987) Studies on the catalytic rate constant of ribosomal peptidyltransferase. Biochim. Biophys. Acta, 923, 275–285. [DOI] [PubMed] [Google Scholar]
- 28.Karahalios P., Amarantos,I., Mamos,P., Papaioannou,D. and Kalpaxis,D.L. (1999) Effects of ethyl and benzyl analogues of spermine on Escherichia coli peptidyltransferase activity, polyamine transport, and cellular growth. J. Bacteriol., 181, 3904–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Döring T., Mitchell,P., Osswald,M., Bochkariov,D. and Brimacombe,R. (1994) The decoding region of 16S RNA; a cross-linking study of the ribosomal A, P and E sites using tRNA derivatized at position 32 in the anticodon loop. EMBO J., 13, 2677–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonner W.M. and Laskey,R.A. (1974) A film detection method for tritium-labeled proteins and nucleic acids in polyacrylamide gels. Eur. J. Biochem., 46, 83–88. [DOI] [PubMed] [Google Scholar]
- 31.Stern S., Moazed,D. and Noller,H.F. (1988) Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol., 164, 481–489. [DOI] [PubMed] [Google Scholar]
- 32.Stahl D.A., Krupp,G. and Stackebrandt,E. (1990) RNA sequencing. In Howe,C.J. and Ward,E.S. (eds), Nucleic Acids Sequencing: A Practical Approach. IRL Press, Oxford, pp. 137–183.
- 33.Dabrowski M. and Nierhaus,K.H. (1998) Synthesis and site-specific binding of thiolated tRNAs to probe ribosome–tRNA interactions. In Marting,R. (ed.), Methods in Molecular Biology; Protein Synthesis: Methods and Protocols. Humana Press Inc., Totowa, Vol. 77, pp. 413–426. [DOI] [PubMed]
- 34.Dresios J., Derkatch,I.L., Liebman,S.W. and Synetos,D. (2000) Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry, 39, 7236–7244. [DOI] [PubMed] [Google Scholar]
- 35.Brimacombe R. (1995) The structure of ribosomal RNA: a three-dimensional jigsaw puzzle. Eur. J. Biochem., 230, 365–383. [PubMed] [Google Scholar]
- 36.Igarashi K. and Kashiwagi,K. (2000) Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun., 271, 559–564. [DOI] [PubMed] [Google Scholar]
- 37.Allain F.H.T. and Varani,G. (1995) Divalent metal ion binding to a conserved wobble pair defining the upstream site of cleavage of group I self-splicing introns. Nucleic Acids Res., 23, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pley H.W., Flaherty,K.M. and Mckay,D.B. (1994) Three-dimensional structure of a hammerhead ribozyme. Nature, 372, 68–74. [DOI] [PubMed] [Google Scholar]
- 39.Schluenzen F., Tocilj,A., Zarivach,R., Harms,J., Gluehmann,M., Janell,D., Basham,A., Bartels,H., Agmon,I., Franceschi,F. and Yonath,A. (2000) Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell, 102, 615–623. [DOI] [PubMed] [Google Scholar]
- 40.Merryman C., Moazed,D., McWhirter,J. and Noller,H.F. (1999) Nucleotides in 16S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol., 285, 97–105. [DOI] [PubMed] [Google Scholar]
- 41.Bullard J.M., van Waes,M.A., Bucklin,D.J., Rice,M.J. and Hill,W.E. (1998) Regions of 16S ribosomal RNA proximal to transfer RNA bound at the P-site of Escherichia coli ribosomes. Biochemistry, 37, 1350–1356. [DOI] [PubMed] [Google Scholar]
- 42.Bhangu R., Juzumiene,D. and Wollenzien,P. (1994) Arrangement of messenger RNA on Escherichia coli ribosomes with respect to 10 16S rRNA cross-linking sites. Biochemistry, 33, 3063–3070. [DOI] [PubMed] [Google Scholar]
- 43.Cate J.H., Yusupov,M.M., Yusupova,G.Z., Earnest,T.N. and Noller,H.F. (1999) X-ray crystal structures of 70S ribosome functional complexes. Science, 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- 44.Moazed D., Samaha,R.R., Gualerzi,C. and Noller,H.F. (1995) Specific protection of 16S rRNA by translational initiation factors. J. Mol. Biol., 248, 207–210. [DOI] [PubMed] [Google Scholar]
