Abstract
Mitochondria are essential for cellular function and viability, serving as central hubs of metabolism and signaling. They possess various metabolic and quality control mechanisms crucial for maintaining normal cellular activities. Mitochondrial genetic disorders can arise from a wide range of mutations in either mitochondrial or nuclear DNA, which encode mitochondrial proteins or other contents. These genetic defects can lead to a breakdown of mitochondrial function and metabolism, such as the collapse of oxidative phosphorylation, one of the mitochondria’s most critical functions. Mitochondrial diseases, a common group of genetic disorders, are characterized by significant phenotypic and genetic heterogeneity. Clinical symptoms can manifest in various systems and organs throughout the body, with differing degrees and forms of severity. The complexity of the relationship between mitochondria and mitochondrial diseases results in an inadequate understanding of the genotype-phenotype correlation of these diseases, historically making diagnosis and treatment challenging and often leading to unsatisfactory clinical outcomes. However, recent advancements in research and technology have significantly improved our understanding and management of these conditions. Clinical translations of mitochondria-related therapies are actively progressing. This review focuses on the physiological mechanisms of mitochondria, the pathogenesis of mitochondrial diseases, and potential diagnostic and therapeutic applications. Additionally, this review discusses future perspectives on mitochondrial genetic diseases.
Subject terms: Diseases, Medical genetics, Molecular medicine
Introduction
Mitochondria, often referred to as the powerhouses of cells, perform their essential function through oxidative phosphorylation (OXPHOS), which generates ATP as a vital energy source.1 Mitochondrial diseases are genetic disorders resulting from abnormalities of mitochondrial function.2 These disorders arise from mutations in either mitochondrial DNA (mtDNA) or nuclear DNA (nDNA), both of which encode subunits of OXPHOS as well as structural or functional mitochondrial proteins.3 These proteins are not only integral to classical mitochondrial metabolism—such as OXPHOS, the Krebs cycle, lipid metabolism, and nucleotide metabolism—but also play key roles in mitochondrial quality control, calcium homeostasis, cell death, and inflammation. Deficiencies in these proteins can lead to mitochondrial dysfunction and subsequent energy failure.4 Given mitochondria’s ubiquitous presence and critical role in cellular metabolism, any tissue in the body can be affected.5 However, organs and tissues with high energy demands, such as the brain, nerve, eye, cardiac, and skeletal muscles, are particularly susceptible to energy failure due to OXPHOS defects, with phenotypes often manifesting in neurological, ophthalmological, and cardiological systems.6 The symptoms of mitochondrial diseases are diverse, with developmental delay, seizure (encephalopathy), hypotonia (myopathy), and visual impairment (retinopathy) being prominent indicators.6,7 Despite recent advances, the molecular mechanisms underlying these diseases remain incompletely understood. The extreme phenotypic and genetic heterogeneity of mitochondrial diseases further complicates diagnosis, making misdiagnosis a common issue.8
Mitochondrial diseases have been recognized as pathway-based diseases rather than merely energy-deficit diseases.7 The variable clinical presentations and tissue specificity suggest that there are contributing factors beyond energy deficit during disease development.9 The reduction of ATP produced from OXPHOS can be compensated by enhanced anaerobic glycolysis, and thus mitochondrial genetic defects may not reduce ATP production.10,11 Furthermore, genetic defects are not always sufficient to cause cellular dysfunction as mitochondria can buffer against mitochondrial lesions, making environmental insults sometimes important to trigger these genetic disorders.12 Recently, the mitochondrial stress responses have gained close attention.9 Mitochondria have a comprehensive quality control system to maintain homeostasis, preventing dysfunction when facing stress. At the molecular level, mitochondria possess the quality control mechanisms of the proteome, such as mitochondrial integrated stress response (mt-ISR).13 At the organelle level, mitochondria can alter their morphology or sub-location through fusion, fission, and transport to adapt to stress or damage. At the cellular level, mitophagy coordinates with mitochondrial biogenesis, controlling the health of the mitochondrial population.14,15 Intercellular mitochondria transfer also plays a role in maintaining mitochondrial homeostasis.16 However, excessive stress can trigger mitochondria-related inflammation or apoptosis as well.17 In the context of mitochondrial diseases, genetic defects can lead to mitochondrial dysfunction. The subsequent responses to the stress induced by mitochondrial dysfunction may aid in understanding these genetic diseases.9,18 Hence, this review concludes the physiological processes of mitochondria and the potential pathogenesis of mitochondrial diseases. Significant progress in diagnosis and treatment is also summarized in this review.
Historical review and Epidemiology of mitochondrial diseases
The history of mitochondrial diseases dates back to 1871 when Theodor Leber documented hereditary and congenital optic nerve diseases, marking the first known description of a genetic mitochondrial disorder, now recognized as Leber hereditary optic neuropathy (LHON).19 The concept of mitochondrial diseases was later introduced in 1962 by Luft et al.20, who identified a young woman with severe hypermetabolism caused by mitochondrial dysfunction due to defective OXPHOS coupling in skeletal muscle mitochondria.20,21 This pivotal discovery brought mitochondrial diseases into the scientific spotlight.
During the 1960s, research primarily focused on mitochondrial myopathies. Milton Shy and Nicholas Gonatas described megaconial and pleoconial congenital myopathies,22,23 hypothesizing that these conditions were linked to mtDNA defects.24 In 1963, Engel et al. introduced an improved Gomori trichrome staining method for muscle histopathology, which enabled the detection of abnormal mitochondrial proliferation as ragged-red fibers, thus advancing histochemical studies of mitochondrial diseases.25 The 1970s saw significant progress in identifying mitochondrial metabolism defects through histochemical assays, including deficiencies in pyruvate dehydrogenase, carnitine, cytochrome c oxidase, and carnitine palmitoyltransferase.26–29 In 1977, Shapira et al. coined the term “mitochondrial encephalomyopathies” to describe a group of neuromuscular disorders characterized by defects in oxidative metabolism.30 A major breakthrough came in 1981 when Anderson et al. successfully mapped the entire mitochondrial genome, establishing a foundation for subsequent mitochondrial research.31
In 1988, the discovery of single large-scale deletions of up to 7 kilobases in patients with mitochondrial myopathies32 and a point mutation in the NADH dehydrogenase subunit 4 gene in families with LHON33 underscored the importance of mtDNA mutations, heralding the beginning of the molecular era in mitochondrial research.34 By 1989, multiple mtDNA deletions had been identified in the muscle tissues of members from a family with autosomal dominant mitochondrial myopathy.35 Further advancements were made in 1991 when Moraes et al. confirmed mtDNA depletion in the affected muscle or liver tissues of infants with autosomal recessive disorders.36 This period also saw increased attention to the role of nDNA in mitochondrial diseases, particularly with the identification of Mendelian mitochondrial disorders. A landmark discovery in 1995 revealed the first nuclear gene mutation causing mitochondrial respiratory chain deficiency in humans: a mutation in the nuclear-encoded flavoprotein subunit gene of succinate dehydrogenase led to complex II deficiency in two sisters with Leigh syndrome.37 The creation of the first comprehensive mtDNA database, MITOMAP, in 1996 further facilitated the study of mitochondrial diseases.38 Soon after, Nishino et al. attributed mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) to a defect in communication between nuclear and mitochondrial genomes.39 The 2000s saw the introduction of next-generation sequencing (NGS) technology in the diagnosis of mitochondrial diseases.40 By the 2010s, transcriptomics and other omics analyses had gained increasing attention, leading to the emergence of multi-omics approaches in the diagnosis of mitochondrial disorders.41
Scientists are actively pursuing potential treatments to address mitochondrial diseases. In 1997, Taylor et al. pioneered the use of peptide nucleic acid (PNA) in gene therapy to selectively inhibit the replication of mutated human mtDNA, thereby increasing the proportion of wild-type mtDNA and correcting defective phenotypes through heteroplasmy alteration.42 In 2006, Spees et al. discovered that intercellular mitochondrial transfer could restore aerobic respiration in mammalian cells.43 By 2009, Tachibana et al. had successfully separated the spindle-chromosome complex from mature metaphase II (MII) oocytes and transferred it into enucleated oocytes, resulting in the birth of healthy primate offspring with nDNA from the spindle donor and mtDNA from the cytoplasmic donor.44 In 2015, idebenone received approval from the European Medicine Agency (EMA) for treating LHON under specific conditions.45 In 2017, Zhang et al. reported the application of the spindle-chromosome complex transfer (ST) method in a woman carrying the m.8993 T > G mutation associated with Leigh syndrome, leading to the birth of a healthy child.46 In 2018, a gene therapy employing an allogeneic expression strategy was tested in a clinical trial for patients with LHON, demonstrating both safety and good tolerability.47 More recently, in 2023, Omaveloxolone became the first drug approved by the Food and Drug Administration (FDA) for treating Friedreich’s ataxia.48
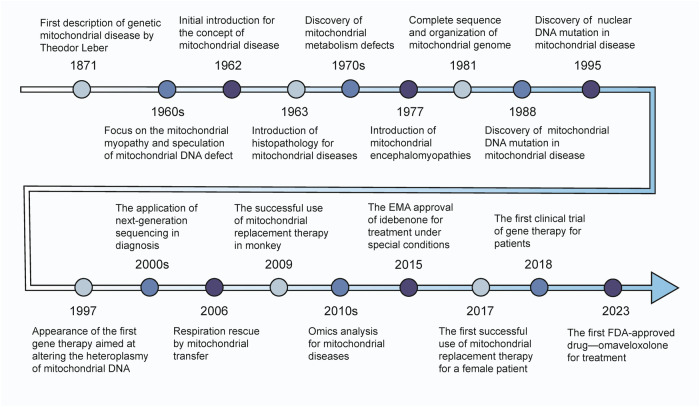
As our understanding of mitochondria deepens, so does our knowledge of the mutant genes and pathogenesis underlying mitochondrial genetic disorders. Figure 1 presents a timeline summarizing key milestones in mitochondrial disease research. Beyond the focus on primary or secondary OXPHOS, significant attention is being directed toward gene mutations that impair mitochondrial structure and function.2,3,41
Fig. 1.
Timeline of Major Historical Events in the Study of Mitochondrial Diseases. From the initial discovery to current advancements, our understanding of the mechanisms underlying mitochondrial diseases has continually deepened. Over time, research explorations and progress have contributed to the development of diagnostic and treatment methods, ultimately providing insights into more efficient and accurate diagnostic and therapeutic strategies
Previous studies estimate the global prevalence of mitochondrial diseases at approximately 1 in 5,000 births,49 with pathogenic mtDNA mutations affecting at least 12.48 per 100,000 individuals.50 Table 1 lists the regional and global incidences of specific mitochondrial diseases.
Table 1.
The prevalence of overall or individual mitochondrial diseases
| Category | Prevalence (95% CI) | Population | Region | Reference |
|---|---|---|---|---|
| Mitochondrial diseases (caused by mtDNA mutation) | 12.48(10.75–14.23)/100,000 |
Female (age<60) Male (age<65) |
Northeast England | 50 |
| 9.2 (6.5–12.7)/100,000 | Age≥18 | Southwest Finland | 881 | |
| Mitochondrial diseases | 12.5 (11.1–14.1)/100,000 |
Female (16<age<60) Male (16<age<65) |
Northeast England | 882 |
| 4.7 (4.1–5.4)/100,000 | Total | New Zealand | 883 | |
| 1.02 (0.81–1.28)/100,000 | Total | Hong Kong, China | 884 | |
| 2.9 (2.8–3.0)/100,000 | Total | Japan | 885 | |
| 2.3 (2.14–2.47)/1,000,000 | Total | Spain | 886 | |
| 7.5 (5.0–10.0)/100,000 | Age≤18 | Northwest Spain | 887 | |
| LHON | 2491 (1996–2986)/126,167,000 | Total | Japan | 888 |
| 3.22 (2.47–3.97)/100,000 | Age<65 | Northeast England | 889 | |
| 2.06 (1.8–2.4)/100,000 | Age≥5 | Finland | 890 | |
| 1/54,000 | Total | Denmark | 891 | |
| 1/68,403 | Age<85 | Australia | 892 | |
| 1/39,000 | Total | Netherlands | 893 | |
| 1.9/1,000,000 | Total | Serbia | 894 | |
| MELAS | 0.18 (0.02–0.34)/100,000 | Total | Japan | 895 |
| 4.7 (2.8–7.6)/100,000 | Age<16 | Western Sweden | 896 | |
| Mitochondrial myopathy | 0.58 (0.54–0.62)/100,000 | Total | Japan | 895 |
| Leigh syndrome | 2.05 (0.72–3.40)/100,000 | Age≤18 | Northwest Spain | 887 |
| Friedreich’s ataxia | 1/176,000 | Total | Norway | 897 |
| 1.2 (0.9–1.6)/100,000 | Total | Italy | 898 | |
| ADOA | 2.87 (2.54–3.20)/100,000 | Total | North England | 899 |
| 1/12,301 | Total | Denmark | 900 | |
| MNGIE | 1–9/1,000,000 | Total | World | 488 |
| MIDD | 0.5–2.8/100 | Diabetic patients | World | 901 |
| Barth syndrome | 1/1,000,000 | Male | World | 902 |
LHON Leber hereditary optic neuropathy, MELAS mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes, ADOA autosomal dominant optic atrophy, MNGIE mitochondrial neurogastrointestinal encephalomyopathy, MIDD maternally inherited diabetes and deafness
Determining the exact global incidence of mitochondrial diseases is challenging due to their rarity, high mortality, and clinical and genetic heterogeneity.51 Additionally, symptoms typically manifest only when a certain mutation threshold is reached—usually 80–90%—though this threshold can vary between different cells and patients.52,53 As a result, the clinical phenotypes of mitochondrial diseases caused by mtDNA mutations can differ significantly among individuals and are influenced by the level of heteroplasmy, making these diseases difficult to diagnose.54 Notably, mtDNA mutations are not exclusive to those with mitochondrial diseases; they are present in the general population as well. At least 1 in 200 healthy individuals carries a pathogenic mtDNA mutation, often with no or only mild symptoms.55 These mutations can be maternally inherited, and it is estimated that nearly 2473 women in the UK and 12,423 women in the US, aged 15 to 44, carry pathogenic mtDNA mutations.56 Interestingly, approximately 80% of mitochondrial diseases in adults are linked to mtDNA mutations, while most mitochondrial diseases in children are associated with nDNA mutations, with only 20–25% stemming from mtDNA mutations.2 These factors underscore the complexity and prevalence of mitochondrial diseases, which are more common and intricate than previously understood. Consequently, further epidemiological studies are essential to improve our understanding and prediction of mitochondrial disease prevalence.
