Abstract
The maintenance of genomic integrity in rapidly proliferating cells is a substantial challenge during embryonic development1–3. Although numerous cell-intrinsic mechanisms have been revealed4–7, little is known about genome-protective effects and influences of developmental tissue microenvironments on tissue-forming cells. Here we show that fetal liver hepatocytes provide protection to haematopoietic stem and progenitor cell (HSPC) genomes. Lineage tracing and depletion in mice demonstrated that delayed hepatocyte development in early fetal livers increased the chromosomal instability of newly colonizing HSPCs. In addition, HSPCs developed tolerance to genotoxins in hepatocyte-conditioned medium, suggesting that hepatocytes protect the HSPC genome in a paracrine manner. Proteomic analyses demonstrated the enrichment of fetuin-A in hepatocyte-conditioned medium but not in early fetal livers. Fetuin-A activates a Toll-like receptor pathway to prevent pathogenic R-loop accumulation in HSPCs undergoing DNA replication and gene transcription in the fetal liver. Numerous haematopoietic regulatory genes frequently involved in leukaemogenic mutations are associated with R-loop-enriched regions. In Fetua-knockout mice, HSPCs showed increased genome instability and susceptibility to malignancy induction. Moreover, low concentrations of fetuin-A correlated with the oncogenesis of childhood leukaemia. Therefore, we uncover a mechanism operating in developmental tissues that offers tissue-forming cell genome protection and is implicated in developmental-related diseases.
Subject terms: Haematopoiesis, Risk factors, Stem-cell niche, Mechanisms of disease
A mechanism is uncovered that results in fetal hepatocytes having a paracrine role in providing genome protection to haematopoietic stem and progenitor cells.
Main
Embryonic development requires the extensive proliferation of three germinal layers that generate various functional cell types. A crucial element of developmental homeostasis is the preservation of genomic integrity during DNA replication and cellular activity through gene transcription1–3. Although a multitude of intrinsic cellular mechanisms for maintaining genome integrity have been extensively examined4–7, little is known about how the tissue microenvironment safeguards tissue-forming cells from DNA damage and genome instability caused by endogenous (for example, metabolic) or exogenous genotoxic agents during development. In this study, we have demonstrated that hepatocytes have a paracrine role in providing genome protection to HSPCs in the fetal liver, the primary site of hepatogenesis and haematopoiesis8–10.
Hepatogenesis in mice begins during embryonic days 9.0–9.5 (E9.0–9.5), as a liver bud composed of proliferating endodermal cells derived from the ventral wall of the foregut within the septum transversum11. Initially, the liver bud forms a solid sheet of epithelial cells, which then proliferate and branch out to form hepatic cords by E10.0–10.5. Endothelial cells originating from vitelline veins, the first vascular connection to the fetal liver, generate capillary-like sinusoids that separate the hepatic cords. By E11.5, the liver primarily consists of hepatic cords and sinusoids. HSPCs from the aorta–gonad–mesonephros region and placenta travel through umbilical vessels when the umbilical veins connect to the liver vascular system and enter the liver at E12.5. From this point forwards, the fetal liver becomes the main organ involved in fetal haematopoiesis12. At E13.5, approximately 75% of the liver is occupied by haematopoietic cells, a plateau that continues until E15.5, when HSPCs start migrating to the bone marrow and haematopoiesis begins to decline. During the plateau stage of haematopoiesis in the liver, hepatoblasts are separated by a high density of haematopoietic cells, but at E15.5–16.5, they return to close contact with each other. As haematopoiesis decreases, hepatogenesis intensifies. By E18.5, the principal sites for haematopoiesis shift from the liver to the bone marrow12. Among the various developmental organs and tissues, the fetal liver is selected as the primary site for haematopoiesis, probably due to its unique microenvironment that supports both haematopoiesis and hepatogenesis. Hepatocytes and HSPCs are mutually dependent on growth and functional maintenance, as supported by our own observations and other studies13,14. Fetal hepatocytes are presumed to have a key role in maintaining the genome integrity of HSPCs. Our current research has revealed that fetal hepatocytes secrete fetuin-A (FetuA; also known as the α2-Heremans–Schmid glycoprotein in humans)15,16, to protect the genome of HSPCs by preventing the accumulation of pathogenic R-loops (three-stranded structures that have an RNA–DNA hybrid). However, this protection is lacking during the early developmental stage of the fetal liver.
HSPC genomes are unstable in the early fetal liver
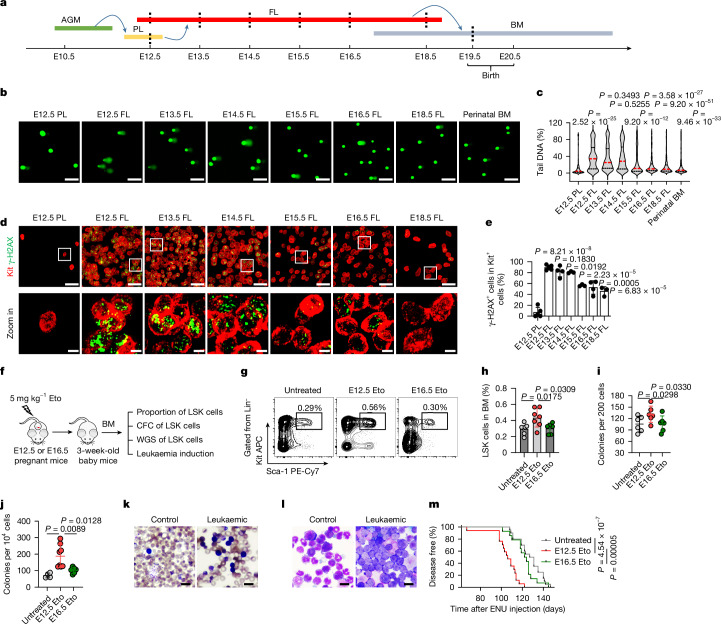
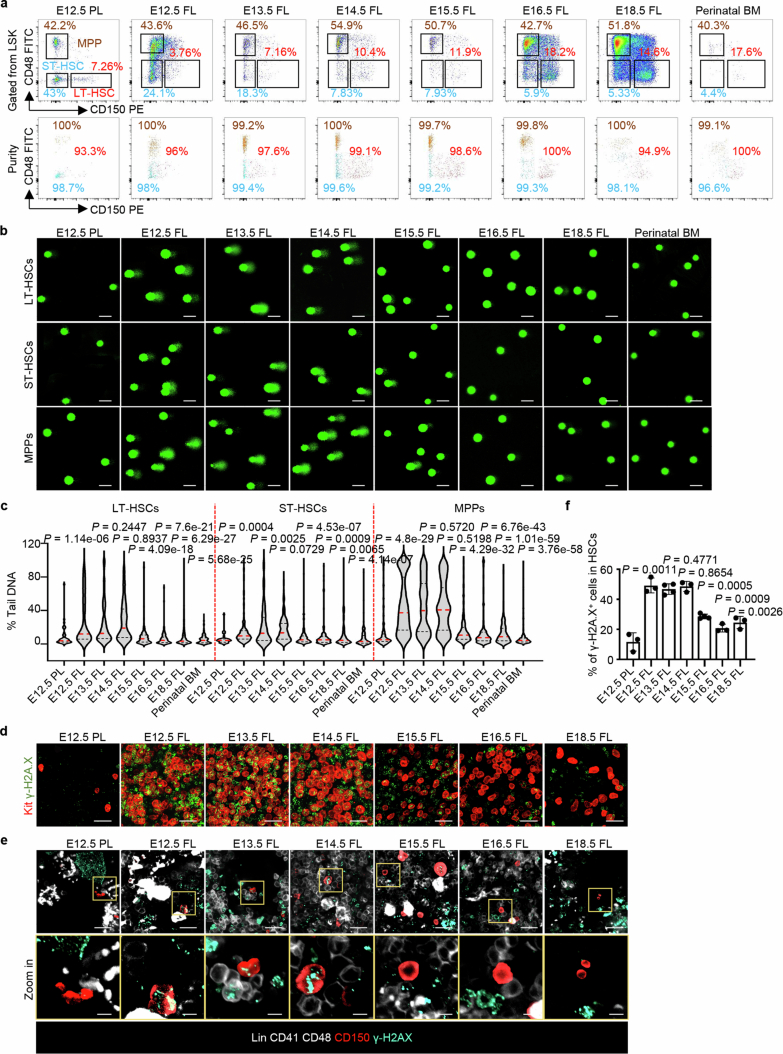
To assess the sensitivity of fetal HSPCs to genotoxic agents such as etoposide (Eto) and ultraviolet light (UV), we harvested HSPCs with negative lineage phenotypes and positive Sca-1 and Kit phenotypes (Lin−, Sca-1+ and Kit+ (LSK)) from E12.5 placentas, E12.5–18.5 fetal livers and perinatal bone marrow. LSK cells are enriched with HSPCs17–20 and share comparable phenotypes among all the examined tissues (Fig. 1a, Extended Data Fig. 1a and Supplementary Fig. 1). Treatment with Eto resulted in a greater number of DNA breaks in the HSPCs of early fetal livers (E12.5–14.5) than in the HSPCs of E12.5 placentas, late fetal livers (E15.5–18.5) and perinatal bone marrow. This was determined using DNA comet assays (Fig. 1b,c). Consistent results were obtained with the γ-H2AX immunofluorescence assay, which detects a protein that responds quickly to DNA breaks. Early fetal liver HSPCs showed more severe DNA damage than other tissues (Extended Data Fig. 1b,c). In an ex vivo experiment, exposure of HSPCs to 500 J of UV induced DNA breaks, with early fetal liver HSPCs exhibiting greater sensitivity to UV than other developing tissues (Extended Data Fig. 1d,e).
Fig. 1. HSPC genomes are unstable in the early fetal liver.
a, Mouse haematopoietic development and tissue harvesting of HSPCs (dashed lines) for the assessment of genome stability. AGM, aorta–gonad–mesonephros; BM, bone marrow; FL, fetal liver; PL, placenta. b,c, Representative comet-tail DNA images (b) and proportions (c) induced by etoposide (Eto) in HSPCs (n = 3). The medians (red dashed lines) and quartiles (black dashed lines) are shown. Scale bars, 50 µm. d,e, Representative in situ fluorescence images (d) and positive proportions (e) of γ-H2AX in Kit+ HSPCs in whole-mount tissues after in vivo Eto treatment (n = 5 for the E12.5 placenta and E12.5 fetal liver; n = 4 for the E13.5 fetal liver and E16.5 fetal liver; and n = 3 for the E14.5 fetal liver, E15.5 fetal liver and E18.5 fetal liver). Scale bars, 30 µm and 5 µm. The white squares represent the areas enlarged in the bottom row. f, The assessment of alterations caused by Eto treatment. CFC, colony-forming cell; WGS, whole-genome sequencing. g,h, Representative flow cytometry images (g) and proportions (h) of 3-week-old bone marrow LSK cells (n = 7 for untreated and E12.5 Eto; and n = 6 for E16.5 Eto). i,j, Plating (i) and replating (j) CFC assays of 3-week-old bone marrow LSK cells (n = 6 per group (i); n = 4 for untreated, n = 8 for E12.5 Eto and n = 6 for E16.5 Eto (j)). k,l, Representative Giemsa-stained images of peripheral blood (k) and bone marrow (l) smears (n = 7 for control and n = 31 for leukaemic (k); n = 7 for control and n = 13 for leukaemic (l)). Scale bars, 100 μm. m, Onset of leukaemia induced in mice (n = 20 for untreated, n = 17 for E12.5 Eto and n = 14 for E16.5 Eto). The n represents independent experiments (b,c), individual fetuses (d,e), mice (g,h,k–m) and dishes (i,j) from three independent experiments. The mean ± s.d. is shown (e,h–j). Unpaired two-sided Student’s t-test (c,e,h,j), unpaired one-sided Student’s t-test (i) and log-rank (Mantel–Cox) test (m) were used.
Extended Data Fig. 1. Flow-sorting of HSPCs and their sensitivity to genotoxic agents ex vivo.
a, Immunophenotypically negative lineage (Lin−), positive Sca-l (Sca-1+) and Kit+ (LSK) cells were sorted by flow cytometry with high purities (mean ± s.d.) from the haematopoietic tissues of the E12.5 placenta (PL) (n = 4), E12.5 fetal liver (FL) (n = 6), E13.5 FL (n = 4), E14.5 FL (n = 4), E15.5 FL (n = 3), E16.5 FL (n = 3), E18.5 FL (n = 5) and perinatal bone marrow (BM) (n = 3) to assay the sensitivity of HSPCs to genotoxic agents. b, c, Representative fluorescence images and positive cell proportions of γ-H2AX etoposide (Eto)-induced ex vivo in HSPCs isolated from PL, FL and BM on different embryonic days (E13.5 FL, n = 4; E15.5 FL, n = 6; other groups, n = 3). Scale bar, 50 µm. d, e, Representative fluorescence images and positive cell proportions of γ-H2AX ultraviolet (UV) light-induced HSPCs isolated from PL, FL and BM on different embryonic days (E12.5 PL, Perinatal BM, n = 4; E12.5 FL, E16.5 FL, n = 5; E13.5 FL, n = 9; E14.5 FL, E18.5FL, n = 3; E15.5 FL, n = 6). Scale bar, 50 µm. The n represents independent experiments. The mean ± s.d. is shown (c, e). Statistical tests: unpaired two-sided Student’s t-test (c, e).
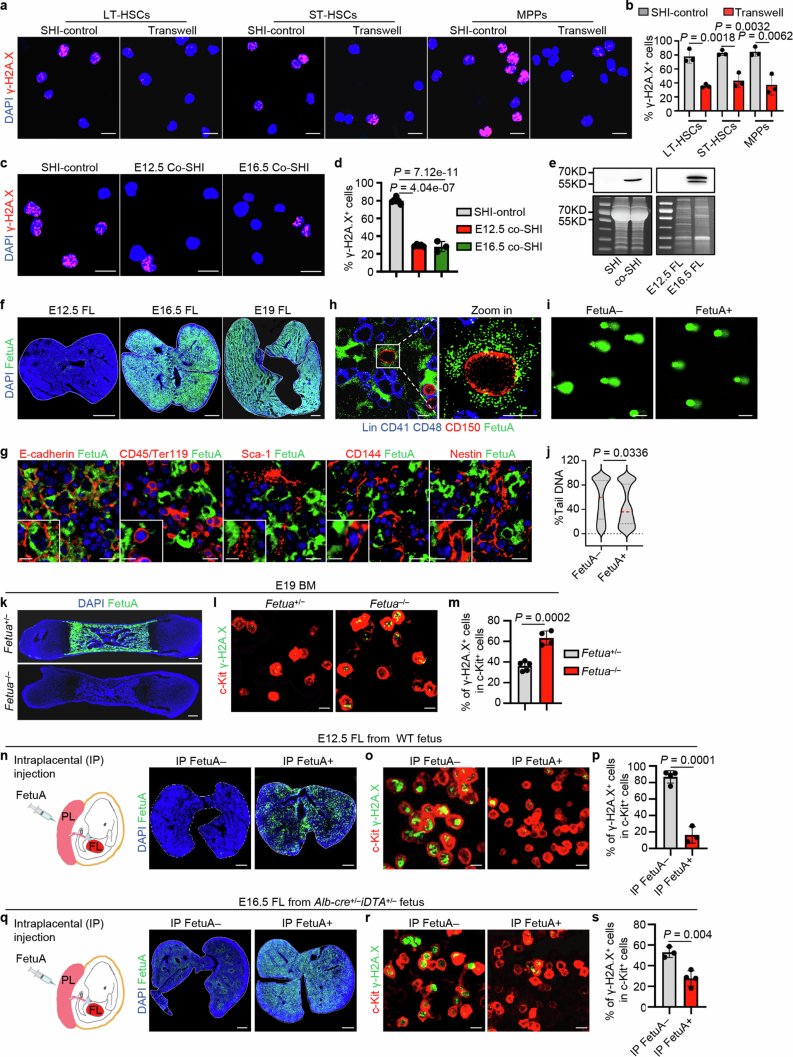
LSK cells were then flow sorted into three fractions based on slam gene expression: long-term haematopoietic stem cells (LT-HSCs), short-term HSCs (ST-HSCs) and multiple-potent progenitors (MPPs; Extended Data Fig. 2a and Supplementary Fig. 1). The sensitivity of these cell fractions to genotoxic agents changed with development. Eto treatment induced more DNA breaks in all three fractions of early fetal liver cells than in the other tissues, and the same trend was observed for the three cell fractions (Extended Data Fig. 2b,c). These ex vivo experimental results indicate that genome fragility varies when HSPCs migrate from the placenta to the fetal liver.
