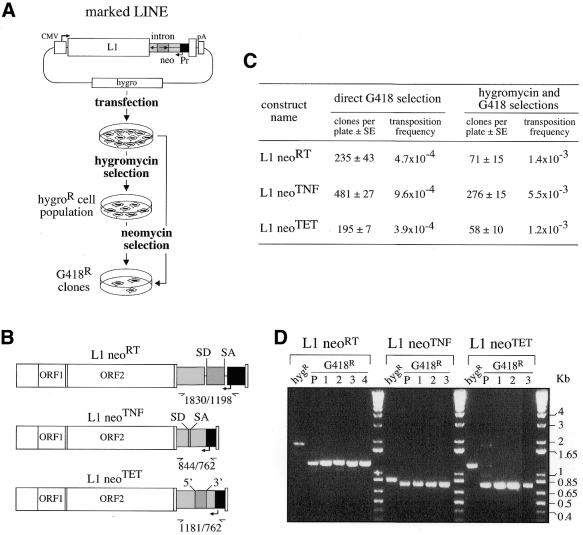
Figure 5.
Quantitative assay for detection of retrotransposition of human LINEs marked with neo-cassettes. (A) Experimental procedures for detection of L1 retrotransposition events. The human L1.2 element (open box) was marked with a neo-cassette (neoTET, neoTNF or neoRT) and introduced into an hygromycin resistance gene-containing episomal vector. Marked L1-containing vectors were introduced into human cells by transfection, and then either cell transformants were isolated upon hygromycin selection and the resulting cell population (hygroR) was thereafter assayed for L1 retrotransposition upon G418 selection or, alternatively, transfected cells were directly submitted to G418 selection. (B) Structure of the three indicator genes (neoRT, neoTNF and neoTET) used for the L1 retrotransposition assay, with the RT and TNF introns being nuclear mRNA introns, and the Tetrahymena TET intron an autocatalytic group I intron. (C) Retrotransposition of the marked L1s. Numbers of foci per plates (5 × 105 and 5 × 104 cells per plate for direct and indirect G418 selections, respectively) are indicated as the mean of four independent transfections assays with the standard errors, together with the corresponding retrotransposition frequencies. (D) PCR analysis of the DNAs from initial and G418R cells assayed for the presence of retrotransposed copies with spliced-out introns, using the same primers bracketting the intronic domain for the three indicator genes. In the BET-labeled agarose gel, the first lane (hygR) for each construct corresponds to the DNA from the transfected hygroR cell population, before G418 selection, and the following lanes correspond to DNA from G418R cell populations (P) and clones (1–4). Bands of the expected size for the spliced-out intron are observed only for the G418R cells. Larger bands, corresponding for each construct to unspliced copies, are observed in the initial hygroR cell populations, before G418 selection.