Molecular basis of mitochondria
General characteristics of mitochondria
The mitochondrion is a double-membrane organelle present in nearly all eukaryotic organisms.57 It is widely believed that mitochondria originated from bacteria, specifically α-proteobacteria.58 The human mitochondrion contains a genome of 16,569 base pairs, distinct from the nuclear genome.31 Notably, fragments of mtDNA can integrate into the nuclear genome, forming nuclear-mitochondrial segments (NUMTs).59 The mtDNA is a circular, double-stranded molecule with multiple copies and is maternally inherited. It encodes 37 genes, including 2 rRNAs, 22 tRNAs, and 11 mRNAs. Of these, 14 tRNAs, 2 rRNAs, and 10 mRNAs are encoded on the heavy (H) strand, while the remaining 1 mRNA and 8 tRNAs are encoded on the light (L) strand.60–62
Mitochondrial non-coding RNAs (ncRNAs), such as microRNAs, long non-coding RNAs, circular RNAs, and piwi-interacting RNAs, have been identified as potential mediators of mitochondrial homeostasis.63 These ncRNAs are key messengers in mito-nuclear communication and have garnered significant attention.64 Most mitochondrial ncRNAs originate from the nuclear genome and are translocated into mitochondria via Ago2, PNPase, or associated mitochondrial proteins. These nuclear-derived ncRNAs can indirectly regulate mitochondrial homeostasis by influencing nDNA-encoded mitochondrial proteins.63 Conversely, mitochondria-derived ncRNAs, which include a limited number of long non-coding RNAs and circular RNAs, can directly regulate mtDNA expression or mitochondrial protein transport.65–67 The biogenesis, processing, and functional mechanisms of ncRNAs encoded by mtDNA remain largely unclear.63 Intriguingly, recent studies suggest the epigenetic inheritance-influenced transfer of mitochondrial tRNA (mt-tRNA) from sperm to oocyte at fertilization, highlighting the potential importance of paternal factors in mitochondrial inheritance.68 Additionally, mt-tRNA fragments have been implicated in mitochondrial diseases.69
Mitochondrial proteomes, comprising approximately 1000 to 1500 proteins, are encoded by both nDNA and mtDNA.70,71 Among these, metabolism-related proteins constitute the largest number and abundance.72 While mtDNA encodes only 13 proteins involved in OXPHOS, the vast majority of mitochondrial proteins (>99%) are encoded by the nuclear genome, synthesized on ribosomes, and subsequently imported into mitochondria.73
Mitochondrial-derived peptides (MDPs) are encoded by short open reading frames within mtDNA, including humanin, small humanin-like peptides 1-6 (SHLP1-6), MOTS-c, and CYTB-187AA.74–76 These MDPs play a pivotal role in cellular protection by maintaining homeostasis and cellular function.75 Each peptide exhibits distinct biological effects; for instance, humanin, SHLP2, and SHLP3 inhibit apoptosis and promote cell viability, whereas SHLP6 induces apoptosis.75,77 SHLP2 and SHLP4 also enhance cell proliferation.75 Humanin is essential for maintaining mitochondrial homeostasis and function by increasing mtDNA copy number and mitochondrial mass and promoting mitochondrial biogenesis.77 Similarly, SHLP2 and SHLP3 contribute to mitochondrial biogenesis, enhancing mitochondrial metabolism and function.74,75,78 MOTS-c, the first MDP discovered to enter the cell nucleus, plays a role in mito-nuclear communication.74 MOTS-c transcripts originate in mitochondria, are exported to the cytosol for translation into peptides, and then return to mitochondria.74 Under stress, AMPK activation triggers the translocation of MOTS-c to the nucleus, where it binds to nuclear DNA and interacts with transcription factors such as NRF2 and ATF1, modulating nuclear gene expression to restore cellular metabolic homeostasis.74,79 Recently, Hu et al. demonstrated that the cytochrome b transcript, encoded by mtDNA, is translated by cytosolic ribosomes using the standard genetic code to produce a 187-amino acid protein, CYTB-187AA.76 CYTB-187AA localizes to the mitochondrial matrix and interacts with SLC25A3 to regulate ATP production.76 Single nucleotide polymorphisms in the mtDNA coding regions for MDPs may facilitate the discovery of new MDPs, such as SHMOOSE.80
Without the protective presence of histones, mtDNA is more susceptible to external factors, leading to a higher likelihood of mutations. These mutations can result in various diseases, given the critical role of mitochondria in nucleated cells. The severity of such conditions often depends on the ratio of mutant to wild-type mtDNA.81 Because mtDNA exists in multiple copies, two scenarios are possible: homoplasmy, where all mtDNA copies are identical, or heteroplasmy, where the copies differ.82
Mitochondrial membrane potential (Δψm) is a fundamental property generated during OXPHOS by the respiratory chain. The stability of Δψm is essential for cell viability. Under normal conditions, Δψm may fluctuate slightly in the short term; however, prolonged changes in Δψm can lead to pathological outcomes. As a result, cells activate mechanisms to eliminate mitochondria with abnormal Δψm.83,84
Mitochondrial metabolism
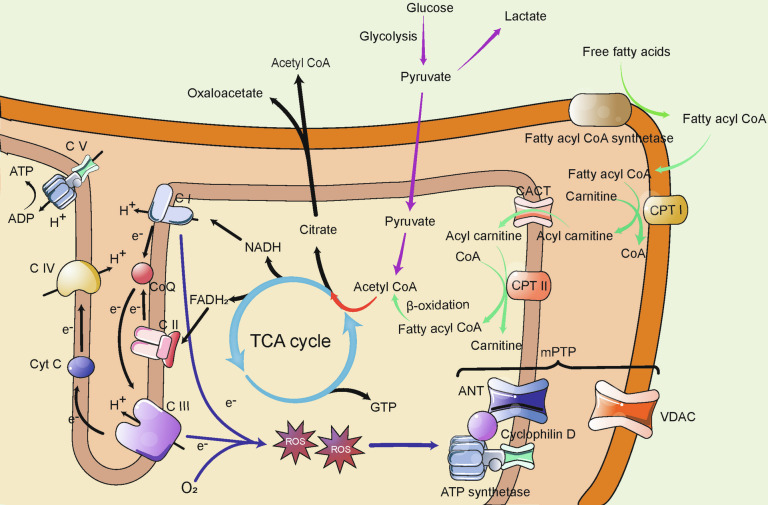
Mitochondria play a central role in substance metabolism, overseeing a vast array of metabolic processes as depicted in Fig. 2.
Fig. 2.
Overview of Mitochondrial Metabolism. As the central hub of bioenergetics, the mitochondrion utilizes NADH and FADH2 produced by the TCA cycle to generate ATP through electron transfer and the H+ gradient across the respiratory chain complexes. Complexes I and III are the primary sources of mtROS, which cause oxidative damage or signaling transduction. The mtROS also can induce the opening of mPTP. Glucose and lipids (via β-oxidation) both contribute to the TCA cycle. Citrate can cross the mitochondrial membrane, allowing acetyl-CoA to be transported into the cytoplasm for various functions. TCA cycle tricarboxylic acid cycle; CACT carnitine-acylcarnitine translocase; CPT I/II carnitine palmitoyltransferase I/II; mtROS mitochondrial reactive oxygen species; ANT adenine nucleotide translocator; VDAC voltage-dependent anion channel; mPTP mitochondrial permeability transition pore
Oxidative phosphorylation
The primary function of mitochondria is energy production, with the majority of ATP being generated through OXPHOS.85 The OXPHOS system, essential for mitochondrial respiration, consists of five multimeric protein complexes located in the cristae of the inner mitochondrial membrane (IMM).86 The respiratory chain complexes (Complexes I–IV), collectively known as the electron transport chain (ETC), facilitate the transfer of electrons from nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FADH2) to oxygen through a series of redox reactions. This process contributes to the formation of an electrochemical (proton) gradient across the IMM, ultimately reducing oxygen to H2O.87 To enhance stability and efficiency, these respiratory chain complexes often assemble into supramolecular structures.88 The proton gradient drives the translocation of protons from the intermembrane space (IMS) to the matrix via ATP synthase (Complex V), which catalyzes the conversion of ADP to ATP.85,89 Of the polypeptides involved in OXPHOS, 13 are encoded by mtDNA, while the remainder are encoded by the nuclear genome.53,89
Glucose metabolism
In glucose metabolism, glucose is initially converted to pyruvate through glycolysis in the cytoplasm. Pyruvate is then either transported into the mitochondria and converted to acetyl-CoA by pyruvate dehydrogenase or converted to lactate by lactate dehydrogenase in the cytoplasm.90 Acetyl-CoA, the principal substrate, enters the tricarboxylic acid (TCA) cycle.91 Within mitochondria, citrate synthase catalyzes the condensation of acetyl-CoA and oxaloacetate to form citrate. Citrate can either proceed through the TCA cycle, generating NADH, FADH2, and guanosine triphosphate (GTP), or be transported to the cytoplasm where it regenerates acetyl-CoA and oxaloacetate.92 When carbohydrate supply is excessive, acetyl-CoA is converted to citrate, which can then exit the mitochondria to participate in lipid synthesis or histone acetylation in the cytoplasm or nucleus.92–94 Additionally, certain gluconeogenesis processes occur in mitochondria, such as the conversion of pyruvate to oxaloacetate, followed by its conversion to malic acid and aspartic acid to facilitate gluconeogenesis.95
Lipid metabolism
The β-oxidation of fatty acids is another major energy source.96 Initially, free fatty acids (FFAs) are activated to fatty acyl-CoA in the cytosol by fatty acyl-CoA synthetase. Fatty acyl-CoA then combines with carnitine to form acylcarnitine before crossing the outer mitochondrial membrane (OMM) and IMM to enter the mitochondrial matrix.97,98 In the mitochondrial matrix, acylcarnitine regenerates into fatty acyl-CoA, which then undergoes β-oxidation to produce NADH/FADH2 and acetyl-CoA.97 The transport of fatty acyl-CoA is facilitated by carnitine palmitoyltransferase I (CPTI) on the OMM, carnitine palmitoyltransferase II (CPTII), and carnitine-acylcarnitine translocase (CACT) on the IMM.99 Some acetyl-CoA generated through β-oxidation is converted into ketone bodies in the liver, which serve as a key energy source.98
Oxidative stress
Oxidative stress occurs when the formation of reactive oxygen species (ROS) exceeds the capacity of the antioxidant defense system, which includes enzymes such as superoxide dismutase (SOD), catalase (CAT), glutathione reductase (GR), and glutathione peroxidase (GPx).100 This imbalance, primarily due to an excess of ROS, leads to cellular damage. ROS includes molecules such as superoxide anion (O2−•), hydrogen peroxide (H2O2), singlet oxygen (1O2), and hydroxyl radicals (OH•),101,102 which can impair mitochondrial function by damaging mtDNA, proteins, membrane lipids, and other cellular components.103
Mitochondria are a major source of ROS, and even under normal conditions, the ETC generates mitochondrial ROS (mtROS) as a byproduct.104,105 However, ROS can also originate from other sources, such as NADPH oxidase (Nox), monoamine oxidase (MAO), p66Shc, α-glycerophosphate dehydrogenase, electron transfer flavoprotein (ETF) and ETF dehydrogenase, and aconitase.102 The contribution of these sources varies depending on the type of ROS, the cell type, and the specific physiological or pathological conditions.106 Despite these other sources, mitochondria remain a significant producer of ROS and therefore merit considerable attention.