Extended Data Fig. 2. Flow-sorting of HSCs and MPPs and DNA damage detection after Eto-treatment ex vivo or in vivo.
a, LT-HSCs (LSKCD150+CD48–), ST-HSCs (LSKCD150–CD48–), and MPPs (LSKCD150–CD48+) were sorted by flow cytometry with high purities from the haematopoietic tissues labelled to assay their sensitivity to genotoxic agents (n = 3 per group). b, c, Representative comet-tail DNA images and proportions induced by Eto-treatment ex vivo in LT-HSCs, ST-HSCs and MPPs isolated from the labelled tissues (n = 3 per group). The medians (red dashed lines) and quartiles (black dashed lines) of each group are shown. Scale bar, 50 µm. d, Representative in situ fluorescence images of Kit and γ-H2AX after Eto-treatment in vivo (E12.5 PL, E12.5 FL, n = 5; E13.5 FL, E16.5 FL, n = 4; E14.5 FL, E15.5 FL, E18.5 FL, n = 3). Scale bar, 30 µm. e, f, Representative in situ fluorescence images and the positive cell proportion of γ-H2AX in HSCs (Lin–CD41–CD48–CD150+) after Eto-treatment in vivo (E13.5 FL, E15.5 FL, n = 4, other groups, n = 3; mean ± s.d.). Scale bars, 20 and 5 µm. The n represents individual samples (a-c) and fetuses (d-f) from 3 independent experiments. Statistical tests: unpaired two-sided Student’s t-test (c, f).
To investigate the effect in vivo, pregnant mice were intraperitoneally injected with Eto (5 mg kg−1) on different embryonic days. At 1 h post-injection, the whole placenta or fetal liver was removed and stained with γ-H2AX and Kit antibodies in situ. Eto treatment caused DNA damages in placental and fetal liver cells including HSPCs (Kit+) and non-HSPCs (Kit−; Extended Data Fig. 2d). γ-H2AX+ in Kit+ cells are shown (Fig. 1d) and their proportions are summarized (Fig. 1e). Eto treatment induced more severe DNA damage in early fetal liver HSPCs at E12.5–14.5 than in E12.5 placental or late fetal liver HSPCs at E15.5–18.5 in vivo. A similar trend was observed in the HSC-enriched fraction when refined immunofluorescence assays were performed using additional markers (CD150+Lin−CD41−CD48−) in situ. Eto treatment induced DNA damage more easily in HSCs in the early fetal liver (Extended Data Fig. 2e,f). These results are consistent with those ex vivo, albeit a possibility of the non-direct effect on HSPCs in vivo.
Given that fetal HSPCs enriched by either LSK or more specific markers consistently exhibit sensitivity to genotoxins both ex vivo and in vivo, HSPCs are referred to as LSK enrichment unless further specified in the paper.
Pregnant mice at E12.5 or E16.5 were exposed to Eto treatment and observed until their offspring were delivered. The goal was to determine whether the treatment resulted in different phenotypical, functional, chromosomal and leukaemia-vulnerable alterations in HSPCs. These alterations could be detected postnatally after the HSPCs migrated into the bone marrow (Fig. 1f). The proportion of LSK populations was greater in the mice treated with Eto at E12.5 than in the mice treated at E16.5 and the untreated group (Fig. 1g,h). In addition, these cells exhibited stronger colony formation ability, as determined by a colony-forming cell assay (Fig. 1i,j). Whole-genome sequencing and chromosome fluorescence in situ hybridization revealed that 3-week-old bone marrow HSPCs treated with Eto at E12.5 carried more mutations than those treated at E16.5 (Extended Data Fig. 3).
Extended Data Fig. 3. Chromosomal alterations in BM HSPCs from Eto-treated mice at the embryonic stage.
a, Experimental design for whole-genome sequencing (WGS) and chromosome fluorescence in situ hybridization (FISH). b, The genome mutation numbers in 3-week-old BM-LSK cells (E12.5 FL, n = 3; E16.5 FL, n = 4). The boxes delimit the minima and maxima, and the horizontal line represents the mean. c, d, Representative fluorescence images and proportions of normal and aberrant chromosomes in cells from mice treated with Eto at E12.5 or E16.5, chromosome 4 (FITC) and chromosome 6 (Texas red); arrows indicate aberrant chromosomes (Untreated, n = 6, E12.5 Eto, n = 15; E16.5 Eto, n = 5; mean ± s.d.). Scale bar, 10 µm. The n represents individual samples (b) and mice (c, d) from 3 independent experiments. Statistical tests: unpaired two-sided Student’s t-test (b), unpaired one-sided Student’s t-test with Welsh’s correction (d).
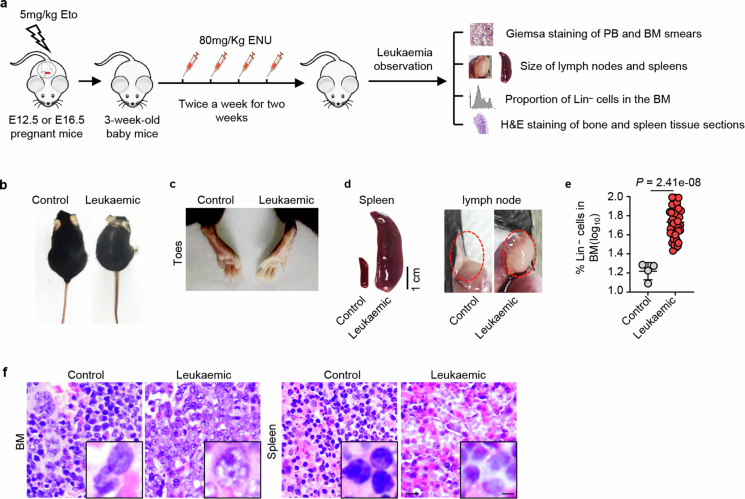
Subsequently, the offspring of mice treated with Eto at E12.5 or E16.5 was exposed to the mutagenic agent N-ethyl-N-nitrosourea at 3 weeks of age to assess their differential vulnerability to leukaemic induction (Extended Data Fig. 4a). The occurrence of leukaemia was assessed through daily observation of leukaemic symptoms and signs, such as white toes, swollen lymph nodes and loss of body weight. In addition, weekly measurements of leukaemic-like cells were conducted through Giemsa staining of peripheral blood smears. Once these symptoms appeared (Fig. 1k and Extended Data Fig. 4b,c), the animals were euthanized, and the presence of leukaemia was confirmed by observing enlarged spleens and lymph nodes (Extended Data Fig. 4d), increased proportions of immature Lin− cells in the bone marrow (Extended Data Fig. 4e) and definitive leukaemic cell infiltration in the bone marrow and spleen (Fig. 1l and Extended Data Fig. 4f). The latency period of induced leukaemia was significantly shorter in Eto-treated mice at E12.5 than in treated mice at E16.5 or untreated mice (Fig. 1m).
Extended Data Fig. 4. Leukaemia induction and observation in mice.
a, Experimental design for leukaemia induction and monitoring in mice. ENU, N-ethyl-N-nitrosourea; PB, peripheral blood; BM, bone marrow. b, c, The physical symptoms of leukaemic and normal control mice. d, Spleens and lymph nodes from leukaemic and control mice. b-d, Control, n = 5, Leukaemic, n = 51. e, The proportion of lineage-negative cells in the BM of leukaemic and control mice (Control, n = 5, Leukaemic, n = 51; mean ± s.d.). f, Representative H&E images of tissue sections (Control, n = 4; Leukaemic, n = 10). Scale bars, 100 and 25 μm. The n represents individual mice. Statistical tests: unpaired two-sided Student’s t-test (e).
In conclusion, HSPCs in the early fetal liver exhibit the greatest susceptibility to genotoxic stresses, leading to a remarkably unstable genome.
HSPCs lack hepatocyte refuge in the early fetal liver
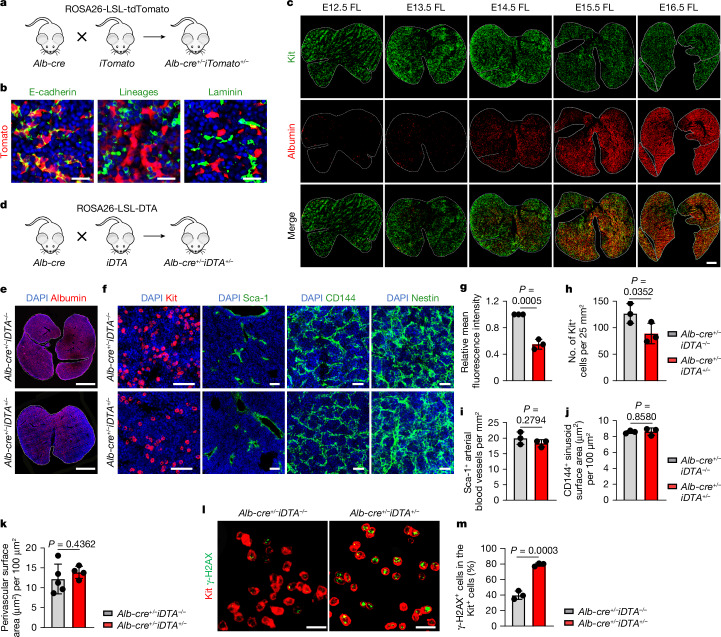
The selection of fetal livers as the primary site for haematopoiesis among developmental organs suggests that hepatocytes have a crucial role in HSPC maintenance, including maintaining genome integrity. Starting at E9.5, hepatocytes express albumin (Alb)21. To track hepatocyte development, Alb-Cre;ROSA26-LSL-tdTomato mice were used (Fig. 2a,b). By lineage tracing, we observed the relative cellular density and dynamics between hepatocytes and HSPCs (Kit+). In the early fetal liver, HSPCs make up the majority of cells with a small number of hepatocytes. However, HSPCs gradually decrease in number and become surrounded by expanding hepatocytes (Fig. 2c). This finding suggested that hepatocyte development lags behind HSPC development in the fetal liver. On the basis of this observation and the high susceptibility of the early fetal liver (E12.5–14.5) to genotoxic damage (Fig. 1), we hypothesized that hepatocytes are crucial for maintaining genomic stability in HSPCs. To test this hypothesis, we manipulated hepatocyte development in Alb-Cre;ROSA26-LSL-DTA mice (Fig. 2d). Compared with that in control Alb-cre mice, the number of hepatocytes in these mice decreased by approximately 50% at E16.5 (Fig. 2e,g). The number of HSPCs (Kit+) was reduced by 30% (Fig. 2f,h), whereas the density and distribution of arterial (Sca-1+) and sinusoid (CD144+) endothelial cells and pericytes (Nestin+) were not significantly altered22–24 (Fig. 2f,i–k). Eto treatment (5 mg kg−1) resulted in more severe DNA damage in the HSPCs of Alb-Cre;ROSA26-LSL-DTA mice than in those of control mice (Fig. 2l,m). These results support a crucial role of hepatocytes in protecting HSPCs from genotoxic insults in the fetal liver, although more subtle modifications of other niche components, and thus indirect effects on HSPCs via their niche, cannot be fully ruled out. These results, together with those of the comparative functional assays across developmental stages (Fig. 1b–e) and of the lineage-tracing observation (Fig. 2a–c), also suggest that HSPCs lack the refuge of fully developed hepatocytes in early fetal livers despite the limitations of our current depletion strategy (that is, possible effects earlier in development).
Fig. 2. HSPCs lack hepatocyte refuge in the early fetal liver.
a, The strategy of hepatocyte tracing using Alb-Cre;ROSA26-LSL-tdTomato mice. b, Alb-expressing cells (red) coexpressed the epithelial marker E-cadherin but not the haematopoietic cell lineage markers (CD3, B220, Gr-1, Mac-1 and Ter119) or the endothelial and mesenchymal marker laminin (n = 3). Scale bars, 25 µm. c, Representative fluorescence images showing tomato fluorescent protein-labelled hepatocytes and HSPCs (Kit (green)) in the developing fetal liver (n = 3). Scale bar, 500 µm. d, The strategy of hepatocyte depletion using Alb-Cre;ROSA26-LSL-DTA mice. e,g, Representative fluorescence images (e) and mean fluorescence intensity (g) of albumin red in the E16.5 fetal liver of Alb-cre+/–iDTA−/− and Alb-cre+/–iDTA+/− mice (n = 3). Scale bars, 1 mm. f,h–k, Representative fluorescence images (f) and the numbers of Kit+ cells (h), the arterial blood vessels (Sca-1+; i), the sinusoid surface area (CD144+; j) and the perivascular surface area (Nestin+; k) in the E16.5 fetal liver of Alb-cre+/–iDTA+/− and Alb-cre+/–iDTA−/− mice (n = 3; n = 5 for Alb-cre+/–iDTA−/−; n = 4 for Alb-cre+/–iDTA+/− (h–k)). Scale bars, 25 µm and 100 µm. l,m, Representative fluorescence images (l) and positive cell proportions (m) of γ-H2AX in Kit+ HSPCs in the E16.5 fetal liver from Alb-cre+/–iDTA+/− and Alb-cre+/–iDTA−/− mice after Eto treatment in vivo (n = 3). Scale bars, 20 µm. The n represents independent experiments (b,c) and individual fetuses from three independent experiments (e–m). The mean ± s.d. is shown (g–k,m). Unpaired one-sided Student’s t-test (h) and unpaired two-sided Student’s t-test (g,i–k,m) were used.
Hepatocytes secrete protective FetuA
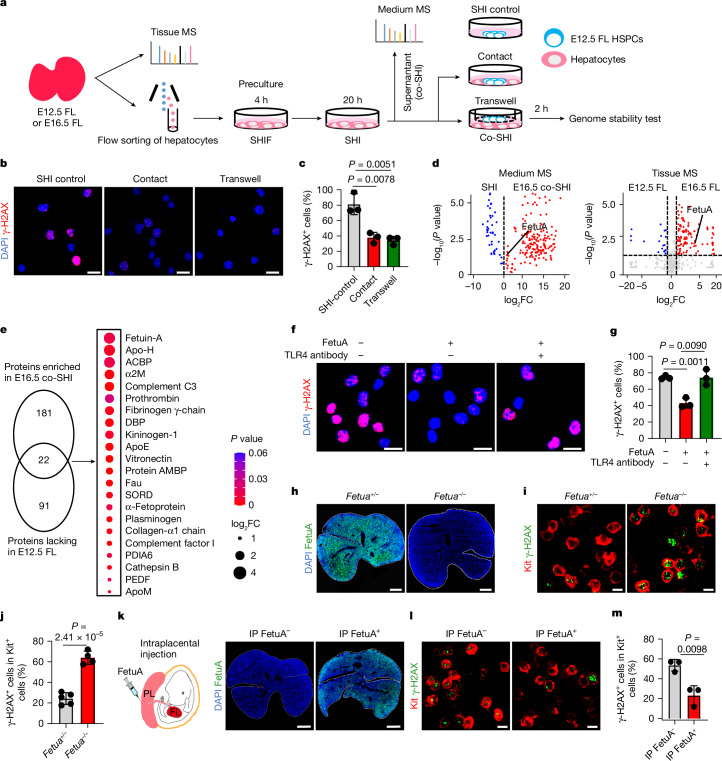
To directly examine the effects of hepatocytes on the genomic protection of HSPCs and determine the underlying mechanism, we isolated Alb–tomato-positive hepatocytes from Alb-Cre;ROSA26-LSL-tdTomato mice at E12.5 or E16.5 (Fig. 3a). The purified hepatocytes were precultured in StemSpan medium supplemented with hepatocyte growth supplement, IL-6 and fetal bovine serum (SHIF) and then cultured in serum-free SHI medium25,26. E12.5 fetal liver HSPCs were then co-cultured in contact with hepatocytes or isolated from hepatocytes using a Transwell system that allows molecular factors to pass through. Genomic stability tests were subsequently conducted (Fig. 3a). Eto treatment resulted in significantly less DNA damage in HSPCs in co-culture, either in contact with or isolated from hepatocytes, than in control cultures without hepatocytes (Fig. 3b,c). Similar results were obtained when co-culturing LT-HSCs, ST-HSCs and MPPs (Extended Data Fig. 5a,b). Therefore, fetal hepatocytes secrete paracrine factors that protect the HSPC genomes.