- 45.Wilson K. and Noller,H.F. (1998) Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell, 92, 131–139. [DOI] [PubMed] [Google Scholar]
- 46.Pioletti M., Schlünzen,F., Harms,J., Zarivach,R., Glühmann,M., Avila,H., Bashan,A., Bartels,H., Auerbach,T., Jacobi,C., Hartsch,T., Yonath,A. and Franceschi,F. (2001) Crystal structures of complexes of the small subunit with tetracycline, edeine and IF3. EMBO J., 20, 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moazed D. and Noller,H.F. (1990) Binding of tRNA to the ribosomal A and P-sites protects two distinct sets of nucleotides in 16S rRNA. J. Mol. Biol., 211, 135–145. [DOI] [PubMed] [Google Scholar]
- 48.Spahn C.M.T. and Prescott,C.D. (1996) Throwing a spanner in the works: antibiotics and the translational apparatus. J. Mol. Med., 74, 423–439. [DOI] [PubMed] [Google Scholar]
- 49.Agalarov S.C., Prasad,G., Funke,P.M., Stout,C.D. and Williamson,J.R. (2000) Structure of the S15, S6, S18-rRNA complex: assembly of the 30S ribosomal central domain. Science, 288, 107–112. [DOI] [PubMed] [Google Scholar]
- 50.Noller H.F. (1991) Ribosomal RNA and translation. Annu. Rev. Biochem., 60, 191–227. [DOI] [PubMed] [Google Scholar]
- 51.Lodmell J.S. and Dahlberg,A.E. (1997) A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science, 277, 1262–1267. [DOI] [PubMed] [Google Scholar]
- 52.Greuer B., Osswald,M., Brimacombe,R. and Stoffler,G. (1987) RNA-protein cross-linking in Escherichia coli 30S ribosomal subunits; determination of sites on 16S RNA that are cross-linked to proteins S3, S4, S7, S9, S10, S11, S17, S18 and S21 by treatment with bis-(2-chloroethyl)-methylamine. Nucleic Acids Res., 15, 3241–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wimberly B.T., Brodersen,D.E., Clemons,W.M., Morgan-Warren,R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- 54.Choli T., Franceschi,F., Yonath,A. and Wittmann-Liebold,B. (1993) Isolation and characterization of a new ribosomal protein from the thermophilic eubacteria, Thermus thermophilus, T. aquaticus and T. flavus. Biol. Chem. Hoppe Seyler, 374, 377–383. [DOI] [PubMed] [Google Scholar]
- 55.Prince J.B., Taylor,B.H., Thurlow,D.L., Ofengand,J. and Zimmermann,R.A. (1982) Covalent cross-linking of tRNAIVal to 16S rRNA at the ribosomal P site: identification of cross-linked residues. Proc. Natl Acad. Sci. USA, 79, 5450–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogle J.M., Brodersen,D.E., Clemons,W.M., Tarry,M.J., Carter,A.P. and Ramakrishnan,V. (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science, 292, 897–902. [DOI] [PubMed] [Google Scholar]
- 57.Lynch S.R. and Puglisi,J.D. (2001) Structural origins of aminoglycoside specificity for prokaryotic ribosomes. J. Mol. Biol., 306, 1037–1058. [DOI] [PubMed] [Google Scholar]
- 58.Murgola E.J., Arkov,A.L., Chernyaeva,N.S., Hedenstierna,K.O.F. and Pagel,F.T. (2000) tRNA functional sites and structures for peptide chain termination. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome Structure, Function, Antibiotics, and Cellular Interactions. ASM Press, Washington, pp. 509–518.
- 59.Douthwaite S., Christensen,A. and Garrett,R.A. (1983) Higher order structure in the 3′-minor domain of small subunit ribosomal RNAs from a gram negative bacterium, a gram positive bacterium and a eukaryote. J. Mol. Biol., 169, 249–279. [DOI] [PubMed] [Google Scholar]