ROS plays a complex and essential role in cellular physiology. Although high concentrations of ROS are typically viewed as harmful byproducts of aerobic metabolism, at physiological levels, ROS function as key secondary messengers. They regulate various signaling pathways, including PI3K, MAPK, AMPK, NRF2, NF-κB, and p53, and influence enzyme activity, gene expression, cell proliferation, differentiation, immune responses, apoptosis, and mitochondrial quality control, allowing cells to adapt to environmental changes.107–109 For example, low ROS levels are critical for maintaining the self-renewal potential of stem cells.110 In addition, moderate ROS concentrations are vital for promoting axonal and dendritic growth, maintaining neuronal function, and supporting the self-renewal of neural stem cells and neurogenesis.107,109
A significant consequence of OXPHOS dysfunction is the increased production of mtROS due to decreased electron transfer efficiency in the ETC, which results in more electrons leaking and interacting with O2.88 Oxidative stress is known to play a role in the pathophysiology of mitochondrial diseases.111 Complexes I and III are the primary sites of mtROS production (Fig. 2). At Complex I, flavin mononucleotide (FMN) and possibly CoQ can transfer an electron to O2, generating O2−•, while at Complex III, ubisemiquinone (CoQH•) in the Qo site also contributes to O2−• production.101 However, the amount of mtROS produced at these sites varies between tissues: the brain primarily generates mtROS at Complex I, while the heart and lungs mainly rely on Complex III.101,104,112 O2−• can be dismutated to H2O2 by SOD or react with NO• to form peroxynitrite. H2O2 can be fully reduced to H2O or partially reduced to OH•.113
ROS disrupts cellular homeostasis by damaging lipids, proteins, and DNA.114 Due to their proximity, mtROS pose a significant threat to mtDNA, which, lacking histone protection, is especially vulnerable. When mtROS accumulate beyond a certain threshold, they can lead to a reduction in Δψm, which in turn may trigger the opening of the mitochondrial permeability transition pore (mPTP).115 The mPTP is a complex structure which spans the IMM and OMM, comprising the voltage-dependent anion channel (VDAC), adenine nucleotide translocator (ANT), ATP synthetase, and cyclophilin D.88,116,117 Additionally, β-tubulin regulates mPTP opening through its interaction with VDAC.118
Transient opening of the mPTP allows mitochondria to release excess ROS and Ca2+, preventing the harmful accumulation of these molecules. However, prolonged mPTP opening can lead to a secondary burst of ROS, a process known as ROS-induced ROS release.102 While RIRR can help eliminate irreversibly damaged mitochondria to maintain cellular homeostasis, it can also result in pathological consequences. The mPTP can also be activated by elevated Ca2+ levels.102,119 Persistent mPTP opening severely disrupts mitochondrial membrane function, becoming an important factor in mitochondrial dysfunction and ultimately leading to the activation of mitochondrial apoptosis.118 Beyond apoptosis, the mPTP plays a role in regulating other forms of cell death.17 The tendency for mPTP opening increases with aging, which further exacerbates the reduction in Δψm.84
Mitochondrial quality control
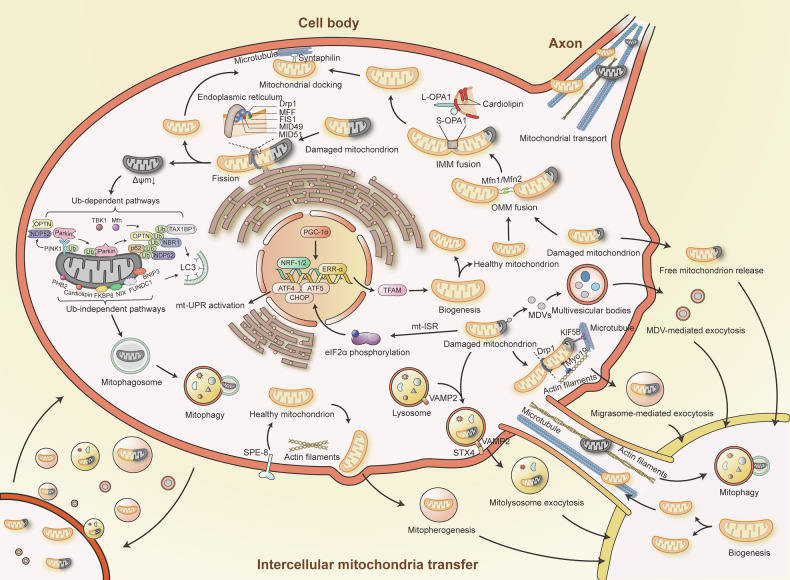
Mitochondrial quality control (MQC) is a complex and integrated network that monitors mitochondrial integrity, responds to damage or stress, and maintains mitochondrial homeostasis. This system coordinates various processes, including mt-ISR, biogenesis, dynamics, mitophagy, and intercellular mitochondria transfer (Fig. 3). MQC plays a pivotal role in a wide range of human diseases, including cancer, cardiovascular diseases, and neurodegenerative disorders.120–124
Fig. 3.
Mitochondrial Quality Control Network. Mitochondria employ both intercellular and intracellular quality control mechanisms to maintain homeostasis and redox balance. These mechanisms include mitochondrial biogenesis, fusion, fission, axonal transport, docking, mitophagy, the mitochondrial integrated stress response, and intercellular mitochondrial transfer. IMM inner mitochondrial membrane; OMM outer mitochondrial membrane; Δψm mitochondrial membrane potential; Ub ubiquitin; mt-ISR mitochondrial integrated stress response; mt-UPR mitochondrial unfolded protein response; MDV(s) mitochondria-derived vesicle(s)
Mitochondrial integrated stress response
The mt-ISR in mammals is a multifaceted mechanism that includes a transcriptional response, metabolic remodeling, and the mitochondrial unfolded protein response (mt-UPR).125 This response can be triggered by various mitochondrial stressors such as OXPHOS defects, reduction in Δψm, increased ROS level, amino acid deprivation, or the accumulation of unfolded proteins.126,127 The mt-ISR is essential for maintaining OXPHOS function and mitochondrial homeostasis through mito-nuclear communication. Key to mt-ISR are the eIF2α kinases—HRI, PKR, PERK, and GCN2—which are activated in response to mitochondrial stress.127,128 Activation of these kinases leads to the phosphorylation of eukaryotic translation initiation factor 2 alpha (eIF2α), which reduces global protein synthesis while selectively enhancing the translation of stress-related transcription factors like ATF4, ATF5, and CHOP, thereby activating the mt-UPR.129–131 This translational reprogramming helps reduce cellular energy consumption and protects cells from mitochondrial dysfunction. However, mt-ISR can also trigger apoptosis to eliminate severely damaged cells under intense stress.128 In the context of mitochondrial diseases, genetically defective respiratory chain complexes or mitochondrial dysfunction can activate mt-ISR, which aims to enhance ATP production and antioxidant capacity, thereby attempting to restore mitochondrial homeostasis before mitophagy is activated.132 While mt-ISR activation could be a common event in mitochondrial diseases, its overactivation may be detrimental, exacerbating the disease condition.125
Mitochondrial biogenesis
Mitochondrial biogenesis is the process by which mitochondria increase in number and size.133 This process is primarily regulated by PGC-1α (PPAR-γ coactivator-1α), a key regulator that activates nuclear respiratory factors NRF-1 and NRF-2, as well as oestrogen-related receptor-α (ERR-α).134 These factors enhance the expression of mitochondrial transcription factor A (TFAM, also known as mtTFA),135,136 which is critical for the replication and transcription of mtDNA and subsequent mitochondrial function.135 PGC-1β, which shares structural and functional similarities with PGC-1α, also promotes mitochondrial biogenesis.133 Additional regulators of this process include AMPK (AMP-activated protein kinase), nitric oxide (NO), SIRTs (sirtuins), TORCs (transducers of regulated CREB-binding protein), CaMK (calcium/calmodulin-dependent protein kinase), calcineurin, p38 MAPK, RIP140 (receptor-interacting protein 140), and Sin3A, which all influence mitochondrial biogenesis through the activation of PGC-1α.15,133
Mitochondrial dynamics
The structure of mitochondria includes the mitochondrial matrix, OMM, IMM, and IMS. The IMM is further divided into the inner boundary membrane and mitochondrial cristae. The shape and size of mitochondria vary and are closely linked to their function.137 Mitochondrial dynamics involve two primary processes: fusion and fission, both of which are essential for maintaining mitochondrial homeostasis and are associated with mtDNA stability, oxidative stress, apoptosis, mitophagy, and cell division.138,139
Fission is the process by which a single mitochondrion divides into two or more daughter mitochondria. This process allows for the segregation of healthy mitochondria from those that are old or damaged, thereby eliminating mitochondria with irreversible mtDNA damage or low Δψm to maintain cellular homeostasis.140 Fission is closely associated with mitophagy, the selective degradation of damaged mitochondria, which will be discussed further in the next section.141 Additionally, fission helps meet increased energy demands by producing more daughter mitochondria.142 In endothelial cells, fission also aids in the localization of mitochondria near cytoskeletal proteins to support metabolic needs.142 Functional fission can mitigate mitochondrial damage in mitochondrial diseases.143 However, excessive fission is linked to pathological outcomes, including impaired mitochondrial bioenergetics as well as induction of oxidative stress and cell death.144 The GTPase dynamin-related protein 1 (Drp1) is a key regulator of fission, and the endoplasmic reticulum also plays a role in this process.145 Drp1 is recruited to mitochondria from the cytoplasm, where it interacts with mitochondrial fission factor (MFF), mitochondrial dynamics protein of 49 kDa (MID49), MID51, and mitochondrial fission 1 protein (FIS1) on the OMM to drive the fission process.146–148 Additionally, FIS1 inhibits mitochondrial fusion by interacting with mitofusins, thereby inhibiting their GTPase activity.149
Mitochondrial fusion is the process where two or more mitochondria come into close contact and merge their IMM and OMM. This process relies on two key GTPases: optic atrophy protein 1 (OPA1) and mitofusins 1 and 2 (Mfn1 and Mfn2).150,151 Fusion begins with the merging of the OMM, driven by Mfn1 and Mfn2, which are localized on the OMM. After the OMM fusion, long OPA1 (L-OPA1) interacts with cardiolipin to facilitate the fusion of the IMM.152,153 There is also a short form of OPA1 (S-OPA1) produced by the proteolytic cleavage of L-OPA.153 The interaction between S-OPA1 and L-OPA1 promotes the fusion of the IMM.154,155 Fusion plays a pivotal role in mitochondrial heteroplasmy, which refers to the co-existence of mutant and wild-type (healthy) mitochondria.156 Defective mitochondria with mutant mtDNA can fuse with healthy mitochondria, compensating for defects by sharing components such as transcripts, thus mitigating the effects of mutations (heteroplasmy). Similarly, two defective mitochondria can fuse to cross-complement each other. Therefore, mitochondrial fusion can rescue certain dysfunctions if the mutation remains within a critical threshold.157,158 Fusion is often viewed as a defensive response, enabling mitochondria to adapt to cellular stress by reducing mtDNA heteroplasmy, bridging Δψm, and exchanging various metabolic intermediates.142
Mitochondrial transport is an ATP-dependent process that is especially critical in neurons. This transport occurs in both anterograde and retrograde directions along microtubules.159 Anterograde transport provides healthy and robust mitochondria from soma for distal axon, while damaged mitochondria in these distal regions are retrogradely transported to the soma for repair and degradation.160 Long-distance mitochondrial transport along microtubules is facilitated by two types of motor proteins: kinesin and dynein. In neurons, axonal microtubules are oriented with their minus ends toward the soma and their plus ends toward the distal axon. The minus-end-directed dynein drives retrograde transport, while the plus-end-directed kinesin (mainly KIF5) controls anterograde transport.161 The kinesin and dynein should interact with their motor adaptors before transport. For anterograde transport, the motor adaptor complex consisting of Miro (an atypical Rho GTPase) and Milton (TRAK in mammals) connects kinesin to mitochondria. Retrograde transport is primarily mediated by dynein and its motor adaptor, dynactin. The initiation of retrograde transport is believed to involve cooperation between the dynein-dynactin complex, VDAC on the OMM, and the Milton-Miro complex.162 Additionally, Drp1, Mfn1, and Mfn2 are thought to play roles in mitochondrial transport.160 There is also short-distance mitochondrial movement along actin filaments within dendritic spines, which is mediated by myosins.162
In contrast to transport, mitochondrial docking ensures that mitochondria remain in place to maintain stable mitochondrial numbers, adequate ATP production, and meet metabolic demands, particularly in regions with high energy requirements and metabolic stress.161 Syntaphilin, an anchoring protein, binds to the OMM and attaches axonal mitochondria to microtubules, resulting in mitochondrial docking.160
Mitophagy
Mitophagy is the process by which damaged mitochondria are delivered to lysosomes for degradation, a concept first introduced by Lemasters as a specific form of organelle autophagy.163 Unlike mitochondrial biogenesis, which generates new mitochondria, mitophagy removes damaged or unnecessary mitochondria to maintain cellular homeostasis, often through a selection process mediated by mitochondrial fission.164 However, excessive mitophagy can lead to a significant loss of mitochondrial content, potentially triggering cell death.165,166 The interaction between Drp1 and Zip1 (a mitochondrial zinc transporter) at the fission site facilitates Zn2+ entry into the mitochondrial matrix, resulting in a localized reduction of Δψm, which subsequently initiates mitophagy in the affected mitochondria.167 Interestingly, mtDNA mutations alone may not be sufficient to trigger mitophagy.168 ROS plays a critical role in activating mitophagy, and in turn, mitophagy helps regulate ROS levels.169 Excessive ROS can induce non-selective autophagy in response to oxidative stress, while mild oxidative stress typically triggers selective mitophagy that is dependent on mitochondrial fission.170 The mitophagy process involves several key steps: the reduction of Δψm, formation of the mitophagosome, delivery of the mitophagosome to the lysosome, and finally, the degradation and recycling of mitochondrial components.171,172
Mitophagy mechanisms are generally categorized into ubiquitin (Ub)-dependent and Ub-independent pathways.