Fig. 3. Hepatocyte-paracrine FetuA protects the HSPC genome.
a, Mechanistic investigation of the effect of hepatocytes on HSPCs. SHIF, SHI and co-SHI are culture media (see Methods). MS, mass spectrometer. b,c, Representative fluorescence images (b) and positive proportions (c) of γ-H2AX Eto induced in E12.5 fetal liver HSPCs in culture (n = 3). Scale bars, 20 µm. d, Volcano plots showing proteins in the media (n = 3 for SHI and n = 4 for co-SHI) and secretory proteins in fetal liver tissues (n = 4). The vertical dashed lines indicate the fold change (FC) threshold (log2FC = 0, log2FC > 1 or log2FC < −1), and the horizontal dashed line indicates the P value threshold (P = 0.05). e, Venn diagram showing proteins enriched in E16.5 co-SHI and lacking in the E12.5 fetal liver (more than twofold change, P < 0.05). The bubble chart and colours show the fold change and P values for the comparison between the E12.5 and E16.5 fetal liver. f,g, Representative fluorescence images (f) and positive proportions (g) of γ-H2AX Eto induced in HSPCs with or without FetuA and TLR4 antibodies in culture (n = 3). Scale bars, 20 µm. h, Representative fluorescence images of FetuA in the E16.5 fetal liver of Fetua+/– or Fetua–/– mice. Scale bars, 500 µm. i,j, Representative fluorescence images (i) and positive proportions (j) of γ-H2AX in Kit+ HSPCs in the E16.5 fetal liver Eto induced in Fetua+/– or Fetua–/– mice (n = 5 for Fetua+/– and n = 4 for Fetua–/–). Scale bars, 10 µm. k–m, Representative fluorescence images of FetuA (k) or γ-H2AX (l) and its positive proportions (m) in Kit+ HSPCs in the E16.5 fetal liver Eto induced in Fetua–/– mice with or without FetuA (n = 3). Scale bars, 500 µm (k) and 10 µm (l). IP, intraplacental. The n represents individual samples (b–g) and fetuses (h–m) from three (b,c,f–m) or four (d) independent experiments. The mean ± s.d. is shown (c,g,j,m). Unpaired two-sided Student’s t-test (c–e,g,j,m) was used.
Extended Data Fig. 5. The hepatocyte-secreted factor FetuA provides genome protection for HSPCs.
a, b, Representative fluorescence images and positive proportions of γ-H2AX induced by Eto in E12.5 FL LT-HSCs, ST-HSCs and MPPs cultured with or without hepatocytes (refer to Fig. 3a) (n = 3). Scale bar, 20 µm. c, d, Representative fluorescence images and positive proportions of γ-H2AX Eto-induced in E12.5 FL-HSPCs in cultures of E16.5 or E12.5 hepatocyte-conditioned medium (co-SHI) or basic medium (SHI) (SHI-control, n = 6, E12.5 Co-SHI, n = 3; E16.5 Co-SHI, n = 5). Scale bar, 20 µm. e, Western-blots analysis of FetuA in SHI and co-SHI media and in E12.5 and E16.5 FL tissues; Coomassie blue was used as a loading control (n = 3). f, Representative fluorescence images of FetuA in E12.5 FL, E16.5 FL and E19 FL (n = 3). Scale bar, 500 µm. g, Representative fluorescence images of FetuA and hepatocytes (E-cadherin+), haematopoietic (CD45/Ter119+), endothelial (Scal-1 or CD144+), mesenchymal (Nestin+) cells in E16.5 FL (n = 3). Scale bars, 10 and 5 µm. h, Representative fluorescence images of FetuA (green) and HSPCs (Lin–CD41–CD48–CD150+) in E16.5 FL (n = 3). Scale bars, 10 and 5 µm. i, j, Representative comet-tail DNA images and the proportions of Eto-induced tail DNA in HSPCs cultured with or without FetuA (n = 3). The medians (red dashed lines) and quartiles (black dashed lines) of each group are shown. Scale bar, 100 µm. k, Representative fluorescence images of FetuA in the E19 bone marrow (BM) of Fetua+/– and Fetua–/– mice. Scale bar, 500 µm. l, m, Representative fluorescence images and positive cell proportions of γ-H2AX (green) in Kit+ cells (red) in the E19 BM of Fetua+/– or Fetua–/– mice (n = 3). Scale bar, 10 µm. n, Representative fluorescence images of FetuA in the E12.5 FL of mice with or without intraplacental (IP) injection of FetuA. Scale bar, 300 µm. o, p, Representative fluorescence images and positive cell proportions of γ-H2AX (green) in Kit+ cells (red) in the E12.5 FL of mice with or without IP injection of FetuA followed by Eto-treatment in vivo (IP FetuA–, n = 4; IP FetuA+, n = 3). Scale bar, 10 µm. q, Representative fluorescence images of FetuA in the E16.5 FL of Alb-cre; ROSA26-LSL-DTA mice with or without IP injection of FetuA. Scale bar, 500 µm. r, s, Representative fluorescence images and positive cell proportions of γ-H2AX (green) in Kit+ cells (red) in the E16.5 FL of Alb-cre; ROSA26-LSL-DTA mice with or without IP injection of FetuA followed by Eto-treatment in vivo (IP FetuA–, n = 3; IP FetuA+, n = 4). Scale bar, 10 µm. The n represents independent experiments (a-j) and fetuses (k-s) from 3 independent experiments. The mean ± s.d. is shown (b, d, m, p, s). Statistical tests: unpaired two-sided Student’s t-test (b, d, j, m, p, s).
To identify the factors involved, the supernatant (conditioned medium (co-SHI)) was collected (Fig. 3a). Culturing of HSPCs in co-SHI, which were conditioned from either E12.5 or E16.5 hepatocytes, increased the resistance of HSPCs to Eto compared with that of HSPCs cultured in basic SHI medium (Extended Data Fig. 5c,d). These results further support that the lack of genomic protection in the early fetal liver is due to the relatively low number of hepatocytes compared with that of haematopoietic cells (Fig. 2c). E16.5 hepatocyte-conditioned co-SHI was used for the identification of genome-protective factors due to its greater availability. The SHI and co-SHI media were analysed by mass spectrometry. A total of 203 proteins were more enriched in co-SHI than in SHI (Fig. 3d). The proteomes of fetal liver tissues at E12.5 and E16.5 were also compared. Exactly 113 secretory proteins were lacking (fold change ≥ 2; P < 0.05) in E12.5 fetal livers compared with E16.5 fetal livers (Fig. 3d), and 22 secretory proteins were enriched in co-SHI (Fig. 3e). FetuA was identified among the significantly altered proteins. The presence and concentration of FetuA in the medium and fetal liver tissues were confirmed (Extended Data Fig. 5e,f and Supplementary Fig. 2). Double immunofluorescence stainings showed the presence of FetuA inside hepatocytes (E-cadherin+) and absence inside haematopoietic (CD45+ or Ter119+), endothelial (Scal-1+ or CD144+) and mesenchymal (Nestin+) cells (Extended Data Fig. 5g). Hence, FetuA is primarily expressed by hepatocytes and so is commonly considered as a hepatokine15,16. FetuA colocalized on the cell surface of HSPCs (CD150+) in the E16.5 fetal liver in situ, indicating that hepatocytes support HSPCs through a paracrine mechanism (Extended Data Fig. 5h). Recombinant FetuA was used to treat E12.5 fetal liver HSPCs in culture, and the resulting HSPCs became more tolerant of Eto (Fig. 3f,g and Extended Data Fig. 5i,j).
To further confirm the genome-protective role of FetuA in the fetal liver and perinatal bone marrow, we treated Fetua-knockout (FKO) mice (Fig. 3h and Extended Data Fig. 5k) with 5 mg kg−1 Eto at E16.5 or at E19. HSPCs from Fetua–/– mice were more sensitive to genotoxins than those from Fetua+/– mice, confirming the protective role of FetuA in the fetal liver (Fig. 3i,j) and perinatal bone marrow (Extended Data Fig. 5l,m).
Finally, we investigated whether the lack of HSPC genomic protection from hepatocytes in mice at the early fetal liver developmental stage, in FKO mice and in Alb-Cre;ROSA26-LSL-DTA mice could be remedied in vivo. Recombinant FetuA was administered via intraplacental injection. At 2 h post-injection, the concentration of FetuA in the fetal liver increased (Fig. 3k and Extended Data Fig. 5n,q). Eto treatment (5 mg kg−1) induced less DNA damage to HSPCs in FetuA-treated fetuses than in untreated fetuses (Fig. 3l,m and Extended Data Fig. 5o,p,r,s). Hence, the HSPC genome was protected from genotoxic attack by FetuA administration in vivo.
In conclusion, the hepatocyte paracrine FetuA provides HSPCs with genomic protection from genotoxic stresses in the fetal liver, although a contribution of FetuA from cells other than hepatocytes cannot be excluded in the absence of tissue-specific deletion of FetuA.
FetuA activates TLR4 to reduce the R-loop
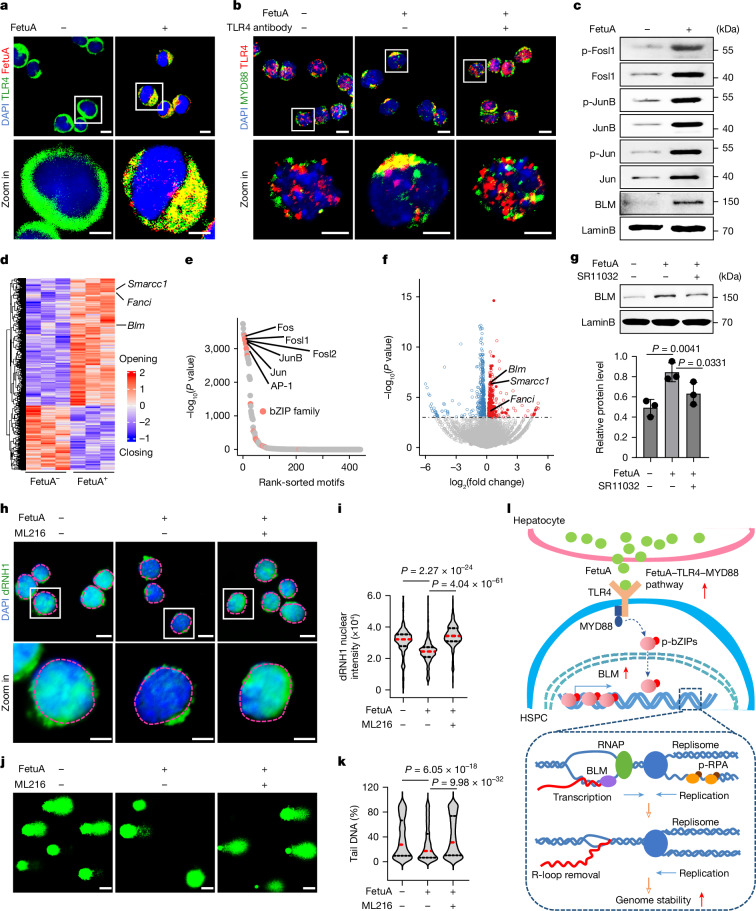
Toll-like receptor 4 (TLR4) serves as the receptor for FetuA27. The protective effects of FetuA on the HSPC genome were nullified by a neutralizing antibody to TLR4 (Fig. 3f,g). The colocalization of FetuA with TLR4 was observed on the surface of cultured HSPCs (Fig. 4a). Upon FetuA treatment, the colocalization and polarization of TLR4 and MYD88 in the cell membrane indicated the formation of a FetuA–TLR4–MYD88 complex and the potential activation of their downstream pathway28 (Fig. 4b). Furthermore, FetuA treatment led to an increase in the expression of basic leucine zippers (bZIPs; Jun, JunB and Fosl1) and phosphorylated bZIPs in HSPCs, as confirmed by western blotting (Fig. 4c and Supplementary Fig. 2).
Fig. 4. FetuA activates the TLR4 pathway to prevent R-loop accumulation.
a, Representative fluorescence images of FetuA and TLR4 on E12.5 fetal liver HSPCs with or without FetuA treatment. Scale bars, 10 µm and 5 µm. The white squares represent the areas enlarged in the bottom row. b, Representative fluorescence images of TLR4 and MYD88 on HSPCs cultured with or without FetuA and TLR4 antibodies. Scale bars, 10 µm and 5 µm. c, Western blots of bZIPs (Fosl1, JunB and Jun), their phosphorylated (p) forms and BLM in E12.5 fetal liver HSPCs cultured with or without FetuA. d, ATAC-seq analysis of E12.5 fetal liver HSPCs with or without FetuA treatment, showing the differential opening and closing peaks and R-loop regulatory genes Smarcc1, Fanci and Blm. e, DNA-binding factors whose motifs were enriched in the opening regions of panel d. f, Volcano plot showing the differential gene expression. The horizontal dashed line shows the P value threshold (P = 0.05). The blue and red spots show the downregulated or upregulated genes, respectively, in FetuA-cultured HSPCs. g, Western blots (top) and signal intensity (bottom) of BLM in E12.5 fetal liver Lin– HSPCs cultured with or without FetuA and the bZIP inhibitor SR11032 (data shown as mean ± s.d.). h,i, Representative fluorescence images (h) and nuclear signal intensity (i) of dRNH1 in E12.5 fetal liver HSPCs cultured with or without FetuA and the BLM inhibitor ML216. The nuclear regions are circled by dotted lines. Scale bars, 10 µm and 5 µm. j,k, Representative comet-tail DNA images (j) and percentages (k) of Eto induced in E12.5 fetal liver HSPCs cultured with or without FetuA and the BLM inhibitor ML216. Scale bars, 20 µm. l, A working model of the FetuA–TLR4 pathway in protecting the HSPC genome. RPA, replication protein A; RNAP, RNA polymerase. n = 3 independent experiments (a–k). The medians (red dashed lines) and quartiles (black dashed lines) are shown (i,k). Two-sided binomial test (e), two-sided Wald test (f), unpaired one-sided Student’s t-test (g) and unpaired two-sided Student’s t-test (i,k) were used.
To investigate the mechanism by which the FetuA-activated TLR4–MYD88–bZIP pathway provides protection to the HSPC genome, we conducted a comparative analysis of the chromatin state and corresponding transcriptome in E12.5 fetal liver HSPCs with or without FetuA treatment. This was achieved through assay for transpose-accessible chromatin using sequencing (ATAC-seq) and RNA-seq. The acquired ATAC-seq data exhibited consistency among the three replicates in each group (Extended Data Fig. 6a). The accessible chromatin in the nucleosome-free and nucleosome-associated regions was readily discernible for further analysis (Extended Data Fig. 6b). The differential opening and closing peaks are depicted (Fig. 4d). Of note, the opening regions post-FetuA treatment exhibited an enrichment of bZIP motifs (Fig. 4e), which aligns with the increase in protein expression and phosphorylation (Fig. 4c). bZIP motifs were enriched in genes involved in the regulation of R-loop formation, such as the helicase Blm, Fanci and Smarcc1 (Fig. 4d and Extended Data Fig. 6c), and these genes exhibited transcriptional upregulation3,29–31 (Fig. 4f). Furthermore, the binding of the transcription factor bZIPs to the Blm gene was identified by Cut&Tag for Jun, JunB and Fosl1 (Extended Data Fig. 6d,e). To further validate the regulatory effect of FetuA on Blm expression through bZIPs, a series of biochemical experiments was performed. BLM protein expression was much lower in E12.5 fetal liver HSPCs than in E16.5 HSPCs (Extended Data Fig. 6f and Supplementary Fig. 2). FetuA-treatment promoted Blm expression in both LSK-enriched and Lin−-enriched E12.5 fetal liver HSPCs (Fig. 4c,g, Extended Data Fig. 6g,h and Supplementary Fig. 2), and this promotional effect was nullified by a specific bZIP inhibitor, SR11032 (Fig. 4g and Supplementary Fig. 2), indicating that FetuA regulates Blm expression by activating the TLR4–bZIP pathway; however, nonspecific effects of the inhibitor cannot be fully ruled out.
