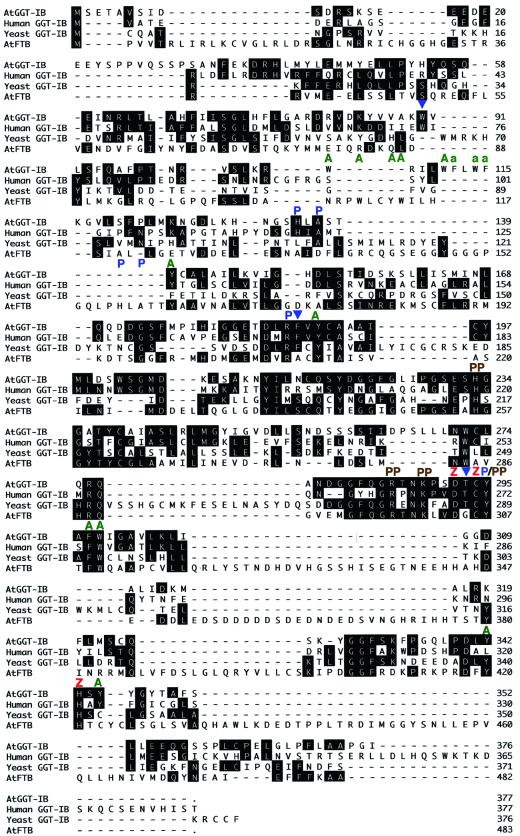
Figure 1.
Amino acid sequence alignment of protein geranylgeranyltransferase β-subunits and the Arabidopsis FTB. Sequence alignments were established by using the John Hein method (Lasergene). Amino acid identities between the Arabidopsis AtGGT-IB and AtFTB proteins, and GGT-I β-subunits from yeast and human, are indicated by black boxes. Dashes denote gaps formed by the alignment algorithm. In some regions the alignment program failed to align the AtFTB sequence correctly because of the divergence from the yeast sequence. Molecular functions were assigned to some of the residues using the crystal structure of the rat FTase as a model. Green A denotes residues in the hydrophobic pocket, blue P denotes residues that interact with the CaaX peptide, brown PP denotes residues that interact with the diphosphate group of FPP (or GGPP), red Z denotes the ligands of the catalytic zinc atom, and blue arrows denote residues that are conserved between all GGT-I but differ in FTBs. AtGGT-IB was isolated as a 1,351-bp clone with 1,128-bp open reading frame. Low stringency DNA-blot analysis indicates that AtGGT-IB exists in a single copy form in the Arabidopsis genome (data not shown). RNA-blot analysis (data not shown) and comparison to the genomic sequence of AtGGT-IB (GenBank accession no. ATAC004218) indicate that AtGGT-IB represents a full-length clone. The GenBank accession nos. for the aligned sequences are as follows: AtGGT-IB, AF311225; human GGT-IB, L25441; yeast (S. cerevisiae) GGT-IB, M74109; and Arabidopsis AtFTB, AF214106.