Among the Ub-dependent pathways, the phosphatase and tensin homolog-induced putative kinase 1 (PINK1)-Parkin pathway is the most extensively studied. In mitochondria with normal Δψm, PINK1 is transported to the IMM, where it is cleaved and degraded.173 However, when Δψm is reduced, PINK1 cannot reach the IMM and instead accumulates on the OMM. There, PINK1 phosphorylates ubiquitin and recruits and phosphorylates the E3 ubiquitin ligase Parkin.174,175 Once phosphorylated, Parkin binds to Ser65-phosphorylated ubiquitin on the OMM, fully activating its E3 ubiquitin ligase activity. This activation amplifies the pathway, promoting mitophagy.176 Mitochondria tagged with phosphorylated poly-Ub chains by Parkin are recognized by ubiquitin-binding receptor proteins such as OPTN, NDP52, SQSTM1/p62, TAX1BP1, and NBR1,177–179 which then bind with LC3 to initiate mitophagy. Additionally, TBK1 and Mfn2 have been shown to participate in this pathway.180,181 Beyond the PINK1-Parkin pathway, there are Parkin-independent, ubiquitin-dependent pathways where PINK1 directly recruits NDP52 and OPTN.177
Mitophagy receptors such as Fun14 domain containing 1 (FUNDC1), BCL2 interacting protein 3 (BNIP3), BCL2 interacting protein 3 like (BNIP3L/NIX), FKBP prolyl isomerase 8 (FKBP8), and ATAD3B can directly interact with LC3 through their LIR (LC3-interacting region) motifs, thereby initiating ubiquitin-independent mitophagy. Additionally, PHB2 and cardiolipin also participate in the ubiquitin-independent pathway due to their translocation to the OMM.171,182,183
Intercellular mitochondria transfer
Intercellular mitochondria transfer, where mitochondria are exchanged between donor and recipient cells, is another key component of MQC. The mitochondria transfer between cells with normal mitochondria and cells with dysfunctional mitochondria can rescue mitochondrial respiration defects.43 Mitochondria transfer is believed to aid in coping with cellular stress by facilitating intercellular communication under both physiological and pathological conditions.16 In this process, stressed donor cells transfer damaged mitochondria to healthy recipient cells. The recipient cells, upon receiving the damaged mitochondria, trigger mitochondrial biogenesis and fission to regenerate healthy mitochondria, which can then be re-transferred to the stressed donor cells. Additionally, stressed donor cells can transfer damaged mitochondria to other cells to initiate transcellular mitophagy (autophagy), especially when the stress or damage exceeds their metabolic capacity.16,184,185 There are three major routes of mitochondria transfer: tunneling nanotubes (TNT), mitochondrial extracellular vesicles (mitoEVs), and free mitochondria release.
Mitochondria can be shuttled across TNTs along either microtubules or actin filaments. Kinesin and its motor adaptor, the Miro-Milton complex, facilitate movement along microtubules, while myosin mediates transfer along actin filaments by interacting with Miro and anchoring mitochondria to the actin filaments.184,185
In addition to conventional extracellular vesicles (EVs), mitochondria produce specialized vesicles known as mitochondria-derived vesicles (MDVs), which encapsulate mtDNA and other mitochondrial components.186 MDVs primarily fuse with multivesicular bodies, such as late endosomes and lysosomes, although a select few are secreted into the extracellular space via a process driven by OPA1 and Snx9.187
Mitolysosome exocytosis, a mitoEVs-related MQC mechanism first observed in flunarizine-induced Parkinsonism-like symptoms, eliminates mitochondria through a mitophagy-independent pathway.188 During this process, mitochondria are directly engulfed by lysosomes and extruded from the cell without the formation of autophagosomes.189 Flunarizine-induced impairment of OXPHOS and the collapse of Δψm are believed to trigger this process. Proteins such as BAX, a mediator of mitochondrial outer membrane permeabilization (MOMP), and NDUFS4, a complex I subunit, may facilitate mitochondrial entry into lysosomes.188 Once inside lysosomes, the extracellular secretion of mitochondria is mediated by a VAMP2 (vesicle-associated membrane protein 2)-STX4 (syntaxin-4)-dependent mechanism.188
Migrasome-mediated exocytosis is also an emerging MQC mechanism involving mitoEVs.190 Migrasomes, defined as vesicles containing cytosolic contents, form on retraction fibers during cell migration.191,192 This process enables cells to clear damaged mitochondria, which may harbor detrimental mutant mtDNA, reduced Δψm, or elevated ROS levels, ensuring mitochondrial quality. Key factors in this process include Myosin19 (Myo19), KIF5B, and Drp1. Damaged mitochondria are transported to the plasma membrane by KIF5B, where Myo19 anchors them to cortical actin before Drp1-driven fission occurs. The reduced recruitment of dynein, the inward motor on microtubules, prevents damaged mitochondria from retracting back, ultimately leading to their incorporation into migrasomes.190 Migrasome formation relies on a reconstituted membrane system rich in tetraspanins and cholesterol.193 Migrasome-mediated exocytosis plays a role in maintaining homeostasis, particularly when the damage is insufficient to trigger mitophagy, and is especially important in migrating cells.
Mitopherogenesis is a specialized form of mitochondria-specific ectocytosis identified in sperm, which functions to control mitochondrial quantity. Unlike other exocytosis processes, mitopherogenesis involves the secretion of healthy mitochondria through EVs, with each vesicle carrying a single healthy mitochondrion. Proper actin-filament dynamics, extracellular protease activity, and the tyrosine kinase SPE-8 significantly influence this process,194 highlighting its potential impact on MQC.
Finally, free mitochondria released without membrane encapsulation occur in cases of mitophagy dysfunction.195 Damaged mitochondria within mitophagosomes lacking mammalian ATG8 conjugation cannot be degraded by lysosomes and instead undergo autophagic secretion, transferring the damaged mitochondria to healthy cells for transcellular degradation.195
Mitochondrial apoptosis
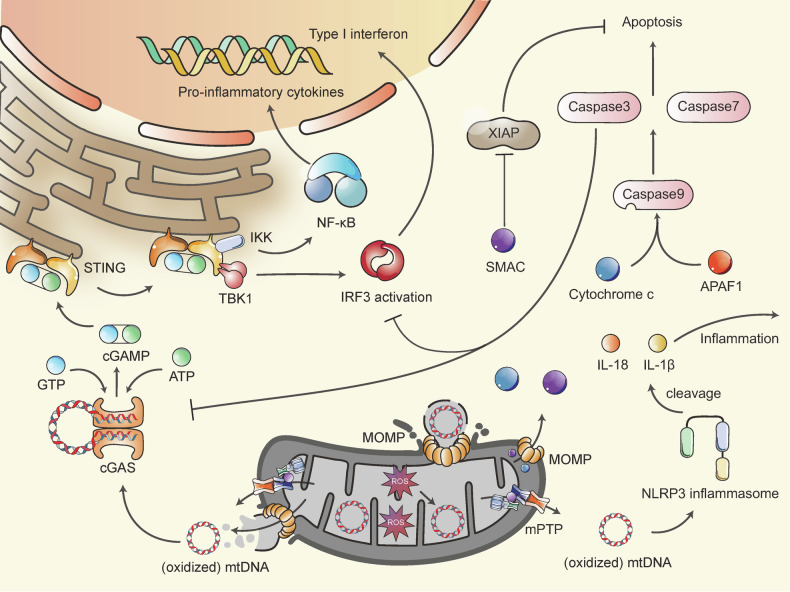
Under stress, cells may initiate apoptosis as a mechanism to manage these adverse situations.196 Apoptosis can occur through mitochondrial pathway (Fig. 4), thereby playing a role in the pathogenesis of mitochondrial diseases.111,197,198
Fig. 4.
Mitochondrial Apoptosis and Inflammation. Components within the IMS or matrix can trigger apoptosis or inflammation if leaked into the cytosol, primarily due to the mPTP and MOMP. Upon MOMP formation and mPTP opening, cytochrome C, SMAC, and mtDNA are released into the cytosol. Cytochrome C interacts with APAF1, activating caspase 9 to initiate the caspase cascade leading to apoptosis. SMAC accelerates this process by inhibiting XIAP. After binding with mtDNA, the cGAS enzyme produces cGAMP from ATP and GTP, activating the cGAS-STING signaling pathway and inducing type I interferon expression and NF-κB activation. The NLRP3 inflammasome can also bind with (oxidized) mtDNA to promote IL-1β and IL-18 cleavage. However, caspase 3 cleaves cGAS and IRF3 during apoptosis, inhibiting inflammation. MOMP mitochondrial outer membrane permeabilization; mPTP mitochondrial permeability transition pore; SMAC second mitochondrial-derived activator of caspases; APAF1 apoptosis protease activating factor 1; XIAP X-linked inhibitor of apoptosis protein; cGAS cyclic GMP-AMP synthase; STING stimulator of interferon genes; cGAMP cyclic guanosine monophosphate–adenosine monophosphate; TBK1 TANK binding kinase 1; IKK IκB kinase; IRF3 interferon regulatory factor 3
The mitochondrial apoptosis pathway is intricately linked to MOMP, which is predominantly regulated by BCL-2 family proteins, though it can also be triggered by the mPTP.116,196 The BCL-2 family is divided into pro-apoptotic and anti-apoptotic members. Pro-apoptotic BCL-2 proteins include BH3-only proteins (such as BID, BIK, and BIM) and effectors like BAK, BAX, and BOX, while anti-apoptotic proteins include BCL-2, BCL-X, BCL-W, A1, and MCL1. BH3-only proteins respond to apoptotic stimuli by activating effectors, typically BAX and BAK.199,200 Once activated, BAX and BAK accumulate on the OMM, dimerize, and form higher-order oligomers that create pores in the OMM, leading to MOMP.201 MOMP results in the release of IMS proteins, such as cytochrome c and the second mitochondrial-derived activator of caspases (SMAC), into the cytoplasm. Cytochrome c binds to apoptosis protease activating factor 1 (APAF1) in the cytoplasm, and together, they activate caspase 9. Activated caspase 9 then activates caspases 3 and 7, which ultimately drive the apoptotic process. Additionally, SMAC enhances the caspase cascade by inhibiting the X-linked inhibitor of apoptosis protein (XIAP).199,202,203 Other forms of programmed cell death, such as necroptosis, pyroptosis, and ferroptosis, are also related to mitochondria.204
Mitochondrial inflammation
Mitochondria can initiate inflammation by releasing damage-associated molecular patterns (DAMPs) due to their evolutionary similarities to bacterial pathogen-associated molecular patterns when subjected to stress or damage. These DAMPs include N-formyl peptides, TFAM, cardiolipin, ATP, ROS, and mtDNA.205 In addition to the inflammatory effects of mtDNA, which will be discussed in detail, other DAMPs have also been recognized for their roles in promoting inflammation.206,207
Under conditions of mitochondrial stress or dysfunction, mtDNA can be released into the cytosol or extracellular space.208,209 MtDNA can trigger pro-inflammatory and type I interferon responses, which vary depending on the cell type and context.210 The release of mtDNA into the cytoplasm occurs via MOMP and the opening of the mPTP.210 It is likely that mPTP and MOMP work together to facilitate mtDNA release.208 The inflammation mediated by mtDNA is primarily driven by the cGAS (cyclic GMP-AMP synthase)-STING (stimulator of interferon genes) signaling pathway (Fig. 4). When cGAS binds to mtDNA, it catalyzes the production of cyclic guanosine monophosphate–adenosine monophosphate (cGAMP) from ATP and GTP.211 cGAMP then activates STING, which subsequently recruits and activates TANK binding kinase 1 (TBK1). TBK1 phosphorylates STING, leading to the activation of transcription factors interferon regulatory factor 3 (IRF3), leading to type I interferon expression.212–214 STING also phosphorylates IκB kinase (IKK) to initiate NF-κB pathway, resulting in increased production of pro-inflammatory cytokines.208 In addition to the cGAS-STING pathway, the mtROS and mtDNA can activate NLRP3 inflammasome to promote IL-1β and IL-18 cleavage.17 Notably, low levels of mtDNA-induced inflammation, caused by minor MOMP, can aid in fighting infections,215 while simultaneous activation of caspase 3 following MOMP can inhibit mtDNA-induced inflammation by cleaving cGAS and IRF3.216,217
A connection between inflammation and mitochondrial bioenergetics has been established, indicating that mitochondrial dysfunction can exacerbate inflammation, which in turn impairs OXPHOS and disrupts MQC.218 An inflammatory transcriptomic profile has been observed in peripheral blood mononuclear cells of patients with mitochondrial diseases.219 Although research on the role of inflammation in mitochondrial diseases is still limited, inflammation is a critical factor in many human diseases, such as neurodegenerative disorders.220 Its potential pathological effects on mitochondrial diseases warrant further investigation.