Extended Data Fig. 6. Effects of FetuA on HSPCs.
a, Scatter-plots showing the consistency of ATAC-seq replicates for each group (n = 3). b, Fragment size distribution of ATAC-seq data (n = 3). c, ATAC-seq signals of Blm, Fanci and Smarcc1 in Integrative Genomics Viewer are shown; the red boxes indicate the peaks where the bZIP motifs are located (n = 3). d, Heatmaps showing the Cut & Tag peak signal intensities (n = 2). e, Cut & Tag sequencing signals of Jun, JunB and Fosl1 on the Blm gene from the Integrative Genomics Viewer are shown (n = 2). Cut & Tag for Jun binding to the Blm promoter. f, Western-blots analysis of the BLM protein in LSK cells from E12.5 FL and E16.5 FL (n = 2). g, h, Western-blots and relative signal intensity of the BLM protein in Lin– cells cultured with or without FetuA at different time points (n = 3; mean ± s.d.). The n represents independent experiments. Statistical tests: unpaired two-sided Student’s t-test (h).
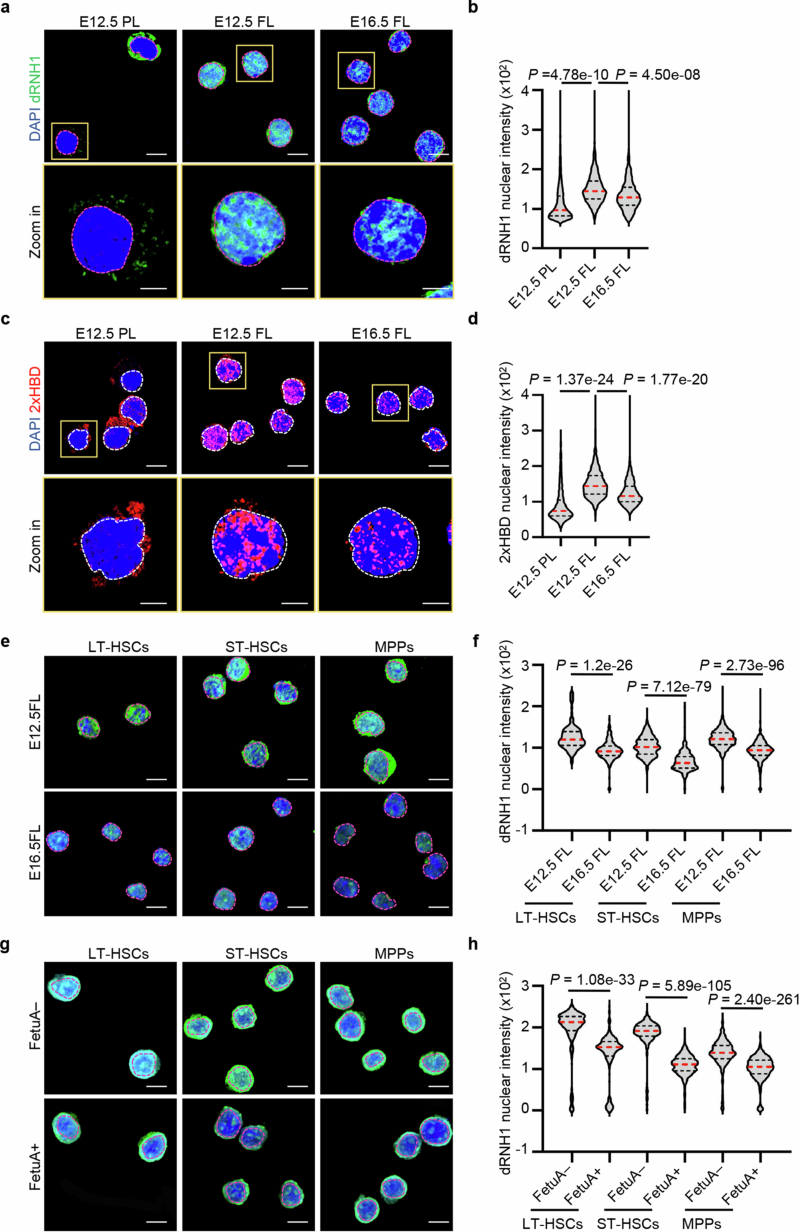
These findings suggest that the accumulation of R-loops is a contributing factor to the increased chromosome fragility in early fetal liver HSPCs. In addition, our study suggested that the FetuA-activated pathway acts as a mechanism to protect the genome by interfering with the formation or removal of R-loops. To support this hypothesis, we conducted an immunofluorescence assay using dRNH1 or 2×HBD antibodies, which specifically bind to DNA-RNA hybrids32,33. We observed a stronger signal in E12.5 fetal liver HSPCs than in the E12.5 placenta and E16.5 fetal liver HSPCs (Extended Data Fig. 7a–d). Furthermore, the dRNH1 signal in E12.5 fetal liver HSPCs was significantly reduced after FetuA treatment (Fig. 4h,i). The specific inhibitor ML216, which targets the BLM helicase, reversed the removal effect of FetuA on R-loops (Fig. 4h,i) and the protective effects on the HSPC genome (Fig. 4j,k). These results demonstrated that FetuA regulates BLM to resolve R-loops and to protect the HSPC genome; however, the nonspecific effects of the inhibitor cannot be fully excluded.
Extended Data Fig. 7. Immunofluorescence assay of dRNH1 and 2xHBD in HSPCs.
a, b, Representative fluorescence images and nuclear intensity of dRNH1 in HSPCs from E12.5 PL, E12.5 FL and E16.5 FL (n = 3). Scale bars, 10 and 5 µm. c, d, Representative fluorescence images and nuclear intensity of 2xHBD in HSPCs from E12.5 PL, E12.5 FL and E16.5 FL (n = 3). Scale bars, 10 and 5 µm. e, f, Representative fluorescence images and nuclear intensity of dRNH1 in HSCs and MPPs from E12.5 FL and E16.5 FL (n = 3). Scale bar, 10 µm. g, h, Representative fluorescence images and nuclear signal intensity of dRNH1 in E12.5 FL HSCs and MPPs cultured with or without FetuA (n = 3). Scale bar, 10 µm. The nuclear regions are circled by dotted lines (a, c, e, g). The medians (red dashed lines) and quartiles (black dashed lines) are shown (b, d, f, h). The n represents independent experiments. Statistical tests: unpaired two-sided Student’s t-test (b, d, f, h).
Moreover, when we evaluated more enriched LT-HSCs, ST-HSCs and MPPs, the dRNH1 signal was stronger in all three populations of E12.5 fetal livers than in E16.5 fetal livers (Extended Data Fig. 7e,f), and the signal in E12.5 fetal liver cells was reduced after FetuA treatment (Extended Data Fig. 7g,h).
Together, our results demonstrate that FetuA protects the HSPC genome by activating the TLR4–bZIP–BLM cascade to prevent R-loop accumulation (Fig. 4l).
Replication and transcription in HSPCs
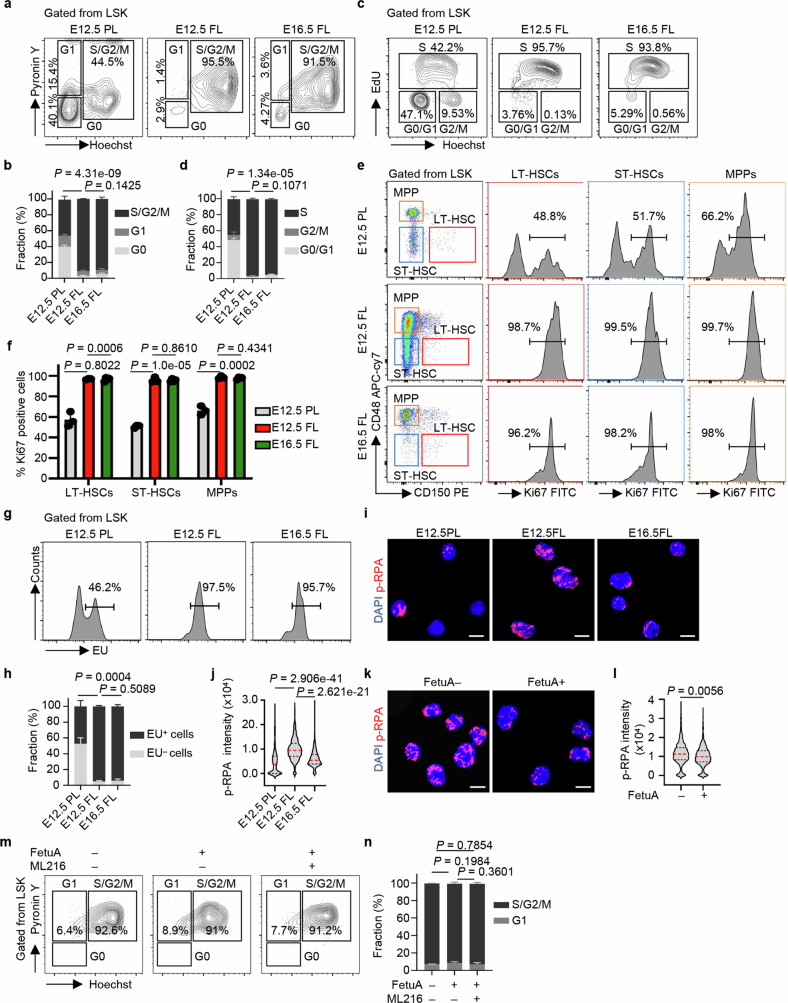
Considering that pathogenetic R-loops are possibly formed by collisions between DNA replication and gene transcription, we conducted comparative analyses of the cell cycle status of HSPCs between E12.5 placentas and E12.5 and E16.5 fetal livers. The results showed that a larger proportion of HSPCs remained in a quiescent state (G0 stage) in the placenta, whereas most of the HSPCs quickly entered the cell cycle upon migration into the fetal liver, as evidenced by DNA–Hoechst and RNA–pyronin Y staining ex vivo (Extended Data Fig. 8a,b). Furthermore, the majority of HSPCs in fetal livers were in the S stage, as indicated by the incorporation of synthetic DNA de novo labelled with 5-ethynyl-2′-deoxyuridine in vivo and measured by flow cytometry (Extended Data Fig. 8c,d). Consistent dynamics were observed in each fraction of LT-HSCs, ST-HSCs and MPPs when analysing the three fractions within LSK cells in the cell cycle (Extended Data Fig. 8e,f). These results suggest that an increased proportion of HSPCs undergo expansion and DNA replication after migrating into the fetal liver.
Extended Data Fig. 8. Fetal liver HSPCs undergo DNA-replication and transcription simultaneously.
a, b, Representative flow cytometry images and the proportions of each cell cycle stage in HSPCs from E12.5 PL, E12.5 FL and E16.5 FL cells stained with Hoechst 33342 and PyroninY (E12.5 PL, E16.5 FL, n = 3; E12.5 FL, n = 6; S/G2/M phase was compared). c, d, Representative flow cytometry images and the proportions of HSPCs in each cell cycle stage at E12.5 PL, E12.5 FL and E16.5 FL after 5-ethynyl-2’-deoxyuridine (EdU) treatment (n = 3 per group; S phase was compared). e, f, Representative flow cytometry images and Ki67-positive cell proportions in HSCs and MPPs from E12.5PL, E12.5FL and E16.5FL (n = 3 per group). g, h, Representative flow cytometry images and the ethyluridine (EU)-positive fraction of HSPCs from E12.5 PL, E12.5 FL and E16.5 FL after EU-treatment (E12.5 PL, E12.5 FL, n = 3; E16.5 FL, n = 6; EU+ population was compared). i, j, Representative fluorescence images and nuclear signal intensity of p-RPA in HSPCs from E12.5 PL, E12.5 FL and E16.5 FL (n = 3). The medians (red dashed lines) and quartiles (black dashed lines) are shown. Scale bar, 10 µm. k, l, Representative fluorescence images and nuclear signal intensity of p-RPA in E12.5 FL-HSPCs treated with or without FetuA (n = 3). The medians (red dashed lines) and quartiles (black dashed lines) are shown. Scale bar, 10 µm. m, n, Representative flow cytometry images and the proportion of each cell cycle stage in HSPCs cultured with or without FetuA and the BLM inhibitor ML216 (n = 3). The mean ± s.d. is shown (b, d, f, h, n). The n represents individual samples from 3 independent experiments (a-h) and independent experiments (i-n). Statistical tests: unpaired two-sided Student’s t-test (b, d, f, h, j, l, n).
Moreover, we performed ethyluridine incorporation experiments to label synthetic RNA de novo. The results revealed a significantly greater proportion of ethyluridine-positive HSPCs in the fetal liver than in the placenta, indicating an increased proportion of HSPCs undergoing gene transcription in the fetal liver (Extended Data Fig. 8g,h).
Simultaneous DNA replication and gene transcription in fetal liver HSPCs may lead to an increase in collisions. An immunofluorescence assay for phosphorylated RPA (p-RPA), which is a marker for stalled replication caused by replication–transcription collisions, revealed that the signal significantly increased in HSPCs in E12.5 fetal livers compared with E12.5 placentas and then decreased again in E16.5 fetal livers (Extended Data Fig. 8i,j). In addition, the p-RPA signal decreased in E12.5 fetal liver HSPCs after FetuA treatment (Extended Data Fig. 8k,l). Therefore, the dynamics of the p-RPA signal were consistent with the changes in R-loop accumulation and genome instability (Fig. 4h–k). However, treatment with FetuA and the BLM inhibitor ML216 did not have a significant effect on the cell cycle of E12.5 fetal liver HSPCs (Extended Data Fig. 8m,n), indicating that FetuA did not protect HSPC genomes by affecting their cell cycle; however, potential off-target effects of the inhibitor cannot be excluded.
In summary, fetal liver HSPCs experience simultaneous DNA replication and gene transcription, leading to increased replication–transcription conflicts, R-loop accumulation and subsequent genomic instability until they receive sufficient genomic protection from the hepatocyte-paracrine FetuA in the fetal liver (Fig. 4l).
Lack of FetuA results in leukaemogenesis
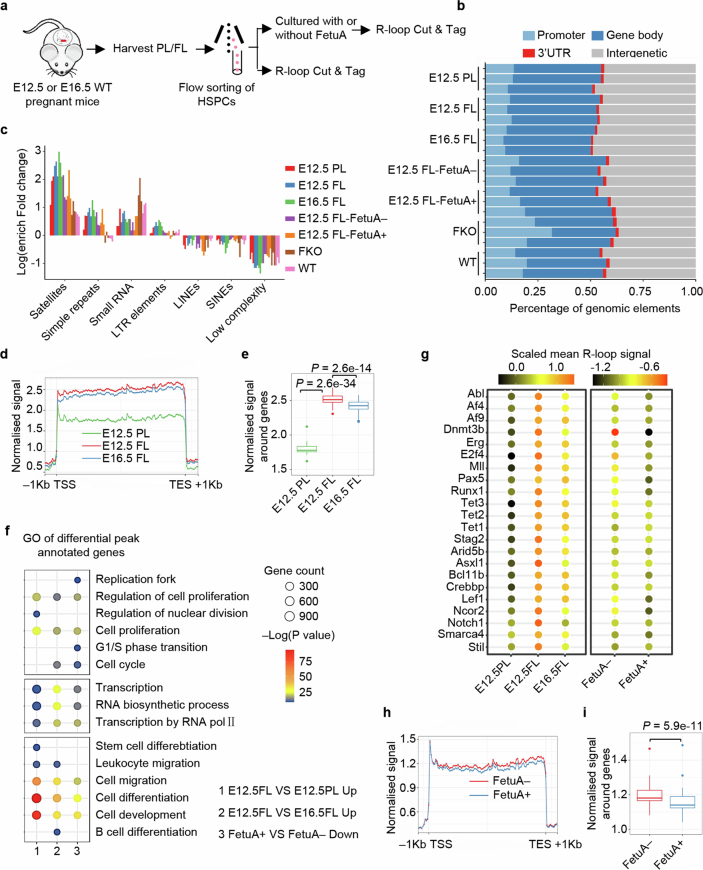
Having established the protective effect of FetuA in reducing R-loop accumulation in HSPC genomes (Figs. 3 and 4), we performed genome-wide R-loop sequencing (Cut&Tag for R-loop) to investigate whether the R-loops enriched in haematopoietic regulatory genes are involved in leukaemogenic mutations34 (Extended Data Fig. 9a). The genomic distribution of R-loop peaks in each cell type and their enrichment in various types of repetitive elements indicated that the data were qualified for further analysis35,36 (Extended Data Fig. 9b,c). Gene metaplots analysis demonstrated greater R-loop enrichment in gene bodies in the E12.5 fetal liver than in the E12.5 placenta and E16.5 fetal liver (Extended Data Fig. 9d,e), which is consistent with the results of dRNH1 and 2×HBD immunofluorescence (Extended Data Fig. 7a–d). Gene Ontology analysis of differentially annotated peak genes revealed that the differential R-loop accumulation peaks in the E12.5 fetal liver compared with the E12.5 placenta and E16.5 fetal liver were associated with DNA replication (cell proliferation), gene transcription and developmental haematopoiesis (Extended Data Fig. 9f), particularly with genes frequently implicated in leukaemogenesis37–40 (Extended Data Fig. 9g). Consistently, after FetuA treatment, the overall R-loop signal was reduced (Extended Data Fig. 9h,i), and less R-loop accumulation was observed in the regulatory gene regions involved in haematopoiesis (Extended Data Fig. 9f,g).
