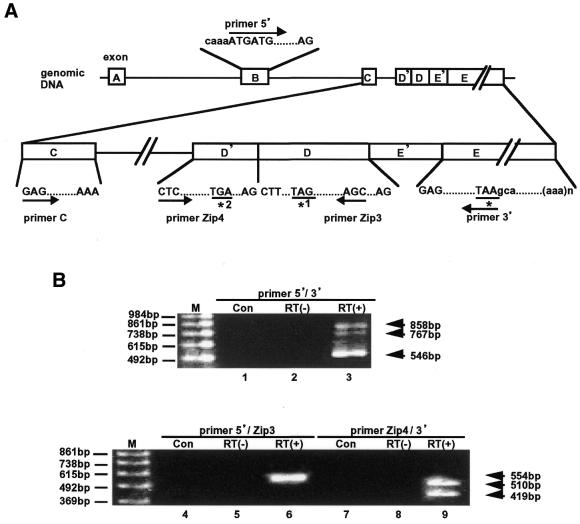
Figure 1.
RT–PCR analysis of ATF3ΔZip2 mRNA in A23187-treated HUVECs. (A) Primers used in this study and their locations in the human ATF3 gene are shown. Exons are indicated by boxes, and labeled as A, B, C, D and E as in Liang et al. (33). Exons D′ and E′ are novel ones described in the text. Initiation codon ATG in exon B is shown under primer 5′. Stop codons for the full-length ATF3 (*), ATF3ΔZip (*1) and ATF3ΔZip2 (*2) are shown. (B) HUVECs (3 × 105 cells) were treated with 10 µM A23187 for 4 h, and total RNA (1 µg) was assayed for RT–PCR using primer 5′ and primer 3′ (lanes 1–3), primer 5′ and primer Zip3 (lanes 4–6), or primer Zip4 and primer 3′ (lanes 7–9). Lanes 1, 4 and 7 are derived from untreated cells; all the other lanes are from A23187-treated cells with (lanes 3, 6 and 9) or without (lanes 2, 5 and 8) RT. Bands of 546, 858 and 767 bp represented products of full-length ATF3, ATF3ΔZip2a and ATF3ΔZip2b, respectively (lane 3). 554 bp was the product derived from ATF3ΔZip2a and b (lane 6); note that the primer set used in this experiment (primer 5′ and primer Zip3) does not distinguish ATF3ΔZip2a from ATF3ΔZip2b. 510 and 419 bp represented ATF3ΔZip2a and ATF3ΔZip2b, respectively (lane 9). M, the 123 bp DNA size marker.