In summary, mitochondria are central to metabolism, stress response, inflammation, and other critical cellular processes. Energy stress, characterized by reduced intracellular ATP levels, is a key feature of many human diseases.221,222 Mitochondrial genetic defects that disrupt energy metabolism lead to protein synthesis defects, ATP insufficiency, and mtROS overproduction.81 Despite the genetic heterogeneity of mitochondrial diseases, the resulting protein, energy, and oxidative stress are consistent.223 Cells initiate various MQC mechanisms to restore normal bioenergetics and redox balance in response to stress.224 However, when damage exceeds the capacity of MQC system, stress-induced mPTP opening and MOMP can release mtDNA or other DAMPs into the cytoplasm, triggering inflammation and cell death.209,225 Understanding how cells respond to stress induced by mitochondrial genetic defects may provide insights into the onset and progression of these diseases.
Molecular mechanisms of mitochondrial diseases
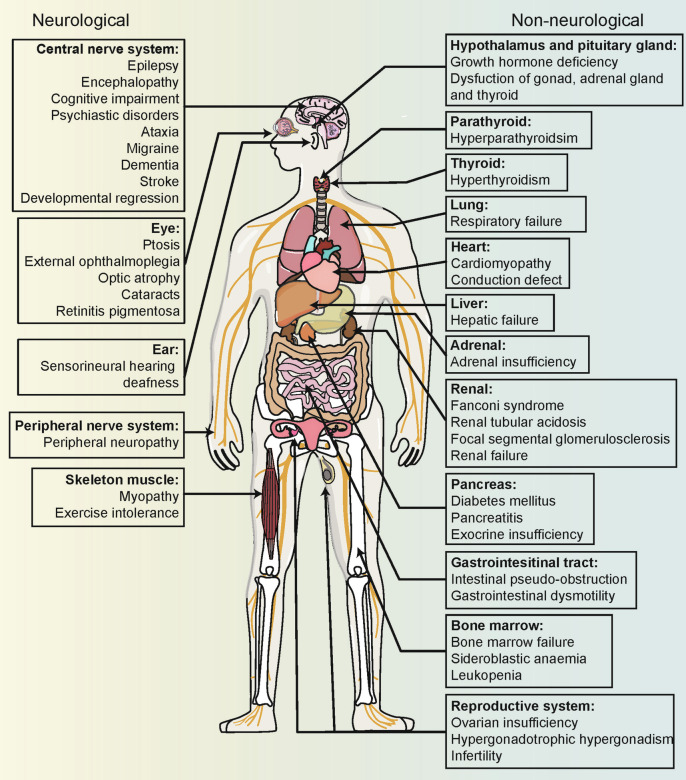
Mitochondrial diseases are characterized by primary or secondary defects in mitochondrial function or structure, resulting from mutations in either nDNA or mtDNA.226 Specifically, 36 pathogenic genes (11%) are encoded by mtDNA, while 302 (89%) are encoded by nDNA.227 Heteroplasmy is present in most healthy individuals without leading to mitochondrial disease.228 However, symptoms manifest only when the proportion of mutant mtDNA surpasses a certain threshold.229 Due to the considerable genetic and clinical heterogeneity, mitochondrial diseases can affect either single or multiple organ systems, leading to a wide range of clinical manifestations (Fig. 5). Tissues with high energy demands are particularly susceptible to energy deficits and are therefore most commonly affected.230 Additionally, the age of onset and severity of these diseases can vary significantly depending on the degree of heteroplasmy and individual differences.4
Fig. 5.
Multisystem Clinical Presentation of Mitochondrial Diseases. Due to the ubiquitous presence of mitochondria, mitochondrial diseases can present in any tissue of the body. Tissues and organs with high energy demands, such as skeletal muscle and brain, are particularly susceptible to oxidative phosphorylation defects, leading to common manifestations like myopathy and encephalopathy in mitochondrial diseases. The diverse and variable symptoms associated with these conditions increase the risk of misdiagnosis
The primary mitochondrial diseases caused by mtDNA mutations
The hallmark of mitochondrial diseases resulting from mtDNA mutations is the primary disturbance of OXPHOS. This disturbance typically leads to energy deficits, increased oxidative stress, and a collapse of the Δψm.
Leber hereditary optic neuropathy
LHON is the most prevalent mitochondrial disease, characterized by maternal inheritance and a pronounced male sex bias.231 It was the first disease definitively linked to mtDNA mutations.33 The primary clinical manifestation of LHON is the bilateral, severe loss of central vision, caused by degeneration of retinal ganglion cells (RGCs).232 The high energy demands of RGCs may explain the cell-specific vulnerability in the eye. Retina is one of the most energy-consuming tissues in the body.233 RGCs are located in the retina, and their axons form the optic nerve. Based on mitochondrial distribution and energy consumption, RGCs can be divided into four subcellular components: dendrites, cell body, unmyelinated axon (including intraocular and optic nerve head segments), and myelinated axon (post-lamina cribrosa).233 RGCs, with their exceptionally long axons and high frequency of action potentials, require substantial energy. Mitochondrial transport along these extensive axons consumes significant amounts of ATP to sustain axonal function.197,233 Additionally, the continuous conduction of action potentials in unmyelinated axons demands more energy than the saltatory conduction in myelinated axons,197,234 making the unmyelinated segments before the lamina cribrosa particularly susceptible to energy deficits.235 Mitochondrial density correlates with energy demand,233 resulting in an uneven distribution where mitochondria are abundant in the cell body and unmyelinated axons but sparse in the myelinated axons.236 Compounding this vulnerability, exposure to light, particularly short-wave or blue light (400–480 nm),237 can negatively impact OXPHOS in RGCs, leading to decreased ATP production and increased ROS levels.237
Over 95% of patients with LHON carry one of three common mtDNA point mutations: m.3460 G > A, m.11778 G > A, or m.14484 T > C.231 These mutations occur in the MT-ND1, MT-ND4, and MT-ND6 genes, respectively, all of which encode subunits of complex I.238 While ATP synthesis deficits are evident, energy failure alone may not be the primary cause of RGC degeneration in LHON.231 Instead, the dysfunctional complex I likely increases mtROS production, which in turn induces cellular apoptosis, playing a critical role in RGC degeneration.235,239 Mitochondrial apoptosis is pivotal in the pathogenesis of LHON.240 For instance, the m.3460 G > A mutation in MT-ND1 is associated with elevated mtROS production and increased levels of pro-apoptotic proteins such as cytochrome c, BAK, BAX, PARP, and cleaved caspases 3, 7, and 9.241 In addition to ROS, energy failure can activate apoptosis-inducing factors and endonuclease G, triggering apoptosis through a caspase-independent pathway.242 Overproduction of mtROS can directly induce cytochrome c release by prolonging mPTP opening or indirectly trigger cytochrome c release through caspase cascade activation and MOMP, leading to apoptosis in RGCs.243 Mito-nuclear communication also plays a role in LHON pathogenesis, as evidenced by the activation of mt-ISR transcripts in LHON, which leads to chronic inhibition of protein synthesis, affecting synaptic and oligodendroglial function, and contributing to disease progression.244,245 Besides, the respiratory chain complex I defect can cause remodeling of one-carbon metabolism through mt-ISR.125,246 This one-carbon metabolic remodeling undermines NADPH production, which sensitizes affected cells to oxidative stress and may facilitate inflammation and cell death.10 However, the anti-apoptotic protein XIAP can prevent RGC apoptosis by inhibiting mitochondrial apoptosis.247
Excessive mitophagy, driven by AMPK activation, has been shown to promote apoptosis and may lead to the widespread and near-synchronous death of RGCs in subacute LHON. In contrast, overexpression of PGC-1α, which facilitates mitochondrial biogenesis, has been found to prevent cell apoptosis.248 Increased mitochondrial biogenesis is thought to contribute to the phenomenon of incomplete penetrance in LHON and serves as a compensatory mechanism to restore mitochondrial turnover in LHON carriers.249 Thus, balancing mitochondrial biogenesis and mitophagy could represent a potential therapeutic target. Additionally, mitochondrial transport, an ATP-dependent process critical for maintaining axonal mitochondrial homeostasis, has been found to be impaired in LHON, potentially contributing to axonal loss.250
Significant progress has been made in developing therapies for LHON. Techniques such as allotopic expression and gene editing hold great promise for future treatments. Currently, Idebenone is the only drug approved by the European Medicines Agency for improving visual impairment in patients with LHON.198,251
Mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes
Mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) manifest as lactic acidosis, headaches accompanied by nausea and vomiting, epilepsy, and stroke-like episodes, all inherited maternally.252 Additional symptoms observed in MELAS include deafness, diabetes mellitus, gastrointestinal disturbances, ataxia, growth failure, myopathy, and cardiomyopathy.253,254 The primary genetic cause of MELAS is the m.3243 A > G mutation in the MT-TL1 gene, which encodes mt-tRNALeu(UUR),255 accounting for over 80% of cases.255 Another mutation, m.3271 T > C in MT-TL1, is responsible for 7–15% of MELAS cases,256,257 while other mtDNA mutations associated with MELAS are rare. Mutations in the MT-TL1 gene can lead to defects in the aminoacylation of mt-tRNALeu(UUR), disrupting the interaction between mRNA and ribosomes.258 This abnormal translation impairs the synthesis of respiratory chain complexes, particularly complexes I and IV.259
Oxidative stress is believed to play a significant role in MELAS pathogenesis.111 Increased levels of ROS, apoptosis, and inflammation have been observed in diseased endothelial cells from patients with MELAS carrying the m.3243 A > G mutation.260 Similar endogenous oxidative stress has been found in fibroblasts and induced pluripotent stem cells (iPSCs) from patients with MELAS.183
In neuronal cells, complex I defects predominate, and active degradation of complex I via mitophagy is a protective response to mitochondrial dysfunction during neuronal differentiation.261 This degradation reduces mtROS production, acting as a protective mechanism.261 However, normal lysosomal function and the sequestration of cytosolic components during autophagy depend heavily on adequate ATP levels.262 In MELAS fibroblasts, although mitophagy is activated, autophagic flux is reduced, and autophagosome elimination is defective, likely due to ATP deficiency.263 This reduction in mitophagy can lead to the accumulation of damaged mitochondria with defective complex I. Interestingly, mt-tRNA fragments, a class of ncRNAs, have been implicated in extracellular lactate accumulation in MELAS, possibly through the downregulation of mitochondrial pyruvate carrier 1.69 Furthermore, the oxidative stress induced by primary tRNALeu(UUR) defects can exacerbate impaired tRNA modification.264 Stress-induced microRNA-9/9* has been shown to post-transcriptionally suppress mt-tRNA-modification enzymes, leading to reduced U34 modification of non-mutant tRNAs and promoting mitochondrial dysfunction.264
Current treatments for MELAS are symptomatic and include the supplementation of antioxidants and cofactors, anti-epileptic drugs, and lactate-lowering agents. However, there remains no consensus on the optimal treatment approach for MELAS.265,266
Maternally inherited diabetes and deafness
Maternally inherited diabetes and deafness (MIDD) is another mitochondrial disease frequently associated with the m.3243 A > G mutation in the MT-TL1 gene.267 The most prominent clinical features of MIDD are diabetes and hearing loss,268 although other complications such as myopathy, neuropathy, oculopathy, cardiac disease, and nephropathy are also observed.269 Approximately 85% of MIDD cases are caused by the m.3243 A > G point mutation.269 Notably, this same mutation is also responsible for MELAS. Interestingly, there have been case reports of MIDD evolving into MELAS over time,270 suggesting that these conditions represent different phenotypes within the same disease spectrum influenced by the level of mtDNA heteroplasmy: MELAS typically occurs with higher levels of mtDNA heteroplasmy (typically >85%), while MIDD is associated with lower levels (typically <45%).271
Insulin production demands significant ATP, and the decreased ATP generation coupled with increased ROS production in pancreatic β-cells with dysfunctional mitochondria leads to a gradual decline in insulin secretion, eventually resulting in insulin deficiency.272 Oxidative stress also plays a critical role in hearing loss,273 with ROS-induced activation of the AMPK-E2F1 pathway promoting apoptosis in the stria vascularis and spiral ganglion neurons.274 Additionally, the role of nuclear factors in the phenotypic variability of the m.3243 A > G mutation has been increasingly recognized.275
Managing blood glucose levels is the primary focus of treatment for MIDD. Since insulin sensitivity is generally preserved, oral hypoglycemic drugs, including insulin secretagogues, should be considered. However, metformin should be avoided due to the increased risk of lactic acidosis.272