Extended Data Fig. 9. Genome-wide R-loop and DNA sequencing assays of fetal HSPCs.
a, Experimental design for genome-wide R-loop sequencing (R-loop Cut & Tag). b, Genomic distribution of R-loop peaks in each sample. c, Enrichment of R-loops in various types of repetitive elements in each sample. d, Gene-metaplots analysis of R-loop Cut & Tag; mean coverages of R-loop signals over gene bodies (±1 kb) in the HSPCs from E12.5 PL, E12.5 FL and E16.5 FL are shown. e, Box-plots showing the R-loop signal over gene bodies (±1 kb) in HSPCs from E12.5PL, E12.5 FL and E16.5 FL. The boxes delimit the lower (25th) and upper (75th) interquartile, and the horizontal line represents the median. f. GO analysis of differentially expressed peak-annotated genes; the circle size shows the number of genes associated with each GO term, and the circle colour denotes the –Log (P value). g, Mean R-loop signal of the genes that are frequently involved in leukemogenesis in HSPCs from E12.5 PL, E12.5 FL, and E16.5 FL and in E12.5 FL HSPCs cultured with or without FetuA. h, Gene-metaplots analysis of R-loop Cut & Tag; mean coverages of R-loop signals over gene bodies (±1 kb) in E12.5 FL HSPCs cultured with or without FetuA are shown. i, Box-plots showing the R-loop signal over gene bodies ( ± 1 kb) in E12.5 FL HSPCs cultured with or without FetuA. The boxes delimit the lower (25th) and upper (75th) interquartile, and the horizontal line represents the median. n = 3 independent experiments (b-i). Statistical tests: two-sided Wilcoxon test (e, i).
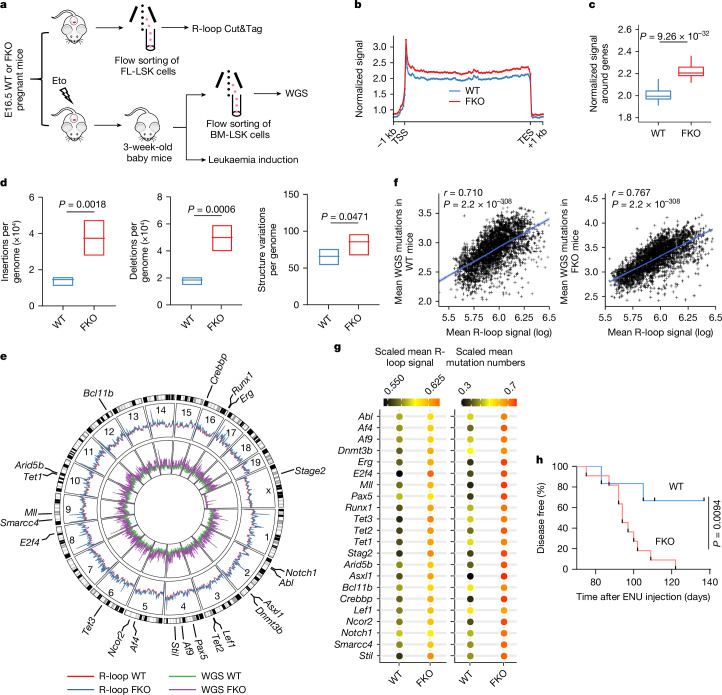
To further determine the effect of FetuA on R-loop accumulation and mutagenesis, we conducted genome-wide R-loop sequencing and whole-genome sequencing in HSPCs from FKO mice and control wild-type (WT) mice (Fig. 5a). As expected, R-loop accumulation (Fig. 5b,c) and mutation numbers (Fig. 5d) were significantly greater in FKO mice than in WT mice. The circus plots displayed the genome-wide distribution of identified R-loop events and mutations (Fig. 5e). The distribution of R-loops was positively correlated with mutations in both the presence and the absence of FetuA (Fig. 5f). Moreover, we observed that numerous important haematopoietic regulatory genes were located in these R-loop-enriched and highly mutated regions (Fig. 5g). These findings suggest that the absence of FetuA protection may lead to vulnerability to leukaemogenesis. When comparing the oncogenesis of leukaemia induced by Eto treatment at E16.5 and N-ethyl-N-nitrosourea treatment postnatally in WT and FKO mice, we found that the latency to induced leukaemia was significantly shorter in FKO mice than in WT mice (Fig. 5h).
Fig. 5. A lack of FetuA increased vulnerability to leukaemogenesis.
a, Experimental design for genome-wide R-loop sequencing (R-loop Cut&Tag), whole-genome sequencing (WGS) and leukaemia induction. b, Gene metaplots analysis of R-loop Cut&Tag. The mean coverages of R-loop signals over gene bodies (±1 kb) in E16.5 fetal liver HSPCs from WT and FKO mice are shown (n = 3). TSS, transcription start site; TES, transcription end site. c, Boxplots showing the R-loop signal over gene bodies (±1 kb) in E16.5 fetal liver HSPCs from WT and FKO mice (n = 3). The boxes delimit the lower (25th) and upper (75th) interquartile, and the horizontal line represents the median. d, The number of chromosomal insertions (left), deletions (middle) and structural variations (right) per genome in bone marrow HSPCs from 3-week-old Eto-treated WT and FKO mice at E16.5 (n = 4). The boxes delimit the minima and maxima, and the horizontal line represents the mean. e, Circos plots showing the genome-wide distribution of the identified R-loop events (outer circles; n = 3 per group) and mutations (inner circles; n = 4 per group) in HSPCs from WT and FKO mice. f, Dot plots revealing the correlation between R-loop signals and mutations in WT (left) and FKO (right) mice (n = 3 for R-loop and n = 4 for WGS). g, Mean R-loop signal (n = 3) and mutation numbers (n = 4) of the genes frequently involved in leukaemia in HSPCs from WT and FKO mice. h, The onset of leukaemia induced in WT and FKO mice by Eto treatment at E16.5 and N-ethyl-N-nitrosourea (ENU) treatment at 3 weeks of age (see Extended Data Fig. 4a; n = 6 for WT and n = 11 for FKO). The n represents individual samples (b–g) and mice (h) from three independent experiments. Two-sided Wilcoxon test (c), unpaired two-sided Student’s t-test (d), two-sided Pearson test (f) and log-rank (Mantel–Cox) test (h) were used.
In summary, FetuA is essential for maintaining genome stability of fetal HSPCs, and a lack of FetuA increases susceptibility to mutagenesis and leukaemogenesis.
Similar protective mechanism in humans
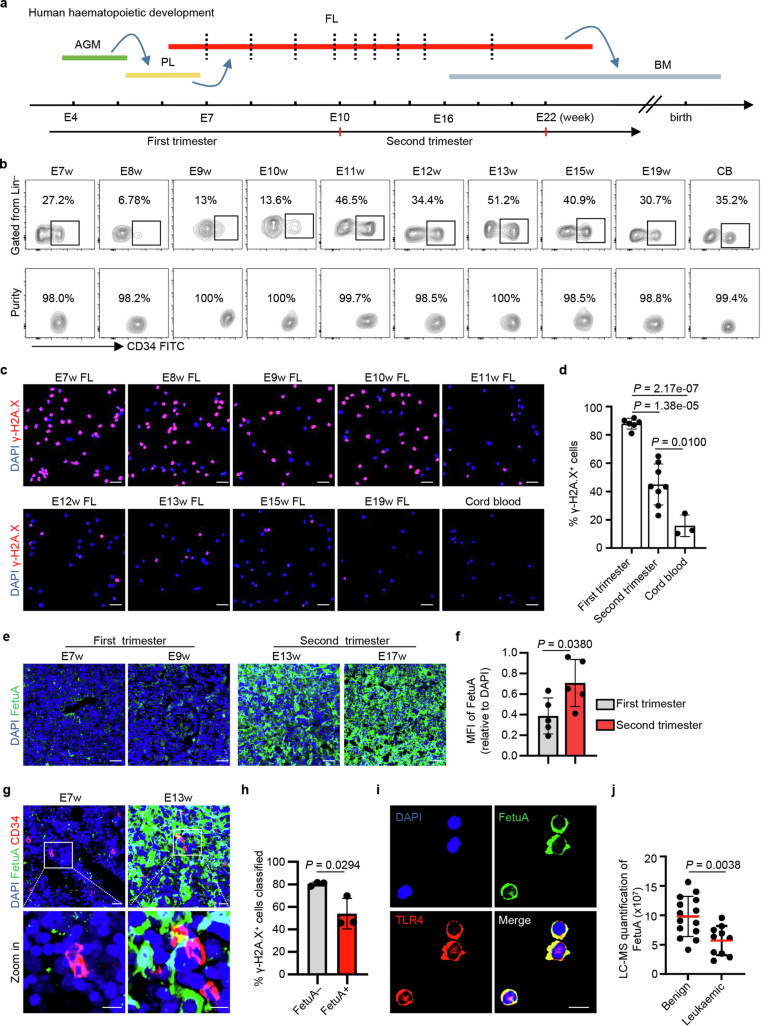
Human samples were examined to determine the conservation between mice and humans (Extended Data Fig. 10a,b). Eto treatment caused more DNA damage in HSPCs (Lin− and CD34+)41 from the early fetal liver of the first trimester (embryonic weeks 7–10 (E7–10w)) than in HSPCs from the later fetal liver of the second trimester (E11–19w) or neonatal cord blood (Extended Data Fig. 10c,d). Similarly, human HSPCs in the early fetal liver were more susceptible to genotoxic insult than those in the later fetal liver or cord blood, consistent with findings in mice.
Extended Data Fig. 10. Similar genome-protective mechanism in humans.
a, Schematic of haematopoietic development and the tissues used for HSPC harvest (red dashed lines) for the assessment of genome stability. b, Immunophenotypically lineage-negative (Lin−) and CD34-positive (CD34+) cells were flow-sorted with high purities from the haematopoietic tissue of E7w fetal liver (FL), E8w FL, E9w FL, E10w FL, E11w FL, E12w FL, E13w FL, E15w FL, E16w FL and neonatal cord blood (CB) to determine the sensitivity of HSPCs to genotoxic agents. c, d, Representative fluorescence images and positive proportions of γ-H2AX induced by Eto in human HSPCs isolated from FL and CB (first trimester, n = 6; second trimester, n = 8; cord blood, n = 3). Scale bar, 50 µm. e, f, Representative fluorescence images and mean fluorescence intensity (MFI) of FetuA (green) in human FL at the indicated developmental stages (n = 5 per group). Scale bar, 50 µm. g, Representative fluorescence images of FetuA and CD34 (red) in FL (n = 3). Upper scale bar, 20 µm; lower scale bar, 10 µm. h, Proportions of γ-H2AX-positive FL-HSPCs cultured with or without FetuA induced by Eto in the first trimester (n = 3). i, Representative fluorescence images of FetuA and TLR4 on HSPCs after FetuA-treatment (n = 3). Scale bar, 20 µm. j, Liquid chromatography-mass spectrometry quantification of FetuA in the bone marrow blood sera of benign and leukaemic infants (Benign, n = 14; Leukaemic, n = 10). The n represents individual samples (c-f, j) and independent experiments (g-i). The mean ± s.d. is shown (d, f, h, j). Statistical tests: unpaired two-sided Student’s t-test (d, f, h, j).
The FetuA concentration in the fetal liver of the first trimester was considerably lower than that in the fetal liver of the second trimester (Extended Data Fig. 10e,f). CD34+ HSPCs were surrounded and bound by FetuA in the fetal liver at E13w but not at E7w (Extended Data Fig. 10g). Ex vivo Eto treatment induced less DNA damage in HSPCs in the presence of FetuA than in the absence of FetuA (Extended Data Fig. 10h). Furthermore, FetuA interacted with TLR4 on the surface of HSPCs (Extended Data Fig. 10i). These findings provide further confirmation that hepatocytes provide genome protection to HSPCs through a paracrine mechanism, and this protection is lacking in the fetal liver of early humans, similar to the fetal liver of mice.
Finally, the concentration of FetuA in the bone marrow blood sera of infant patients with leukaemia and benign samples of patients with non-leukaemia was measured. The concentration of FetuA was significantly lower in leukaemic sera than in non-leukaemic sera (Extended Data Fig. 10j), indicating that the absence of FetuA protection is a risk factor for childhood leukaemia.
Overall, we observed a similar genome-protective mechanism in humans as that previously identified in mice. The lack of this protection is correlated with the onset of childhood leukaemia.
Discussion
During embryonic development, there is a significant expansion of highly proliferative cells in the three germinal layers to produce a wide range of functional cell types and form tissues and organs. It is a considerable challenge to maintain genomic integrity in these tissue-forming cells during DNA replication and normal cellular activity with specific gene transcription. However, further studies are needed to uncover the mechanisms by which developmental organ tissues provide genome protection for tissue-forming cells and how the absence or pathological loss of this protection leads to DNA damage and genome instability in the presence of endogenous (for example, metabolic) or exogenous genotoxic agents. In this study, we discovered a genome-protective mechanism in the fetal liver: hepatocyte-paracrine FetuA protects the HSPC genome by preventing R-loop accumulation. We also found that this genome-protective mechanism is absent in the early fetal liver due to the delayed development of hepatocytes, which exposes newly colonized HSPCs to genotoxic insults in unprotected environments lacking the hepatocyte-paracrine factor FetuA. Within this specific time period, HSPCs become highly susceptible to genotoxic insults. In pathological processes, this unprotected time window is either extended or protection is lost, leading to persistent DNA damage and genome instability throughout the developmental stage and postnatal period, thereby facilitating the initiation and accumulation of mutations during this prolonged genome-unstable period.
Therefore, the findings of this study will have implications in the following: (1) mechanical revelation of the establishment of genetic and epigenetic homeostasis during development; (2) identification of developmental features that are vulnerable to development-related diseases; and (3) exploitation of effective approaches to prevent these diseases. Further studies are required to determine whether such a mechanism is universal or an analogous mechanism that also appears in other developmental organs or tissues and to screen out the druggable substitutes of FetuA or the agonists or antagonists of its activated pathway for the prevention of development-related diseases, such as childhood cancer.
Methods
Human tissue acquisition
Human fetal liver tissues and cord blood samples were obtained from Tongji Hospital, Tongji University School of Medicine, Shanghai, China, with written informed consent from the parents and approval from the Medical Ethics Committee (k-w-2010-010) of Tongji Hospital. Fetal developmental age was estimated from measurements of crown–rump length and compared with a standard growth chart42. Plasma from infant patients with leukaemia and benign patients was obtained from Shanghai Children’s Medical Center, Shanghai Jiao Tong University School of Medicine, Shanghai, China, with written informed consent from guardians of the patients and approval from the Medical Ethics Committee (SCMCIRB-K2024163-1) of the Shanghai Children’s Medical Center.
