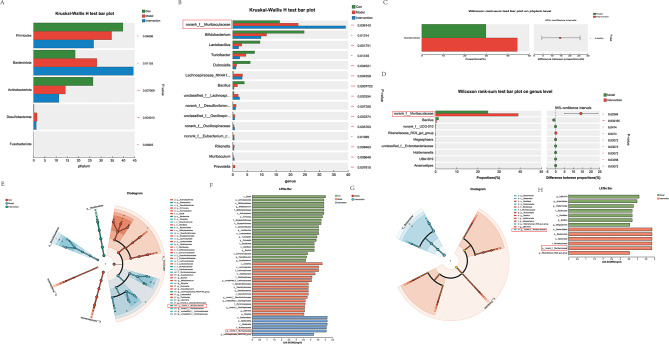
Fig. 4.
MSC-EVs modulate the gut microbiota and appear to increase the abundance of the norank_f__Muribaculaceae genus. Fecal bacterial DNA from control mice, Con A + PBS-treated mice, and Con A + MSC-EV-treated mice (n = 6–7 individuals/group) was analyzed via 16 S rRNA gene sequencing. (A) The gut microbiota differed among the indicated groups at the phylum level. Compared with that in the control group, the relative abundance of Bacteroidia in the Con A + PBS and Con A + MSC-EV groups increased. (B) The gut microbiota differed among the indicated groups at the genus level. Compared with that in the control group, the relative abundance of norank_f__Muribaculaceae in the Con A + PBS and Con A + MSC-EV groups increased. (C) The gut microbiota differed among the indicated groups at the phylum level. The relative abundance of Bacteroidia was greater in the Con A + MSC-EV group than in the Con A + PBS group. (D) The gut microbiota differed among the indicated groups at the genus level. The relative abundance of norank_f__Muribaculaceae increased in the Con A + MSC-EV group compared with the Con A + PBS group. (E, F) Cladograms generated by LEfSe depict taxonomic associations between microbiome communities from control, Con A + PBS, and Con A + MSC-EV mice (LDA > 3.5). Red, blue, and green bars indicate taxa predominant in the control, Con A + PBS, and Con A + MSC-EV groups, respectively. (G, H) Cladograms generated by LEfSe depict taxonomic associations between microbiome communities from Con A + PBS and Con A + MSC-EV mice (LDA > 3.5). Red bars indicate taxa predominant in Con A + PBS-treated mice, and blue bars indicate taxa predominant in Con A + MSC-EV-treated mice