Myoclonic epilepsy with ragged red fiber
Myoclonic epilepsy with ragged red fibers (MERRF) is a mitochondrial disorder defined by the presence of progressive myoclonus epilepsy and ragged-red fibers (RRF) observed in muscle biopsies.276 This disease also manifests with symptoms like ataxia, cardiomyopathy, lipomatosis, and dementia.277 In 1990, a significant association was identified between MERRF and the m.8344 A > G point mutation in the MT-TK gene, responsible for encoding mt-tRNALys(UUR).278 This mutation, m.8344 A > G, is the most prevalent pathogenic variant, accounting for over 80% of cases.279 It hinders the N1-methyladenosine (m1A) modification at position 58 in mt-tRNALys(UUR),280 leading to defects in mt-tRNALys(UUR), which subsequently disrupts the synthesis of respiratory chain complexes, primarily complexes I and IV, resulting in mitochondrial dysfunction.281 Recently, mutations in the MT-RNR1 and MT-RNR2 genes, which encode 12S rRNA and 16S rRNA, respectively, have also been linked to MERRF.282 These rRNAs are integral to mitochondrial ribosomes and the mtDNA-encoded mRNAs.282
The neurological defects observed in MERRF may be related to compromised synaptic plasticity in excitatory neurons, a consequence of mitochondrial dysfunction and synaptic impairment.283 The study utilizing human iPSCs derived from patients harboring the m.8344 A > G mutation reveals mitochondrial fragmentation, reduced oxygen consumption, and elevated ROS production.284 Furthermore, neurons induced from patients with MERRF exhibit smaller, rounded, and fragmented mitochondria with decreased Δψm, increased ROS. The activated mitophagy and impaired autophagy flux are also present in these neurons.285 Despite the activation of autophagy and mitophagy in MERRF, their flux remains impaired, which is associated with ATP deficiency.286 Treatment with coenzyme Q10 (CoQ10) has been shown to enhance mitochondrial function in MERRF fibroblasts and cybrids by promoting autophagy flux,286 indicating that reduced mitophagy and autophagy flux might be critical contributors to MERRF pathogenesis. Notably, no apoptotic changes are observed in the muscles of mitochondrial encephalomyopathies, possibly due to sarcoplasmic expression of XIAP.287,288 However, the exact mechanisms underlying this apoptosis suspension require further investigation. Importantly, defective tRNAs can lead to the accumulation of damaged and unfolded mitochondrial proteins,289,290 suggesting that targeting mt-ISR could be a potential therapeutic strategy for MERRF and other mitochondrial disorders caused by tRNA defects. Continued research into the molecular etiology of these conditions is essential.
Neurogenic muscle weakness, ataxia, and retinitis pigmentosa syndrome
Neurogenic muscle weakness, ataxia, and retinitis pigmentosa (NARP) syndrome is a maternally inherited disorder characterized by a range of symptoms including muscle weakness, sensory neuropathy, ataxia, seizures, dementia, retinitis pigmentosa, optic atrophy, and developmental delay.291
Mutations m.8993 T > G or m.8993 T > C in the MT-ATP6 gene are implicated in the pathogenesis of NARP.291–293 These mutations are also associated with Leigh syndrome,294,295 with the specific phenotype determined by the level of heteroplasmy. A heteroplasmy level exceeding 85% for the m.8993 T > G mutation is predominantly linked to childhood-onset Leigh syndrome, whereas a level of 60–70% typically results in adult-onset NARP. Both diseases may occur with heteroplasmy levels between 70–85%.234,296 The m.8993 T > G mutation leads to the substitution of a highly conserved leucine with arginine (while m.8993 T > C replaces leucine with proline) in the ATPase6-encoded proton channel, impairing the proton translocation mechanism of ATP synthase (complex V) and subsequently disturbing OXPHOS.234
In addition to role in OXPHOS, the ATP synthase subunits e and g are important to the formation of mitochondrial cristae.297 Dissolution of mitochondrial cristae has been observed in patients with the m.8993 T > G mutation.298 Additionally, ATP synthase is a component of mPTP.88 Aberrant mitochondrial cristae and mPTP opening may lead to the release of mtDNA and IMS proteins, such as cytochrome c, thereby triggering mitochondrial apoptosis and inflammation.299 This hypothesis warrants further investigation. In yeast cells harboring defective MT-ATP6, IMM fusion is inhibited. It is plausible that dysfunctional IMM fusion could be a common feature in all diseases with genetically defective OXPHOS, potentially initiating mitophagy to eliminate defective mtDNA.300 The molecular basis underlying cerebellar atrophy in patients with the m.8993 T > G or m.8993 T > C mutations remains unclear. However, in a mouse model of inherited Purkinje cell degeneration, increased mitophagy and autophagy have been associated with Purkinje cell loss.301
Progressive external ophthalmoplegia
Progressive external ophthalmoplegia (PEO), also known as chronic progressive external ophthalmoplegia (CPEO), is a prevalent clinical syndrome within mitochondrial diseases, characterized by progressive bilateral ptosis and diffuse, symmetric ophthalmoparesis.302 PEO can be classified into three phenotypes: pure PEO (isolated occurrence), Kearns-Sayre syndrome, and PEO-plus syndrome, where myopathy or other extraocular symptoms are present.303 The most common cause of PEO is a single large-scale mtDNA deletion,303 placing it within the category of mitochondrial diseases caused by mtDNA mutations.
Typically, single large-scale mtDNA deletions arise sporadically rather than through maternal inheritance, resulting from the amplification of a single mutation associated with spontaneous errors in DNA polymerase γ during early embryonic development.304,305 The age of onset and severity of the disease are correlated with the size of the deletion, the level of heteroplasmy, and the specific region affected within the mtDNA.306 These deletions, whether singular or multiple, can impair the function of one or more mtDNA-encoded proteins, resulting in OXPHOS disturbance.
PEO can also be attributed to multiple mtDNA deletions or depletion, secondary to nDNA mutations, which follow either autosomal recessive or dominant inheritance patterns and involve genes such as POLG, POLG2, SLC25A4, C10orf2, SPG7, DNA2, RNASEH1, TOP3A, TK2, DGUOK, RRM2B, GMPR, LIG3, and RRM1.307 Generally, mtDNA depletion is associated with early-onset disorders that typically have a fatal course, whereas adult-onset disorders are more commonly linked to multiple mtDNA deletions.308
The minimal mitochondrial replisome consists of polymerase γ, Twinkle, the mitochondrial single-stranded DNA-binding protein (mtSSB), and the mitochondrial RNA polymerase.309 DNA polymerase γ, the only polymerase responsible for mtDNA replication,310 comprises three subunits encoded by two nuclear genes: the p140 catalytic subunit encoded by POLG (POLG1) and the p55 accessory subunit encoded by POLG2.311 Mutations in POLG or POLG2 genes can lead to mtDNA defects. Specifically, mutations in POLG compromise mitochondrial genetic integrity, resulting in multiple deletions that contribute to PEO.310 Similarly, POLG2 mutations impair the proper stimulation of p140, disrupting mtDNA replication.312 The deletion pattern associated with POLG mutations may result from faulty strand displacement replication, initiated by replication fork stalling.313
Twinkle, encoded by C10orf2, is essential for nascent H-strand synthesis in the D-loop and is thus indispensable for mtDNA replication, despite the existence of other potential mtDNA helicases.314 A PEO mouse model with a C10orf2 defect has been established,315 demonstrating that type IIB fibers of extraocular muscles are particularly vulnerable to mtDNA deletions, likely due to their relatively low mitochondrial content, which allows even a few mutant mtDNA to surpass the onset threshold.316 The endosomal-mitophagy pathway, involving ATAD3, VPS35, SAMM50, and BAK/BAX, plays a role in mtDNA deletion caused by C10orf2 mutations.317 Furthermore, persistent activation of the mTORC1-mt-ISR pathway due to mtDNA replication defects disrupts cellular metabolic homeostasis, thereby contributing to disease progression.125 Thymidine kinase 2 (TK2) is a mitochondrial enzyme responsible for catalyzing the conversion of deoxycytidine and deoxythymidine nucleosides to their nucleoside monophosphates, which are then converted into deoxynucleoside triphosphates.318 Mutations in the TK2 gene disrupt the maintenance of the mitochondrial deoxyribonucleotide pool, leading to mtDNA depletion or multiple mtDNA deletions, and subsequently resulting in PEO.318,319
For patients with pure PEO, surgery remains the primary treatment option, whereas symptomatic treatment is recommended for extraocular symptoms in PEO-plus syndrome.302
Kearns-Sayre syndrome
Kearns-Sayre syndrome was first identified in 1958 by Kearns and Sayre, characterized by the triad of retinitis pigmentosa, PEO, and complete heart block.320 It is classified as one of the mitochondrial encephalomyopathies and presents with additional common symptoms, including cerebellar ataxia, cerebrospinal fluid protein levels exceeding 100 mg/dL, deafness, dementia, diabetes, delayed puberty, and amenorrhea.321,322 An onset before the age of 20 is a defining feature of Kearns-Sayre syndrome.234
The syndrome is predominantly caused by spontaneous single large-scale deletions of mtDNA,323,324 with the 4977 bp deletion being the most prevalent.325 Symptom manifestation requires the accumulation of mtDNA deletions beyond a pathogenic threshold, implying that the defective mtDNA must retain replication capability. This likely explains why most deletions occur within the long arc between heavy and light strands but preserve the replication sites.326 Additionally, during cellular differentiation, mtDNA deletions are preferentially replicated over wild-type mtDNA, potentially due to a kinetic advantage.327 These deletions often result in the loss of genes encoding respiratory chain proteins and tRNAs essential for translation, leading to ATP deficiency and multi-tissue dysfunction.328 For instance, the common 4977 bp deletion typically disrupts complexes I, III, IV, and mt-tRNA.323
Oxidative stress is evident in cells harboring the 4977 bp deletions.329 Respiratory chain complex defects may be independent of the deletion sites, with aberrant translation playing a key role in the pathogenesis of single large-scale mtDNA deletions.289 Increased oxidative damage and misfolded mitochondrial proteins inhibit both the ubiquitin-proteasome system and the OXPHOS system.330 Inhibition of ubiquitin-proteasome leads to decreased amino acid salvage, which triggers eIF2-α phosphorylation and induces mt-ISR. Under conditions of energy deficit and oxidative stress, genes involved in mt-ISR and autophagy are upregulated.330 Prolonged mt-ISR activation propagates and maintains mtDNA deletions, exacerbating the disease condition.331 Moreover, amino acid depletion, combined with ATP insufficiency, collectively inhibits the mTOR pathway, thereby increasing autophagy.330 The protein synthesis inhibition and autophagy increase, reducing mitochondrial contents, could be pathomechanisms of Kearns-Sayre syndrome.330
Notably, alterations in tau protein levels are observed in the cerebrospinal fluid of patients with Kearns-Sayre syndrome.332 Tau protein plays a role in ROS generation, mitochondrial dynamics, and mPTP opening,333 making its impact on Kearns-Sayre syndrome particularly intriguing. Cardiomyocytes in patients with Kearns-Sayre syndrome display increased and enlarged mitochondria.334 Arrhythmias in mitochondrial diseases are closely linked to dysfunctional ion channels, transporters, and membrane excitability caused by ATP deficiency, excessive ROS production, and Δψm collapse.335,336 Future research should focus on ionic dysregulation mediated by the mitochondrial Ca2+ uniporter complex, uncoupling proteins, and mPTP.