Mice
B6.129P2-Gt(ROSA)26Sortm1(DTA)Lky/J, B6.Cg-Tg(Alb-cre)21Mgn/J, B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, C57BL/6J (B6-Ly5.2) and C57BL/6JGpt-Fetua-knockout (cas9) mice were maintained and bred in a pathogen-free facility in ventilated cages, a maximum of six mice per cage, on a 12-h day–night cycle, at 20–26 °C and 30–70% humidity, in compliance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals. For embryo collection, 8–10-week-old male and female mice were mated at night and then separated the next morning; the time of separation was considered E0.5. For colony-forming cell assays, whole-genome sequencing and leukaemic models, 3-week-old mice were used. Male and female mice were used in all experiments. Mice were placed into groups depending on their gestational days and genotypes; when possible, mice were randomized and the group allocation was blinded. No sample size calculation was performed. All animal experiments were approved by the Institutional Animal Care and Use Committees of Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Isolation of human HSPCs
Liver tissues were processed immediately after isolation. The tissues were dissected into single-cell suspensions, and mononuclear cells were then separated using Ficoll density gradient centrifugation. Lineage-positive cells were depleted using the MagniSort Human Haematopoietic Lineage Depletion Kit (8804-6836-74, Thermo Fisher). Lineage-negative (Lin−) cells were incubated with a combination of biotin-labelled lineage antibodies (to CD2, CD3, CD10, CD11b, CD14, CD16, CD19, CD56, CD123 and CD235a; 8804-6836-74, Invitrogen) and FITC-labelled CD34 (581; 555821, BD). After 15 min at 4 °C, the cells were washed with PBS, suspended in magnetic bead selection buffer (MACS; PBS, 2 mM EDTA and 0.5% BSA), and then incubated with streptavidin–phycoerythrin (12-4317-87, Thermo Fisher). After 10 min at room temperature, the cells were washed and suspended in IMDM (12440053, Thermo Fisher) supplemented with 1% BSA. Lin− and CD34+ HSPCs were subjected to flow cytometry on an Aria III flow cytometer (BD), and the data were collected using BD FACSDiva (V8.0.3). The HSPCs were cultured in StemSpan medium (09650, Stem Cell) supplemented with 10 ng ml−1 IL-6 (200-06, PeproTech), 10 ng ml−1 IL-3 (200-03, PeproTech), 10 ng ml−1 stem cell factor (SCF; 300-07, PeproTech) and 10 ng ml−1 Flt3 (300-19, PeproTech) at a concentration of 1 × 105 per millilitre for further experiments.
Isolation of mouse HSPCs
Pregnant mice were anaesthetized and euthanized by cervical dislocation. Placentas, fetal livers or bone tissues were then dissected into single-cell suspensions. Subsequently, the cells were incubated with biotin-labelled monoclonal antibodies targeting haematopoietic lineage markers (including B220, CD3, Gr-1 and Ter119; 88-7774-75, Thermo Fisher). After incubation, the cells were washed with MACS buffer and stained with streptavidin-conjugated magnetic beads (558451, BD). Following a 20-min incubation at 4 °C, the cells were washed again and resuspended in MACS buffer. Lineage-positive cells were depleted using a magnetic system. The Lin− cells were then incubated with biotin-labelled lineage markers (88-7774-75, Thermo Fisher), phycoerythrin–Cy7-labelled Sca-1 (D7; 25-5981-82, Thermo Fisher), APC-labelled Kit antibodies (2B8; 17-1171-82, Thermo Fisher), phycoerythrin-labelled CD150 (mShad150; 12-1502-82, Thermo Fisher) and FITC-labelled CD48 antibodies (HM48-1; 11-0481-82, Thermo Fisher). After a 15-min incubation at 4 °C, the cells were washed with PBS and resuspended in MACS buffer. Streptavidin–phycoerythrin (12-4317-87, Thermo Fisher) or streptavidin–APC–Cy7 (405208, BioLegend) was added to the cells, which were then incubated for 10 min at room temperature. The cells were then washed and resuspended in IMDM supplemented with 1% BSA. LSK cells, lineage-negative, Sca-l-positive, Kit-positive, CD150-positive and CD48-negative cells (LT-HSCs), lineage-negative, Sca-l-positive, Kit-positive, CD150-negative and CD48-negative cells (ST-HSCs), and lineage-negative, Sca-l-positive, Kit-positive, CD150-negative and CD48-positive cells (MPPs) were flow-sorted according the gating strategy in Supplementary Fig. 1 in the Aria III flow cytometer (BD), and the data were collected using BD FACSDiva (v8.0.3). The HSPCs were cultured in StemSpan medium (09650, Stem Cell) supplemented with 10 ng ml−1 IL-6 (216-16, PeproTech), 10 ng ml−1 IL-3 (213-13, PeproTech) and 10 ng ml−1 SCF (250-03, PeproTech) at a concentration of 1 × 105 per millilitre for further experiments.
Isolation and culture of mouse fetal hepatocytes
To obtain Alb-Cre;ROSA26-LSL-tdTomato fetuses, we cross-mated B6.Cg-Tg(Alb-cre)21Mgn/J and B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J mice. Fetal livers were then removed at E12.5 or E16.5 and digested with 0.6 mg ml−1 collagenase IV (17104019, Thermo Fisher) in Hank’s balanced salt solution for 20 min at 37 °C. The digestion reaction was halted using cold PBS, and the mixture was subsequently centrifuged at 500 rpm for 5 min. Magnetic cell sorting (558451, BD) was used to remove blood cells expressing Ter119, B220, CD3, Gr-1 and Mac-1. Tomato-positive hepatocytes were flow-sorted using an MoFlo Astrios flow cytometer, and the data were collected using Summit (v6.3.1.16945; Beckman Coulter). The isolated hepatocytes were precultured in StemSpan medium (09650, Stem Cell) supplemented with hepatocyte growth supplement (1:100; 5201, ScienCell), 2 ng ml−1 IL-6 and 5% FBS (F2442, Sigma; referred to as SHIF) for 4 h. After removing the medium and non-adherent cells, the adherent hepatocytes were cultured in the same medium without FBS (referred to as SHI) for 20 h. The supernatant from the cultured fetal hepatocytes (referred to as conditioned SHI or co-SHI) was collected for further experiments.
Comet assay
The HSPCs at different development stages and HSPCs cultured with or without FetuA (100 μg ml−1; 10318-H08H, SinoBiological) or ML216 (25 mΜ; S0469, Selleck) were treated with 20 μM etoposide (341205, Merck Millipore) for 30 min and then collected for the comet assay. A CometAssay kit from TREVIGEN (4250-050-k, R&D) was utilized to assess DNA damage. The alkaline method was utilized using the following steps: (1) 5,000 cells were mixed with 50 µl of melted LMAgarose (4250-050-02, R&D) at 37 °C and then pipetted onto a CometSlide; (2) the cells were then gelled for 3–5 min at 4 °C in the dark, followed by lysis with 4 °C lysis solution for 1 h; (3) the CometSlide was immersed in an alkaline unwinding solution (200 mM NaOH and 1 mM EDTA, pH > 13) for 1 h at 4 °C in the dark, and electrophoresis was conducted in the same solution at 20 V and 300 mA for 30 min at 4 °C); (4) the slides were washed twice with ddH2O for 5 min each, followed by washing with 70% ethanol for 5 min, and the slides were then dried at room temperature overnight; and (5) the dried agarose was stained with SYBR Green I (A25742, Thermo Fisher) nucleic acid gel stain for 30 min. Images were acquired with Las X (v4.7) on a Leica TCS Stellaris8 STED Microscope at ×20 resolution and analysed using OpenComet software (v1.3.1).
Immunofluorescence staining of cells
Human HSPCs treated with 80 µM etoposide (341205, Merck Millipore) for 30 min; mouse HSPCs treated with 20 µM etoposide for 30 min, 500 J m−2 ultraviolet radiation B (UVB) irradiation; human HSPCs cultured with or without FetuA (100 μg ml−1; 10318-H08H, SinoBiological) for 2 h and then treated with 80 µM etoposide; mouse HSPCs cultured with co-SHI or hepatocytes for 2 h and then treated with 20 µM etoposide; and mouse HSPCs pre-treated with or without TLR4 antibody (1:50; 53-9041-80, Thermo Fisher) at 4 °C for 30 min and then cultured with or without FetuA (100 μg ml−1; 50093-M08H, SinoBiological) for 2 h and thereafter treated with 20 µM etoposide were collected. These cells were then spun onto slides and fixed in 4% PFA for 10 min. After treatment with PBS containing 2% serum, 1% BSA and 0.2% Triton X-100, the cells were directly stained with primary antibodies overnight at 4 °C. Subsequently, the sections were stained with secondary antibodies for 1 h at room temperature. Phosphor-histone H2AX (Ser13; 20E3) rabbit monoclonal antibody (1:400; 9718S) was acquired from Cell Signaling Technology (CST), and TLR4 antibody (1:200; ab13556), mouse FetuA antibody (EPR17839-163; 1:100; ab187051) and p-RPA (phospho S33; 1:200; ab211877) were obtained from Abcam. Human FetuA antibody (1F6B9; 1:100; 66094-1-Ig) was obtained from Proteintech. The MYD88 (E11; 1:400; sc-74532) antibody was acquired from Santa Cruz. Images were acquired with ZEN (v2.3) on a Zeiss 880 Microscope at ×20 resolution or ×63 resolution, and analysed using ImageJ (v1.52p) and HALO (v3.6.4134).
Immunofluorescence staining of whole-mount tissues
Fetal livers and placentas were fixed in 4% PFA for 30 min, washed with PBS for 2–3 h, and stained with primary antibodies (diluted in PBS containing 1% BSA, 2% FCS and 0.5% Triton X-100) for 1–3 days. The tissues were then incubated with secondary antibodies for 2 h. Anti-mouse CD117 (Kit, ACK2; 1:50; 14-1172-85) was acquired from Thermo Fisher, and phospho-histone H2AX (Ser139; 20E3; 1:400; 9718S) rabbit monoclonal antibody was obtained from CST. Images were acquired with Las X (v4.7) on a Leica TCS Stellaris8 STED Microscope at ×20 resolution, and analysed using Image J (v1.52p) and Imaris (v9.0.1).
Immunofluorescence staining of tissue sections
Human or mouse placenta, fetal liver and bone tissues were fixed in 4% PFA for 30–60 min at room temperature (placenta and fetal liver tissues) or 5 h at 4 °C (bone tissues), dehydrated in 15% and 30% sucrose, and embedded in optimal cutting temperature compound at −20 °C. The tissues were then sectioned (20–25 μm) using a cryostat. The tissue sections were stained with primary antibodies for 6–12 h at 4 °C in PBS containing 1% BSA, 2% FCS and 0.5% Triton X-100. The sections were incubated with secondary antibodies for 1 h at room temperature. CD48–FITC (HM48-1; 1:100, 11-0481-82) was acquired from Thermo Fisher. APC-anti-lineage (Ter119 (1:400, 116212), Gr-1 (1:400, 108412), Mac-1 (1:400, 101212), B220 (1:400, 103212), CD3 (1:100, 100236), CD150-BV421 (SLAM; 1:100, 115925) and CD41-APC (MWReg30; 1:400, 133913)) were acquired from BioLegend. Kit goat monoclonal antibody (Gln25–Thr519; Ala207Glu; 1;400, AF1356) and anti-human serum albumin antibody (MAB1455; 1:200, 188835) were acquired from R&D. E-cadherin rabbit monoclonal antibody (24E10; 1:200, 3195T) and phospho-histone H2AX (Ser139; 20E3; 1:400, 9718S) rabbit monoclonal antibodies were acquired from CST. Laminin monoclonal antibody (1:200, ab11575), mouse FetuA antibody (EPR17839-163; 1:400, ab187051) and CD34 antibody (EP373Y; 1:100, ab81289) were acquired from Abcam. CD45–FITC (104; 1:100, MCD45201) was acquired from Invitrogen. Nestin antibody (1:50, AN205-1) was acquired from Beyotime. E-cadherin antibody (DECAM-1; 1:200, sc-59778) was acquired from Santa Cruz. CD144 antibody (1:200, 550548) and Sca-1 antibody (D7; 1:200, 557403) were acquired from BD. Human FetuA antibody (1F6B9; 1:200, 66094-1-Ig) was acquired from Proteintech. Images were acquired with ZEN (v2.3) on a Zeiss 880 Microscope at ×20 resolution or ×63 resolution, and analysed using ImageJ (v1.52p), Imaris (v9.0.1) and HALO (v3.6.4134).
Colony-forming cell assay
The assay was performed in a semi-solid methycellulose medium (03434, Stem Cell Technologies) following the technical manual. In brief, the sorted HSPCs were plated in methycellulose in a 35-mm dish (200 cells per dish). Cultures were incubated at 37 °C in a humidified incubator (more than 95%) with 5% CO2 in the air. The colonies were scored under a microscope 10–12 days post-plating. Replating was performed by pooling total cells from primary cultures and inoculating 104 cells into fresh methycellulose medium.
Metaphase chromosome preparation and FISH
Bone marrow Lin− cells were incubated with 0.05 μg ml−1 colcemid for 1 h at 37 °C and then centrifuged at 400g for 10 min. The cells were suspended in 1 ml of hypotonic solution (0.075 M KCl) for 30 min at 37 °C, and the reaction was stopped by the addition of freshly prepared fixative solution (3:1 methanol:glacial acetic acid). The cells were then subjected to three rounds of fixative changes. After that, the cells were dropped onto slides and allowed to dry at room temperature. Two-colour FISH was performed using whole-chromosome probes for mouse chromosome 4 (FITC; D-1404-050-FI, MetaSysterms) and chromosome 6 (Texas red; D-1406-050-OR, MetaSysterms), and counterstaining was performed with DAPI-Antifade solution. Images were acquired with Las X (v4.7) on a Leica TCS Stellaris8 STED Microscope at ×20 resolution and analysed using Image J (v1.52p).
Mouse models of leukaemia
Three-week-old mice were intraperitoneally injected with 80 mg kg−1 N-ethyl-N-nitrosourea (ENU; N3385, Sigma) four times, which was administered twice a week, and the mice were monitored by daily observation of leukaemic symptoms and signs including fired hair, white toes, swollen lymph nodes and loss of body weight, and weekly measurement of leukaemia-like cells by Gimsa staining of peripheral blood smears. Once these appeared, the animals were killed, and the disease of leukaemia was determined by exhibition of enlarged spleens and lymph nodes, increased proportions of immature Lin− cells in the bone marrow and leukaemic cell infiltration in spleens and bone marrow shown by haematoxylin and eosin staining of the tissue sections.
Western blot
After treatment with or without FetuA (100 μg ml−1; 50093-M08H, SinoBiological) for 2 h, mouse HSPCs (LSK) were lysed and blotted with bZIP antibody and their phosphorylated forms. After treatment with or without FetuA (100 μg ml−1; 50093-M08H, SinoBiological) and the bZIP inhibitor SR11032 (2 mM; HY-15870, MedChemExpress) for 6 h, mouse Lin– haematopoietic cells were lysed and blotted with BLM antibody. The cells were lysed using SDS lysis buffer. The lysates were then separated on SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad). Western blotting was carried out using the following primary antibodies: FetuA (1:2,000), JunB (1:1,000), phosphorylated JunB (p-JunB; 1:1,000), Jun (1:1,000), p-Jun (1:1,000), Fosl1 (1:10,000), p-Fosl1 (1:1,000), BLM (1:500) and laminB1 (1:2,000). The membranes were incubated overnight at 4 °C with primary antibodies. After rinsing to remove any unbound primary antibody, the membranes were exposed to a horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h. The secondary antibody was detected using chemiluminescence (WBKLS0500, Merck Millipore). The following primary antibodies were used: anti-p-JunB (Thr102/Thr104; 8053S) and anti-p-Fosl1 (S265; 3880S) from CST and anti-JunB (EPR6518; ab128878), anti-Fosl1 (ab232745), anti-p-Jun (phospho S63; ab32385), anti-Jun (EP693Y; ab40766), anti-laminB1 (EPR8985; ab133741) and anti-FetuA (EPR17839-163; ab187051) from Abcam, and anti-BLM (B-4; sc-365753) from Santa Cruz. The intensity of bands was measured using ImageJ (v1.52p). For gel source data, see Supplementary Fig. 2.