Furthermore, nDNA mutations in RRM2B, which encodes the ribonucleotide reductase p53R2 subunit, can lead to multiple mtDNA deletions in Kearns-Sayre syndrome through defective ribonucleotide reductase assembly.337 This defective assembly disrupts deoxynucleotide provision and the maintenance of dNTP pools.338
For patients with heart block, pacemaker implantation or implantable cardioverter defibrillators are recommended.339 In cases where heart failure develops, heart transplantation has been employed.340 The long-term safety and feasibility of human retinal progenitor cell transplantation for retinitis pigmentosa have been demonstrated.341 Gene therapy also holds promise as a potential treatment for retinitis pigmentosa associated with Kearns-Sayre syndrome.342
Pearson syndrome
Pearson syndrome, a fatal multisystem mitochondrial disorder, was first identified in 1979 by Pearson, who described it as a condition primarily affecting the bone marrow and exocrine pancreas.343 This disease is linked to defects in OXPHOS caused by sporadic single large-scale deletions (or rearrangements) of mtDNA.344,345 These deletions vary in size and location, ranging from 1.3 to 10 kb, with the size of the deletion potentially serving as a predictor for disease progression.306,346 Approximately 40–50% of patients with Pearson syndrome harbor the “common deletion,” which is 4977 bp in length.345,347 Sideroblastic anemia is typically the first and most prominent symptom of Pearson syndrome.348 Additional symptoms may include intracerebral bleeding, pancreatic exocrine insufficiency, lactic acidosis, and congenital malformations.348,349 The prognosis for Pearson syndrome is poor, with an average age of death being 5.44 years in individuals aged 0–15 and 7.41 years in those aged 0–19.350 Patients with the 4977 bp deletion have a higher mortality risk.349
Single large-scale mtDNA deletions exhibit phenotypic heterogeneity and contribute to a spectrum of diseases, including Pearson syndrome, Kearns-Sayre syndrome, and PEO.351 It is hypothesized that the timing of the mtDNA deletion during fetal development influences the clinical phenotype: late-stage deletions may result in PEO,4 while earlier-stage deletions may manifest as Kearns-Sayre syndrome or Pearson syndrome, affecting multiple systems.4 Interestingly, Pearson syndrome can evolve into Leigh syndrome or Kearns-Sayre syndrome over time,352,353 indicating that the phenotype of an mtDNA deletion disorder may change with age and is influenced by the concentration of mtDNA with deletions.354 Patients with Pearson syndrome typically have a higher proportion of mtDNA deletions compared to those with Kearns-Sayre syndrome or PEO.347,355
Spontaneous hematological recovery is observed in some Pearson syndrome cases, with a decrease in the amount of deleted mtDNA in blood cells corresponding with an improvement in anemia.356 This recovery is attributed to the positive selection of hematopoietic stem cells (HSCs), where HSCs with a higher load of deleted mtDNA are hard to survive, while those with a lower load are selected for survival.347,348,357 This concept aligns with findings from a mouse model study, which showed a decrease in the mtDNA deletion load with age in affected tissues, such as peripheral blood and liver.358 However, the study also indicated that mtDNA deletions may accumulate in muscle and other tissues, potentially leading to the development of Kearns-Sayre syndrome.358 This tissue-specific change in mtDNA deletion load partially explains the progression from Pearson syndrome to Kearns-Sayre syndrome.
In addition to ATP deficiency and oxidative stress caused by mtDNA deletions, iron accumulation may play a significant role in Pearson syndrome. The abnormal iron deposit is a feature in patients with Pearson syndrome, while the molecular mechanisms behind abnormal iron metabolism remain unclear.359 In a mouse model of large-scale mtDNA deletion, Pearson syndrome-like anemia worsened with the knockout of Drp1, and Drp1 knockout alone also caused anemia.359 Drp1 knockout decreases the pathogenic threshold of mtDNA deletion in erythrocytes,359 which drives us to think about the role of MQC in this anemia. Loss of Drp1 leads to HSC quiescence, reducing their regenerative potential.360 Interestingly, mitochondria with impaired fission ability are retained and accumulate with HSC divisions, potentially preventing unlimited self-renewal of HSCs.360 During erythropoiesis, mitochondria exhibit a specific pattern: increased fusion at early stages and heightened fission at later stages, associated with mPTP opening.361 Besides, the quiescence of HSCs is partly mediated by the regulation of mitochondrial content and activity.362 Thus, mitochondrial content and activity, which are influenced by mitophagy, are closely linked to HSC differentiation.363 A low Δψm is a key trigger for mitophagy, and there is a distinct difference in Δψm between quiescent and cycling-primed HSCs, with quiescent HSCs exhibiting low Δψm.362 Moreover, HSC quiescence is supported by an abundance of large lysosomes, and maintaining this quiescence requires restrained lysosomal activity.362 Interestingly, while suppressed mitophagy may specifically impair terminal erythrocyte maturation without affecting erythroid progenitor differentiation, hyperactivated mitophagy can hinder the differentiation of erythroid lineages.364 Thus, the fine regulation of mitophagy appears crucial during HSC differentiation, highlighting the potential importance of MQC in the anemia associated with Pearson syndrome. Further investigation is needed to determine whether and how energy and oxidative stress lead to MQC alterations, impacting anemia in Pearson syndrome.
Despite the possibility of spontaneous recovery from anemia in some patients with Pearson syndrome, others may require transfusions during infancy and early childhood.347 Hematopoietic stem cell transplantation is a potential option for those with persistent transfusion dependency or severe neutropenia.347,365 Additionally, mitochondrial augmentation therapy—where autologous CD34+ hematopoietic cells are augmented with maternally derived healthy mitochondria—has shown promising outcomes for mtDNA deletion syndromes like Pearson syndrome and Kearns-Sayre syndrome. This therapy has been observed to increase mtDNA content and improve aerobic function, suggesting it may be a viable treatment option.366
The primary mitochondrial diseases caused by nDNA mutations
Given that the majority of mitochondrial proteins are encoded by nDNA, mitochondrial diseases resulting from nDNA mutations encompass not only disturbances in OXPHOS but also defects in various structural or functional proteins essential for mtDNA maintenance, mitochondrial function and structure, and mito-nuclear communication. Consequently, the pathogenesis of mitochondrial diseases caused by nDNA mutations is inherently more complex than those arising from mtDNA mutations.
Autosomal dominant optic atrophy
Autosomal dominant optic atrophy (ADOA) is marked by progressive bilateral vision loss and color vision deficits, typically manifesting before the age of 20.231 The primary histopathological features of ADOA include the degeneration and demyelination of RGCs.367 Mutations in the OPA1 gene, which encodes a dynamin-related GTPase involved in IMM fusion, are the most common cause of ADOA.197 In many cases, haploinsufficiency is the primary pathogenic mechanism, as these mutations often result in premature translation termination.368 OPA1 mutations are typically heterozygous, as bi-allelic homozygous mutations, which lead to a complete loss of OPA1 function, are likely embryonically lethal.369,370 Although no cases of bi-allelic homozygous OPA1 mutations have been reported, bi-allelic compound heterozygous mutations have been observed and are associated with ADOA-plus (or Behr syndrome),371,372 which presents with additional multisystemic features beyond the optic neuropathy.372 Other genes involved in mitochondrial dynamics, such as OPA3, Drp1, and Mfn2, can also cause ADOA or ADOA-plus.197
The specific vulnerability of RGCs to OPA1 mutations remains unclear. However, research has shown that OPA1 mutations lead to defective differentiation and impaired mitochondrial function in RGCs, as demonstrated in human retinal organoids.373 The distinct mitochondrial morphology observed in RGCs and optic nerves in mouse models may provide some insights. In the unmyelinated segments of RGC axons, mitochondria are typically round before the lamina cribrosa, whereas they become elongated after crossing this structure in the myelinated segments, suggesting dynamic changes during mitochondrial transport.197 Elongated mitochondria are generally associated with enhanced ATP production, decreased fission, or increased fusion.374 It is hypothesized that OPA1 mutations may impair mitochondrial fusion after crossing the lamina cribrosa, leading to mitochondrial fragmentation. The clustering of fragmented mitochondria could cause traffic jams, obstructing the axonal transport of mitochondria.375 This theory aligns with observations that small RGCs with thin axons, which have limited mitochondrial transport capacity, are the first to be lost in ADOA.197 This suggests that disrupted mitochondrial transport may contribute to the RGC-specific susceptibility to OPA1 mutations. OPA1 also plays a role in intercellular mitochondria transfer.187 A significant proportion of mitochondria in RGC axons are not degraded by lysosomes within the RGC soma but are instead transferred to astrocytes at the optic nerve head for transcellular degradation.376 The potential impairment of mitochondrial transfer and transport due to OPA1 mutations should be considered as a contributing factor to the vulnerability of RGCs. Further research is needed to verify whether these mitochondrial properties specifically facilitate the susceptibility of RGCs to OPA1 mutations.
The OPA1 mutation has been shown to increase Drp1 expression, thereby promoting mitochondrial fission. Inhibiting Drp1 can help balance mitophagy and improve mitochondrial abnormalities associated with ADOA.377 Mitophagy, which can be activated by AMPK, is another critical factor; inhibiting both AMPK and mitophagy has been found to preserve mitochondrial content in RGC axons and mitigate visual loss caused by OPA1 mutations.378 Additionally, OPA1 mutations can lead to an increase in PINK1-Parkin-independent mitophagy, which may be directly driven by the excessive presence of fragmented mitochondria.375 This overactivation of mitochondrial fission and mitophagy could be central to the pathogenesis of ADOA caused by OPA1 mutations. Chronic inhibition of mitochondrial fusion due to OPA1 loss results in mtDNA depletion, exacerbating mitochondrial dysfunction,379 and excessive mitophagy further contributes to this depletion.375 The haploinsufficient OPA1 mutations not only impact mtDNA maintenance but also downregulate nuclear genes encoding mitochondrial components, implying the potential role of mito-nuclear communication in ADOA.373 Given OPA1’s role in mediating intercellular mitochondrial transfer via MDVs,187 the ability to secrete damaged mitochondria into the extracellular space might be impaired in ADOA. The dysfunctional MQC resulting from OPA1 loss reduces mitochondrial content and leads to secondary mtDNA depletion, disrupting OXPHOS.
Beyond its influence on mitochondrial fusion, OPA1 also plays a key role in preventing remodeling of cristae structure and mobilization of cytochrome c during apoptosis.380,381 Loss of OPA1 disturbs the structure and integrity of the IMM, leading to the release of cytochrome c in a which is mainly sequestered within the tight cristae junctions.380–382 Cytochrome c release is a well-known trigger of apoptosis. Excessively fragmented mitochondria can induce mtROS overproduction,144 making oxidative stress correlated with ADOA.383,384 The secondary mtDNA depletion may exacerbate OXPHOS disturbance, increasing the production of mtROS. Consequently, OPA1 mutations might facilitate stress-induced cytochrome c release by promoting the remodeling of cristae and mobilization of cytochrome c, thereby leading to RGCs degeneration.380,381,383,385
Given the critical role of mitochondrial cristae architecture in preventing mtDNA release and inflammation,299 mitochondrial inflammation in ADOA warrants significant attention. OPA1 loss has been associated with muscle inflammation due to mitochondrial dysfunction,386 even in the absence of mtDNA leakage into the cytosol. In this context, OPA1 deficiency induces NF-κB-mediated inflammation through the TLR9 pathway.386 Additionally, OPA1 defects reduce muscle mass and lead to premature death.379 The stress-induced mt-ISR, triggered by OPA1 loss, also contributes to muscle loss, and the reduction of FGF21 (a downstream hormone of mt-ISR) can mitigate this muscle wasting.379 Interestingly, while OPA1 loss impairs mitophagy in muscle cells,386 it increases mitophagy in RGCs. Understanding these tissue-specific differences in response to OPA1 mutations could help explain the phenotypic heterogeneity and tissue-specificity observed in ADOA.