Mass spectrometry analysis
SHI and co-SHI media, E12.5 and E16.5 fetal liver tissues and bone marrow plasma samples from infants were collected for data-independent acquisition tandem mass spectrometry (DIA MS/MS) analysis. The medium samples were treated with 2% SDS buffer containing 50 mM dithiothreitol for 20 min at room temperature and then boiled at 100 °C for 5 min. The protein samples were alkylated in the dark at room temperature for 1 h by adding 200 mM iodoacetamide. To precipitate the proteins, a 5× volume of pre-cooled acetone was used overnight at −20 °C. The tissue samples were diluted with 50 mM NH4HCO3 and centrifuged three times at 20 °C and 14,000g using YM-10 filter units. The protein lysates were reduced for 1 h at room temperature with a final concentration of 10 mM dithiothreitol and then alkylated for 1 h in the dark at room temperature with a final concentration of 55 mM iodoacetamide. The protein mixtures were exchanged with 50 mM NH4HCO3 by centrifugation at 20 °C and 14,000g three times. The protein precipitates were digested overnight at 37 °C at a protein-to-enzyme ratio of 50:1 with trypsin. Tryptic peptides were collected by centrifugation at 20 °C and 14,000g for 20 min. The peptides were then treated with 1% trifluoroacetic acid, purified using C18 Ziptips and eluted with 0.1% trifluoroacetic acid in 50–70% acetonitrile. The eluted peptides were dried using a SpeedVac (Thermo Savant) and resuspended in 1% formic acid and 5% acetonitrile. Before analysis, indexed retention time (iRT) peptides (Biognosys) were spiked into the samples following the manufacturer’s instructions. The pooled digestates were dried using a SpeedVac (Thermo Savant) and resuspended in 5% ACN in 0.05 M ammonium formate. The digested peptides were fractionated using high-pH reversed-phase separation on a Dionex ultra-high-performance liquid chromatography (Thermo Scientific) with a 2.1 × 150 mm ethylene-bridged hybrid (BEH) C18 3-μm column at 40 °C, with a flow rate of 0.2 ml min−1 and a 60-min ACN gradient (5–30%) in 5 mM ammonium formate (pH 10). Fractions were collected at 1-min intervals and pooled at various intervals, resulting in up to 12 fractions. The samples were dried and resuspended in 1% formic acid and 5% acetonitrile. Data-dependent acquisition (DDA) analysis was conducted on an Orbitrap Fusion LUMOS mass spectrometer (Thermo Fisher Scientific) connected to an Easy-nLC 1200 via an Easy Spray (Thermo Fisher Scientific). The peptide mixtures were loaded onto a self-packed analytical PicoFrit column (75 μm × 40 cm) with an integrated spray tip (New Objective) packed with ReproSil-Pur 120A C18-AQ 1.9 μm (Dr. Maisch GmbH). The peptides were separated using a 120-min linear gradient from 95% solvent A (0.1% formic acid, 2% acetonitrile and 98% water) to 28% solvent B (0.1% formic acid and 80% acetonitrile) at a flow rate of 250 nl min−1 at 50 °C. The mass spectrometer was operated in positive-ion mode and used the data-dependent mode with a specialized cycle time (3S) to automatically switch between MS and MS/MS scans. A full MS scan from 350 to 1,500 m/z was acquired at a resolution of R = 120,000 (defined at m/z = 400). MS/MS scans were performed at a resolution of 30,000, with an isolation window of 4 Da and higher-energy collisional dissociation fragmentation with a collision energy of 30 ± 5%. Dynamic exclusion was set to 30 s. Sequences were identified using the mouse UniProt fasta database (53,099 entries, downloaded on 4 November 2018) with default parameters. The digestion enzyme used was a specific trypsin enzyme with two missed specialized cleavages. Carbamidomethyl of cysteine was set as a fixed modification, and oxidation of methionine was set as a variable modification. The iRTs derived from median iRTs across all DDA runs were calculated. Fragment ions for the targeted data analysis were selected from 300 to 1,800 m/z, with a minimal relative intensity set to more than 5% and a fragment ion number greater than 3. The false discovery rate (FDR) was set to 1% for protein and peptide spectrum matches. Protein inference was performed using the ID Picker algorithm integrated within the Spectronaut software. DIA MS/MS acquisition was performed using the same liquid chromatography-MS systems and liquid chromatography linear gradient method as DDA. For MS/MS acquisition, the DIA method was set with 50 variable isolation windows based on the full-width at half-maximum and constructed using the respective DDA data. The full scan was set at a resolution of 1,200,000 over a m/z range of 350–1,500, followed by DIA scans at a resolution of 30,000. The collision energy (CE), auto gate control (AGC) and maximal injection time were set to 30 ± 5%, 1 × 106 and 54 ms, respectively. The DIA raw files were analysed using Spectronaut X (Biognosys). The retention time prediction type was set to dynamic iRT, and a correction factor was applied for window 1. Interference correction at the MS2 level was enabled. Systematic variance was normalized using a local normalization strategy. The FDR was estimated using the mProphet approach and set to 1% at the peptide precursor and protein levels. Protein intensity was calculated by summing the intensities of their respective peptides, which were measured using the peak areas of their fragment ions in MS2 and multiplied by a factor based on the total sample volume of each sample. All the results were filtered with a Q value cut-off of 0.01 (corresponding to a 1% FDR).
Intraplacental injection of FetuA
The pregnant mice were anaesthetized using 3.5% chloral hydrate and then secured onto a heating pad with all four legs immobilized. The abdominal surface was shaved and disinfected with 75% alcohol. A longitudinal incision measuring 1–1.5 cm in length was made on the abdominal skin, and the peritoneum was cut. Cotton gauze was placed around the incision. One uterine horn was carefully exposed and pulled out using blunt forceps onto gauze soaked in PBS. The uterus was held in place with blunt forceps, and recombinant FetuA (10 μg per 15 μl each) was injected into the placenta. After the injection, the uterus was carefully returned to the abdomen, ensuring that it was positioned exactly as before. The peritoneum was closed with a haemostat. Two hours later, 5 mg kg−1 etoposide was administered through intraperitoneal injection. The fetuses were harvested after 1 h and fixed for immunofluorescence assays.
ATAC-seq library preparation and sequencing
ATAC-seq libraries were prepared as previously described43. In brief, HSPCs were lysed using lysis buffer containing 10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2 and 0.1% IGEPAL CA-630 for 10 min before being spun at 4 °C to obtain nuclear preparations. The supernatant was discarded, and the nuclei were then incubated with Tn5 transposome and tagmentation buffer at 37 °C for 30 min (Vazyme). The resulting tagmentation products were purified and amplified using PCR. PCR amplification involved ten cycles under the following conditions: 72 °C for 5 min; 98 °C for 30 s; thermocycling at 98 °C for 10 s, 63 °C for 30 s and 72 °C for 1 min; and 72 °C for 5 min. The libraries were purified using a PCR purification kit (28004, Qiagen), and the fragments were enriched using 0.5× and 1.0× VAHTS DNA Clean Beads (N412-01, Vazyme) after amplification.
ATAC-seq data processing
All reads were aligned to the mm10 genome using the Burrows-Wheeler Aligner (BWA-MEM) after trimming the adapter sequences with Trim_Galore (v0.6.7). Low-quality reads were filtered out, whereas PCR duplicates and reads mapped to the mitochondria or the Y chromosome were discarded. The remaining reads on the left were shifted (+4/−5) to correct Tn5 enzyme insertions based on the read strands. Peak calling was performed using MACS2 (v2.2.6) with the following options: -f BAM, -g mm, --nomodel, --shift -100 and --extsize 200. The samples were normalized using the bamCoverage function from deepTools (v3.5.1) to visualize the signal in IGV (v2.7.0).
Quality control of ATAC-seq data was conducted, and correlation analysis was performed using deepTools (v3.5.1). The fragment distribution was generated using ATAC-seq QC (v1.14.4). The peak atlas was obtained by expanding the peak summit by ±500 bp, and differential peaks were identified using DESeq2 (v1.26.0). DNA-binding factor motifs were analysed by determining the motifs in the differential peaks using HOMER (v4.11). ATAC signals were visualized as a heatmap using the complexHeatmap package (v2.2.0).
RNA-seq and analysis
Total RNA was extracted from cultured or non-cultured HSPCs using Tri Pure Isolation Reagent (11667157001) from Roche following the manufacturer’s instructions. The quality of the RNA was assessed using the Fragment Analyser platform. High-quality samples were chosen for library construction using the Illumina TruSeq RNA Prep Kit (20015949). The libraries were subsequently sequenced on the Illumina HiSeq4000 platform, generating 2 × 150 bp paired-end reads. To process the raw data, Trim Galore (v0.6.7) was used with the following parameters: ‘--quality 20 --fastqc --length 20 --stringency 1’. The resulting clean reads were then aligned to the mouse reference genome (mm10) using hisat2 (v2.2.1). GENCODE annotations (gencode.vM25.annotation.gtf; downloaded in April 2021) and HTSeq-count (v0.13.5) were used to assign the aligned reads to genes. Subsequently, the counts were normalized to fragments per kilobase of transcript per million mapped reads (FPKM), and log(FPKM + 1) was utilized to analyse the overall similarity or dissimilarity between the samples.
Cut&Tag library preparation and sequencing
The Hyperactive Universal Cut&Tag Assay Kit for Illumina Pro (TD904, Vazyme) was used in this study. In summary, 1 × 105 LSK cells treated with or without FetuA were collected and washed in 500 µl of wash buffer. The cells were then resuspended in 100 µl of wash buffer. Subsequently, 10 µl of concanavalin A-coated magnetic beads were activated and added to 1 × 105 cells. The cells were incubated at room temperature for 10 min, after which the supernatant was removed. The resulting bead-bound cells were resuspended in 50 µl of antibody buffer. Next, 1 µl of Jun rabbit monoclonal antibody (60A8; 9165T, CST), JunB rabbit monoclonal antibody (C37F9; 3753S, CST) or Fosl1 mouse monoclonal antibody (C-12; sc-28310, Santa Cruz) was added and incubated with the bead-bound cells overnight at 4 °C with rotation. The supernatant was then removed, and the bead-bound cells were resuspended in 50 µl of dig-wash buffer containing goat anti-rabbit IgG antibody (Ab207-01-AA, Vazyme) or goat anti-mouse IgG antibody (Ab208-01-AA, Vazyme; diluted 1:100). This mixture was incubated at room temperature for 1 h. The bead-bound cells were washed three times with 200 µl of dig-wash buffer to remove any unbound antibodies. Next, 2 µl of the pA-Tn5 adapter complex was diluted in 98 µl of dig-300 buffer and mixed with the bead-bound cells. The mixture was subjected to rotation at room temperature for 1 h. The bead-bound cells were washed three times with 200 µl of dig-300 buffer to eliminate any unbound pA-Tn5 protein. The cells were then resuspended in 50 µl of tagmentation buffer and incubated at 37 °C for 1 h. To terminate the tagmentation reaction, 2 µl of SDS was added to the cells and incubated for an additional 10 min at 55 °C. The tagmentation products were purified using DNA Extract Beads Pro and eluted in 15 µl of nuclease-free water. For generation of the sequencing libraries, the DNA tagments were mixed with a universal i5 primer and a unique i7 primer and amplified using 2× Cut &Tag amplification mix. The resulting PCR products were purified using VAHTS DNA lean beads (N411, Vazym), and subsequently analysed using an Agilent 2100 Bioanalyzer and Illumina Novaseq 6000.
Cut&Tag data processing
Cut&Tag reads were aligned to the mm10 genome with Bowtie2 (v2.3.5.1) using the following parameters: --end-to-end --very-sensitive --no-mixed --no-discordant --phred33 -I 10 -X 700. Duplicate reads were removed with Picard (v2.25.5). The track files were made with the bamCoverage command from deepTools (v3.5.1). Cut&Tag peaks were called using MACS2 (v2.2.6). The distribution of Cut&Tag peaks was annotated with the R package ChIPseeker (v1.22.1).
R-loop staining
HSPCs from E12.5 placenta, E12.5 fetal liver, E16.5 fetal liver or E12.5 FL-HSPCs with or without FetuA (100 μg ml−1; 50093-M08H, SinoBiological) or ML216 (25 mΜ; S0469, Selleck) treatment for 2 h were collected. The cells were spun onto slides, fixed in 4% PFA for 10 min and washed three times with PBS. After permeabilization with PBS containing 0.3% Triton X-100 for 10 min at room temperature, the cells were washed three times with PBS and then blocked with PBS containing 2% serum, 1% BSA and 0.2% Triton X-100 for 1 h at 37 °C. Immunofluorescence experiments with the dRNH1 protein were performed as previously described33. In brief, the cells were incubated with 30 μl dRNH1 (0.24 mg ml−1) for 1 h at 37 °C, followed by three washes with PBS. The cells were then incubated with 1 μg ml−1 DAPI for 10 min. For immunofluorescence experiments with the GST-His6-2×HBD protein32, the cells were incubated with 30 μl GST-His6-2×HBD (2 μg ml−1) overnight at 4 °C. After three washes with PBS, the cells were stained with an anti-HisTag monoclonal antibody (AMC0149; 1:400; AE003, ABclonal) for 1 h at room temperature, followed by three washes with PBS. The cells were then stained with a rabbit anti-mouse IgG antibody (1:400; SPA231, Solarbio) for 1 h at room temperature. After three washes with PBS, the cells were incubated with 1 μg ml−1 DAPI for 10 min. Images were acquired using a Zeiss 880 microscope, and the signal intensity was measured using HALO (v3.6.4134).
R-loop Cut&Tag library preparation and sequencing
The R-loop Cut&Tag library was prepared following protocols previously described32 with minor modifications. For this experiment, the Hyperactive Universal Cut&Tag Assay Kit for Illumina (TD903, Vazyme) was utilized. In brief, 1 × 105 cells were gently pipetted and washed twice in 500 µl of wash buffer. Then, 10 µl of concanavalin A-coated magnetic beads was activated and added to the 1 × 105 cells, followed by incubation at room temperature for 10 min. The supernatant was then removed, and the bead-bound cells were resuspended in 90 µl of antibody buffer. Subsequently, 10 µl of recombinant GST-His6-2×HBD (0.2 mg ml−1) protein was added and the mixture was incubated with the bead-bound cells overnight at 4 °C with rotation. After two washes with dig-wash buffer, the samples were incubated with an anti-HisTag monoclonal antibody (AMC0149; 1:400; AE003, ABclonal) for 1 h at room temperature, followed by incubation with a rabbit anti-mouse IgG antibody (final concentration, 10 µg ml−1; SPA231, Solarbio) for 1 h at room temperature. Unbound antibodies were removed by three brief washes with 200 µl of dig-wash buffer. To facilitate tagmentation, 2 µl of a pA-Tn5 adapter complex was diluted in 98 µl of dig-300 buffer and mixed with bead-bound cells, which were then rotated at room temperature for 1 h. After three washes in 200 µl of dig-300 buffer to remove unbound pA-Tn5 protein, the cells were resuspended in 50 µl of tagmentation buffer and incubated at 37 °C for 1 h. The tagmentation reaction was stopped by adding 1.8 µl of 0.5 M ethylenediaminetetraacetic acid, 0.6 µl of 10% SDS, 5 µl of nuclease-free water and 1 µl of proteinase K (20 mg ml−1), and further incubated at 55 °C for 60 min. Following purification with 1× DNA clean beads (Vazyme Biotech), the resulting tagmentation products were eluted in 10 µl of nuclease-free water. For the strand displacement reaction, the eluent was mixed with 10 U of Bst 2.0 WarmStart DNA polymerase (M0538, NEB) in 1× Q5 polymerase reaction buffer and incubated at 65 °C for 30 min. The reaction was then halted by incubation at 80 °C for 20 min. To generate the sequencing libraries, the mixture was combined with a universal i5 primer and a uniquely barcoded i7 primer and subsequently amplified using Q5 high-fidelity master mix (M0491, NEB). The libraries were size selected with 0.56–0.85× DNA clean beads and subjected to analysis using an Agilent 2100 Bioanalyzer and Illumina PE150 sequencing.
R-loop Cut&Tag data processing
The Cut&Tag data were processed as previously described32. In summary, Cut&Tag reads were aligned to the mm10 genome using Bowtie2 (v2.3.5.1), allowing for uniquely mapped reads with up to two mismatches. The aligned reads were normalized to the total number of reads (reads per million). Subsequently, track files were generated using the bamCoverage command from deepTools (v3.5.1). Cut&Tag peaks were called using MACS2 (v2.2.6), and the distribution of Cut&Tag peaks was annotated using the R package ChIPseeker (v1.22.1). The average coverage was used to create metaplots within the indicated windows. Gene Ontology enrichment analysis was performed using clusterProfiler (v3.14.3), and circus-plots were generated using circlise (v0.4.8).
Whole-genome sequencing of mouse HSPCs
Genomic DNA was extracted using the QIAamp DNA Mini Kit (51304, Qiagen) following the manufacturer’s protocol. Whole-genome sequencing was conducted as previously described44. In brief, short-insert 350-bp genomic libraries were constructed following Illumina library protocols, and sequencing was performed on an Illumina NovaSeq 6000 platform using 150-base paired-end reads, achieving an average coverage of 30×. The sequence data were mapped to the mouse genome reference mm10 using the BWA-MEM algorithm. Unmapped reads and PCR-derived duplicates were excluded from the analysis. Insertions and deletions (indels) and structural variants were called using the Pindel and BreakDancer algorithms, respectively, as described elsewhere45,46. The group treated with saline was used as germline control.
Cell cycle analysis of mouse HSPCs by combining Hoechst and pyronin Y
The cells were harvested from E12.5 placenta, E12.5 fetal liver and E16.5 fetal liver samples, and lineage-positive cells were depleted using a magnetic system. The Lin− cells or Lin− cells cultured with or without FetuA (100 μg ml−1; 50093-M08H, SinoBiological) or ML216 (25 mM; S0469, Selleck) for 2 h were collected. The cells were then suspended in 1 ml of StemSpan medium (09650, Stem Cell). Hochest33342 (10 µg ml−1; b2261-25mg, Sigma) and verapamil (50 µM; M14204, AbMole) were added to the cell suspension. The mixture was thoroughly mixed and incubated at 37 °C for 60 min in the dark. Subsequently, 5 µl of 100 µg ml−1 pyronin Y (213519-1g, Sigma) was directly added to the cells, followed by continuous incubation at 37 °C for another 15 min in the dark. After centrifugation at 300g for 5 min at 4 °C, the cells were suspended in MACS buffer and incubated with biotin-labelled lineage markers (88-7774-75, Thermo Fisher), FITC-labelled Sca-1 (E13-161.7; 122506, BioLegend) and APC-labelled Kit antibodies (2B8; 17-1171-82, Thermo Fisher). Following a 15-min incubation at 4 °C, the cells were washed with PBS, suspended in MACS buffer and incubated with streptavidin–APC–Cy7 (405208, BioLegend). After another 15 min at 4 °C, the cells were washed and resuspended in MACS buffer. The samples were analysed using a Beckman cytoFLEX LX, and the data were analysed using FlowJo (v10).
DNA synthesis analysis of mouse HSPCs by EdU
Pregnant mice were administered intraperitoneal injections of 100 mg kg−1 EdU (CX000, CellularLab) 2 h before being killed. Cells were collected from E12.5 placenta, E12.5 fetal liver and E16.5 fetal liver and subjected to lineage-positive cell depletion using a magnetic system. The cells were then incubated with biotin-labelled lineage markers (88-7774-75, Thermo Fisher), phycoerythrin–Cy7-labelled Sca-1 (D7; 25-5981-82, Thermo Fisher) and APC-labelled Kit (2B8; 17-1171-82, Thermo Fisher) antibodies. After 15 min at 4 °C, the cells were washed with PBS, suspended in MACS buffer and incubated with streptavidin–phycoerythrin. EdU was detected using the EdU Cell Proliferation Kit with Alexa Fluor 488 (CX002, CellularLab) following the manufacturer’s instructions. The samples were analysed on a Beckman cytoFLEX LX, and the data were analysed using FlowJo (v10).
Cell cycle analysis of mouse HSCs and MPPs by Ki67
Harvested cells from E12.5 placenta, E12.5 fetal liver and E16.5 fetal liver were subjected to lineage-positive cell depletion using a magnetic system. The cells were incubated with biotin-labelled lineage markers (88-7774-75, Thermo Fisher), phycoerythrin–Cy7-labelled Sca-1 (D7; 25-5981-82, Thermo Fisher), APC-labelled Kit (2B8; 17-1171-82, Thermo Fisher), phycoerythrin-labelled CD150 (mShad150; 12-1502-82, Thermo Fisher) and APC–Cy7-labelled CD48 (HM48-1; 103431, BioLegend) antibodies. Following a 15-min incubation at 4 °C, the cells were washed with PBS, suspended in MACS buffer and then incubated with streptavidin–Percpcy5.5. After another 15 min at 4 °C, the cells were fixed and permeabilized with Foxp3/Transcription Factor Staining Buffer Set Kit (00-5523-00, Invitrogen) according to the manufacturer’s instructions. Subsequently, the samples were incubated with FITC-labelled Ki67 (SolA15; 11-5698-80, Thermo Fisher). After a 30-min incubation at 4 °C, the cells were washed with permeabilization buffer and analysed using a Beckman CytoFLEX LX, and the data were analysed using FlowJo (v10).
RNA synthesis analysis of mouse HSPCs by ethyluridine
Pregnant mice were injected intraperitoneally with 50 mg kg−1 ethyluridine (2469–25 mg, Lumiprobe) 1 h before being killed. Cells were collected from E12.5 placenta, E12.5 fetal liver and E16.5 fetal liver and subjected to lineage-positive cell depletion using a magnetic system. The cells were then incubated with biotin-labelled lineage markers (88-7774-75, Thermo Fisher), FITC-labelled Sca-1 (E13-161.7; 122506, BioLegend) and APC-labelled Kit (2B8; 17-1171-82, Thermo Fisher) antibodies. After 15 min at 4 °C, the cells were washed with PBS, suspended in MACS buffer and incubated with streptavidin–APC–Cy7 (405208, BioLegend). The ethyluridine was stained using the Cell-Light EU Apollo567 RNA Imaging Kit (C10316-1, RIBOBIO) according to the manufacturer’s instructions. The samples were analysed on a Beckman cytoFLEX LX, and the data were analysed using FlowJo (v10).
Statistics
Statistics analyses were performed using R (v.3.6.3) and GraphPad Prism (v.9.5). n Denotes biological replicates. For violin plots in all panels, the median and quartiles are shown. For boxplots, the mean ± s.d. is shown. For boxplots in Fig. 5c and Extended Data Fig. 9e,i, the median and quartiles are shown. For boxplots in Fig. 5d and Extended Data Fig. 3b, the boxes delimit the minima and maxima, and the horizontal line represents the mean. For survival analysis, the long-rank test was used to compare the difference between groups. For correlation analysis, the Pearson test was used. For comparing two groups, the unpaired Student’s t-tests and Wilcoxon tests were used. P < 0.05 was considered to be significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-024-08307-x.
Supplementary information
The flow-cytometric gating strategy for LSK cells, HSCs and MPPs. The flow-cytometric gating strategy for LSK cells (Lin−Sca-1+Kit+), LT-HSCs (Lin−Sca-1+Kit+CD150+CD48−), ST-HSCs (Lin−Sca-1+Kit+CD150−CD48−), and MPPs (Lin−Sca-1+Kit+CD150−CD48+) in samples from E12.5 PL, E12.5FL, E13.5FL, E14.5FL, E15.5FL, E16.5FL, E18.5FL, perinatal BM. PL, placenta; FL, fetal liver; BM, bone marrow.
Uncropped western blots associated with Fig. 4 and Extended Data Figs. 5, 6. The fully scanned images of all the western blots in this paper are shown in uncropped form. The molecular weight markers and loading controls are indicated. The blue dotted lines delineate the regions cropped for the final figures.
Source data
Acknowledgements
We thank G.-L. Xu for discussing the data and critical reading of the manuscript; L.-J. Hui for assisting with the culture of the fetal hepatocytes; and the Core Facility of Basic Medical Science in Shanghai Jiao Tong University School of Medicine for technical support. This work was supported by grants from the National Natural Science Foundation of China (81920108005, U23A20417, 81730007, 31872842, 91442106, 81120108006, 90919055 and 81721004) and from the CAMS Innovation Fund for Medical Sciences (CIFMS; 2019-I2M-5-051).
Extended data figures and tables
Author contributions
X.-L.G. designed the experiments and composed the figures. Lei Chu and H.Z. supplied the human samples. X.-L.G., Y.-D.W., Y.-J.L., Y.H., R.-Y.W., H.-Y.X., F.K. and C.-Y.X. performed the experiments. Y.-D.W., R.-Y.W. and J.Y. analysed the ATAC-seq data. Y.-D.W., S.-P.L. and R.-Y.L. analysed the RNA-seq data. Y.-D.W. analysed the R-loop Cut&Tag data. Y.-D.W. and Y.-J.L. analysed the whole-genome sequencing data. G.-H.L. and Lei Chen provided advice for the ATAC-seq analysis. Liang Chen provided advice for the R-loop Cut&Tag analysis. Lei Chen, G.-Q.C., Liang Chen, F.B., T.E., G.-H.L. and H.-F.L. discussed the data and the manuscript. D.-L.H. conceived and supervised the project, analysed the data and wrote the manuscript.
Peer review
Peer review information
Nature thanks John Chute, Stephan Hamperl and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The ATAC-seq, RNA-seq, R-loop Cut&Tag sequencing, bZIP Cut&Tag sequencing and whole-genome sequencing data reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under the study accession number PRJCA026810. The reference genome used was mm10 (https://hgdownload.soe.ucsc.edu/goldenPath/mm10/bigZips/mm10.fa.gz). The annotation dataset used in the RNA-seq analysis was gencode.vM25.annotation.gtf (http://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M25/gencode.vM25.annotation.gtf.gz). Source data are provided with this paper.
Code availability
This paper did not generate new code and new software.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiao-Lin Guo, Yi-Ding Wang, Yan-Jun Liu
Change history
2/18/2025
A Correction to this paper has been published: 10.1038/s41586-025-08714-8
Extended data
is available for this paper at 10.1038/s41586-024-08307-x.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-024-08307-x.
References
- 1.Saxena, S. & Zou, L. Hallmarks of DNA replication stress. Mol. Cell82, 2298–2314 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaillard, H. & Aguilera, A. Transcription as a threat to genome integrity. Annu. Rev. Biochem.85, 291–317 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Aguilera, A. & Garcia-Muse, T. Causes of genome instability. Annu. Rev. Genet.47, 1–32 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Hoeijmakers, J. H. Genome maintenance mechanisms for preventing cancer. Nature411, 366–374 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Branzei, D. & Foiani, M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol.11, 208–219 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Dion, V. & Gasser, S. M. Chromatin movement in the maintenance of genome stability. Cell152, 1355–1364 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Papamichos-Chronakis, M. & Peterson, C. L. Chromatin and the genome integrity network. Nat. Rev. Genet.14, 62–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crawford, L. W., Foley, J. F. & Elmore, S. A. Histology atlas of the developing mouse hepatobiliary system with emphasis on embryonic days 9.5–18.5. Toxicol. Pathol.38, 872–906 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, C. et al. Delineating spatiotemporal and hierarchical development of human fetal innate lymphoid cells. Cell Res.31, 1106–1122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, S. et al. Identification of HSC/MPP expansion units in fetal liver by single-cell spatiotemporal transcriptomics. Cell Res.32, 38–53 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ober, E. A. & Lemaigre, F. P. Development of the liver: insights into organ and tissue morphogenesis. J. Hepatol.68, 1049–1062 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Mikkola, H. K. & Orkin, S. H. The journey of developing hematopoietic stem cells. Development133, 3733–3744 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Chou, S. & Lodish, H. F. Fetal liver hepatic progenitors are supportive stromal cells for hematopoietic stem cells. Proc. Natl Acad. Sci. USA107, 7799–7804 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamiya, A. et al. Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. EMBO J.18, 2127–2136 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jirak, P. et al. Clinical implications of fetuin-A. Adv. Clini. Chem.89, 79–130 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Icer, M. A. & Yıldıran, H. Effects of nutritional status on serum fetuin-A level. Crit. Rev. Food Sci. Nutr.60, 1938–1946 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Okada, S. et al. In vivo and in vitro stem cell function of c-Kit- and Sca-1-positive murine hematopoietic cells. Blood80, 3044–3050 (1992). [PubMed] [Google Scholar]
- 18.Morrison, S. J., Hemmati, H. D., Wandycz, A. M. & Weissman, I. L. The purification and characterization of fetal liver hematopoietic stem cells. Proc. Natl Acad. Sci. USA92, 10302–10306 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ottersbach, K. & Dzierzak, E. The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev. Cell8, 377–387 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Guo, X. L. et al. Recipient bone marrow assimilates the myeloid/lymphoid reconstitution of distinct fetal hematopoietic stem cells. Oncotarget8, 108981–108988 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyajima, A., Tanaka, M. & Itoh, T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell14, 561–574 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Khan, J. A. et al. Fetal liver hematopoietic stem cell niches associate with portal vessels. Science351, 176–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itkin, T. et al. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature532, 323–328 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunisaki, Y. et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature502, 637–643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Begum, S. et al. Characterization and engraftment of long-term serum-free human fetal liver cell cultures. Cytotherapy12, 201–211 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Zhang, K. et al. In vitro expansion of primary human hepatocytes with efficient liver repopulation capacity. Cell Stem Cell23, 806–819.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Pal, D. et al. Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nat. Med.18, 1279–1285 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Fitzgerald, K. A. & Kagan, J. C. Toll-like receptors and the control of immunity. Cell180, 1044–1066 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang, E. Y. et al. RECQ-like helicases Sgs1 and BLM regulate R-loop-associated genome instability. J. Cell Biol.216, 3991–4005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang, Z. et al. Binding of FANCI–FANCD2 complex to RNA and R-loops stimulates robust FANCD2 monoubiquitination. Cell Rep.26, 564–572.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayona-Feliu, A., Barroso, S., Munoz, S. & Aguilera, A. The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription-replication conflicts. Nat. Genet.53, 1050–1063 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Wang, K. et al. Genomic profiling of native R loops with a DNA–RNA hybrid recognition sensor. Sci. Adv.7, eabe3516 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crossley, M. P. et al. Catalytically inactive, purified RNase H1: a specific and sensitive probe for RNA–DNA hybrid imaging. J. Cell Biol.220, e202101092 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enver, T. et al. Developmental impact of leukemic fusion genes on stem cell fate. Ann. N. Y. Acad. Sci.1044, 16–23 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Chen, L. et al. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol. Cell68, 745–757 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niehrs, C. & Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol.21, 167–178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong, D. et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science319, 336–339 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Li, Z. et al. Leukaemic alterations of IKZF1 prime stemness and malignancy programs in human lymphocytes. Cell Death Dis.9, 526 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brady, S. W. et al. The genomic landscape of pediatric acute lymphoblastic leukemia. Nat. Genet.54, 1376–1389 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papaemmanuil, E. et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med.374, 2209–2221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boiers, C. et al. A human IPS model implicates embryonic B-myeloid fate restriction as developmental susceptibility to B acute lymphoblastic leukemia-associated ETV6–RUNX1. Dev. Cell44, 362–377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corton, M. M., Leveno, K. J., Bloom, S. L. & Hoffman, B. L. Williams Obstetrics 24th edn (McGraw Hill, 2014).
- 43.Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol.109, 21–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Behjati, S. et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature513, 422–425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raine, K. M. et al. cgpPindel: identifying somatically acquired insertion and deletion events from paired end sequencing. Curr. Protoc. Bioinformatics52, 11–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen, K. et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods6, 677–681 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The flow-cytometric gating strategy for LSK cells, HSCs and MPPs. The flow-cytometric gating strategy for LSK cells (Lin−Sca-1+Kit+), LT-HSCs (Lin−Sca-1+Kit+CD150+CD48−), ST-HSCs (Lin−Sca-1+Kit+CD150−CD48−), and MPPs (Lin−Sca-1+Kit+CD150−CD48+) in samples from E12.5 PL, E12.5FL, E13.5FL, E14.5FL, E15.5FL, E16.5FL, E18.5FL, perinatal BM. PL, placenta; FL, fetal liver; BM, bone marrow.
Uncropped western blots associated with Fig. 4 and Extended Data Figs. 5, 6. The fully scanned images of all the western blots in this paper are shown in uncropped form. The molecular weight markers and loading controls are indicated. The blue dotted lines delineate the regions cropped for the final figures.
Data Availability Statement
The ATAC-seq, RNA-seq, R-loop Cut&Tag sequencing, bZIP Cut&Tag sequencing and whole-genome sequencing data reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under the study accession number PRJCA026810. The reference genome used was mm10 (https://hgdownload.soe.ucsc.edu/goldenPath/mm10/bigZips/mm10.fa.gz). The annotation dataset used in the RNA-seq analysis was gencode.vM25.annotation.gtf (http://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M25/gencode.vM25.annotation.gtf.gz). Source data are provided with this paper.
This paper did not generate new code and new software.