Alpers-Huttenlocher syndrome
Alpers-Huttenlocher syndrome is the most prevalent mitochondrial disease caused by nDNA mutations. It is an autosomal recessive hepatocerebral syndrome with early onset,387 typically presenting with a triad of intractable seizures, developmental regression, and liver dysfunction.388 Notably, hepatic dysfunction can be aggravated by exposure to valproate, sometimes leading to a misdiagnosis of valproate hepatotoxicity.389 The disease is primarily caused by mutations in the POLG gene, such as p.A467T, p.W748S, and p.G848S, which result in mtDNA depletion.387,390 Alpers-Huttenlocher syndrome is likely the most common POLG-related disorder.391 Recessive POLG mutations can be homozygous or compound heterozygous, with compound heterozygous mutations in trans often associated with a more severe and earlier-onset phenotype, whereas homozygous recessive mutations tend to result in a milder and later-onset presentation.392 Additionally, ecogenetic structural nucleotide variants can influence the clinical phenotype.393 It’s worth noting that compound heterozygous mutations in C10orf2, which encodes the mitochondrial replicative helicase Twinkle, can also cause mtDNA depletion and present as Alpers-Huttenlocher syndrome.394 Mutations in C10orf2 disrupt mtDNA maintenance, leading to secondary mtDNA depletion and subsequent mitochondrial dysfunction.309
Another POLG-related disorder, Myocerebrohepatopathy spectrum (MCHS), presents with a triad of hypotonia, developmental delay, and hepatopathy.395 Other clinical manifestations may include renal tubulopathy, choreoathetosis, neuropathy, ataxia, and cataracts.2,396 MCHS is the earliest POLG-related disorder with mtDNA depletion, with a median onset age of 4.7 months.391,396 A case report documented the clinical progression of a child from infantile MCHS to Alpers-Huttenlocher syndrome, suggesting that MCHS and Alpers-Huttenlocher syndrome may represent different stages or severities of the same disorder.395
The histopathological features of mitochondrial encephalopathies include astrocyte activation, cortical degeneration, and neuron loss.397 In iPSC-derived neural stem cells with compound heterozygous POLG mutations, BNIP3-mediated mitophagy is increased, likely due to elevated ROS levels and a low NAD+/NADH ratio. Concurrently, decreased SIRT1 signaling and increased UCP2 signaling contribute to neuronal senescence.398 The combination of mitochondrial dysfunction and senescence leads to neuron loss in POLG-related disorders.398 Astrocytes, which play a pivotal role in supporting neurons,399 also suffer from dysfunction due to mtDNA defects, contributing to the development of mitochondrial encephalopathy. Indeed, astrocyte dysfunction has been demonstrated in patients with Alpers-Huttenlocher syndrome,400 and astrocytic neurotoxicity caused by mitochondrial dysfunction associated with POLG mutations also has been observed.401 The failure of mtDNA maintenance due to POLG mutations results in the loss of complexes I and IV in astrocytes,402 impairing their ability to proliferate and respond effectively to neuronal damage.403 POLG mutations can also disrupt mitochondrial biogenesis, as well as mitophagy, leading to astrocytic neurotoxicity and a loss of supportive functions.402 Similarly, mtDNA depletion caused by C10orf2 mutations leads to chronic astrocyte activation and dysfunction, which may produce neurotoxic factors and promote neuronal morphology changes and progressive spongiotic encephalopathy.404
Valproic acid is contraindicated in Alpers-Huttenlocher syndrome due to its hepatotoxicity, which is linked to increased apoptotic sensitivity through mPTP opening.405 In iPSC-derived hepatocytes from patients with Alpers-Huttenlocher syndrome, mitochondria exhibit reduced mtDNA content, cristae disorganization, and OXPHOS disturbances.405 However, the full extent of functional changes in hepatocytes remains unknown.
Currently, there are no effective treatments for Alpers-Huttenlocher syndrome. Symptomatic treatments, such as anti-epileptic therapy (excluding valproic acid and divalproex) and ventilation support, should be considered.393 Due to the multisystem involvement, liver transplantation alone is contraindicated.393 Supportive therapies, such as a ketogenic diet, may provide some benefit.406
Ataxia neuropathy spectrum
Ataxia Neuropathy Spectrum encompasses a group of mitochondrial disorders, including sensory ataxia, neuropathy, dysarthria, and ophthalmoplegia (SANDO), as well as mitochondrial recessive ataxia syndrome (MIRAS). SANDO was first identified as a syndrome involving multiple mtDNA deletions in muscle and peripheral nerve tissues by Fadic et al. in 1997.407 It is primarily caused by recessive POLG mutations and is considered part of the PEO-plus syndrome spectrum.408,409 While SANDO is most commonly associated with multiple mtDNA deletions due to POLG mutations, it can also be caused by mutations in the C10orf2 and RNASEH1 genes.410–413 RNase H1, which cleaves the RNA component of RNA: RNA hybrids as an endonuclease,414 plays a critical role not only in nDNA replication but also in mtDNA replication.415 In mitochondria, RNase H1 is essential for directing RNA primer formation for origin-specific initiation of mtDNA replication and for removing primers at the origin of replication to complete mtDNA replication.416,417 The absence of RNase H1 activity leads to defective mtDNA replication, resulting in linear deletions and depletion of mtDNA.418
MIRAS, an adult-onset mitochondrial disease, is characterized by clinical manifestations including ataxia, headache, axonal neuropathy, late-onset ophthalmoplegia, partial epilepsy in the occipital lobe, and a high risk of status epilepticus.410 Mutations in the POLG gene, such as W748S and A467T, have been identified in patients with MIRAS exhibiting multiple mtDNA deletions.410,419–421 Recent research has shown that POLG also plays a role in antiviral defense, and mutations in this gene can compromise antiviral tolerance, leading to epilepsy and liver disease in MIRAS and other POLG-related disorders.422
The spectrum of POLG-related disorders includes conditions such as myoclonic epilepsy myopathy sensory ataxia (MEMSA, characterized by epilepsy, myopathy, and ataxia without ophthalmoplegia), MCHS, Alpers-Huttenlocher syndrome, SANDO, MIRAS, and PEO.423 Despite being linked by common POLG mutations, these disorders differ in their onset age and the specific mtDNA defects they involve, leading to distinct clinical presentations.423 For example, MCHS typically manifests earliest, in neonates or infants, Alpers-Huttenlocher syndrome appears in infancy or childhood, and the remaining disorders are more likely to present in adolescence or adulthood.396 Early-onset and juvenile-onset POLG-related disorders generally result from biallelic pathogenic variants with autosomal recessive inheritance, whereas late-onset disorders (mainly PEO) may arise from a heterozygous POLG pathogenic variant with autosomal dominant inheritance.424
Another mitochondrial ataxia, infantile-onset spinocerebellar ataxia (IOSCA), is an autosomal recessive disorder characterized by sensory axonal neuropathy and progressive atrophy of the cerebellum, brain stem, and spinal cord.425–427 IOSCA is caused by two point mutations in the C10orf2 gene, which encodes the mitochondrial helicase Twinkle, as well as a rarer splice variant known as Twinky.428 These mutations result in a preponderance of messenger RNAs encoding Y508C polypeptides, leading to a Y508C alteration in the helicase domain of Twinkle or Twinky.428,429 IOSCA is classified as a mtDNA depletion syndrome, as patients’ brains show significant mtDNA depletion without mtDNA deletions or an increased number of mtDNA point mutations.429
The pathogenesis of POLG-related disorders has been discussed previously, but it’s important to note that different mutant genes involved in mtDNA replication may lead to distinct patterns of mtDNA deletions, varying in size and location. These variations can affect mitochondrial function differently depending on the nature of the mutations.313 Mitochondrial dysfunction in cerebellar tissue, mediated by impaired mitophagy, has been confirmed as a key factor in the development of ataxic diseases.430 Further research is needed to elucidate the molecular and functional changes in mitochondria that underlie mitochondrial ataxias.
As with most mitochondrial diseases, there are no specific treatments or cures for MIRAS, SANDO, IOSCA, and MEMSA. Treatment is primarily supportive and symptomatic, with clinical management focused on alleviating symptoms and improving quality of life.
Barth syndrome
Barth syndrome, an X-linked mitochondrial disorder, is characterized by a clinical triad of cardiomyopathy, skeletal myopathy, and neutropenia, along with aberrant cristae morphology and respiratory chain abnormalities.431 The gene responsible for Barth syndrome, TAZ (also known as G4.5), was identified as an X-linked gene encoding an acyl-specific phospholipid transacylase involved in remodeling cardiolipin acyl chains within mitochondrial membranes.432,433 In highly oxidative tissues such as the heart and skeletal muscle, tetralinoleoyl cardiolipin is the predominant form.434,435 Mutations in TAZ lead to a deficiency of tetralinoleoyl cardiolipin and an accumulation of monolysocardiolipins, which lack a linoleoyl acyl group.436,437 The elevated ratio of monolysocardiolipins to tetralinoleoyl-cardiolipin represents not only a biochemical hallmark but also the underlying molecular mechanism of Barth syndrome.438 This cardiolipin imbalance disrupts the phospholipid composition of the IMM, compromising the function of respiratory chain complexes and other mitochondrial proteins.439 Cardiolipin is essential for the stability of respiratory chain supercomplexes, which are crucial for the efficient operation of OXPHOS.88 In Barth syndrome, TAZ mutations disrupt supercomplex formation, resulting in reduced ATP production.440 Moreover, cardiolipin imbalances may interfere with the coupling of respiration to ATP synthesis, further diminishing energy production.439 The loss of respiratory chain supercomplexes also contributes to increased ROS, a phenomenon observed in Barth syndrome.441 Cardiolipin also plays a pivotal role in mitophagy, acting as a mediator in the process.442 Defective cardiolipin remodeling due to TAZ mutations hinders the initiation of mitophagy, leading to dysfunctional OXPHOS and heightened oxidative stress.443 Restoration of mitophagy through mTOR complex 1 inhibition has been shown to alleviate cardiomyopathy in Barth syndrome.444 The failure of dysfunctional mitophagy to remove and recycle mitochondria with defective components may exacerbate mitochondrial stress.
Current treatment strategies for Barth syndrome focus on managing heart failure, cardiac arrhythmias, and neutropenia.445 Physical therapy and nutritional support are also important considerations. Beyond gene or cell therapy, lipid replacement therapy aimed at restoring mitochondrial cardiolipin levels presents a promising therapeutic approach.445
Friedreich’s ataxia
Friedreich’s ataxia is the most common spinocerebellar ataxia with autosomal recessive inheritance.446 The disease is classically characterized by progressive and unremitting ataxia of the limbs and trunk, typically presenting before the age of 25.447 Additional symptoms include dysarthria, absent tendon reflexes in the lower extremities, loss of deep sensation, scoliosis, and cardiomyopathy.447 This condition is most commonly associated with a homozygous unstable guanine-adenine-adenine (GAA) trinucleotide expansion in the first intron of the Frataxin gene (X25) on chromosome.446,448 The age of onset is correlated with the number of GAA repeats.449 Some cases also result from a compound heterozygous expansion combined with a point mutation or deletion.450,451 Atypical phenotypes of Friedreich’s ataxia have also been reported.450 The GAA expansion within the intron silences the Frataxin gene, leading to reduced production of Frataxin protein and the associated disease phenotypes.452 The silencing mechanism may involve the formation of sticky DNA (a novel DNA structure) and epigenetic modifications.452,453
Frataxin is a mitochondrial protein essential for maintaining mitochondrial iron homeostasis.454 Its critical role in the assembly or transport of iron-sulfur (Fe-S) clusters means that Frataxin deficiency leads to aconitase and mitochondrial Fe-S respiratory enzyme deficiencies (respiratory chain complexes I, II, and III), resulting in mitochondrial iron accumulation.455–457 The Fe-S clusters mediate electron transfer and ROS production in complexes I, II, and III.458 Specifically, in response to Frataxin deficiency, the activation of iron-responsive element binding protein 1 (Fe-S protein) increases cellular iron uptake, with the iron being translocated into mitochondria via mitochondrial iron transporters in an attempt to compensate for impaired Fe-S cluster biogenesis.459 However, due to the Frataxin defect, mitochondrial iron cannot be effectively utilized, leading to iron accumulation and oxidation, which in turn causes severe oxidative stress and subsequent cell death.459,460
The development of Friedreich’s ataxia within the nervous system is believed to be driven by oxidative stress, iron neurotoxicity, and neuroinflammation.461 In Drosophila models, reduced Frataxin expression impairs mitochondrial transport in neuronal regions, potentially affecting axonal function.462 Other MQC processes appear robust, with increased mitochondrial turnover and dynamics observed in the hearts of Frataxin-knockout mouse models, suggesting an effort to re-establish mitochondrial energetic and redox homeostasis.463 The interplay of oxidative stress and disordered iron metabolism raises questions about the role of ferroptosis, a form of cell death linked to iron-dependent lipid peroxidation, in Friedreich’s ataxia. While ferroptosis is indeed implicated in this disease,464,465 the precise contribution of mitochondrial dysfunction to ferroptosis in Friedreich’s ataxia remains unclear.
Fatty acid oxidation disorder
Fatty acid oxidation disorder represents a spectrum of syndromes primarily caused by defects in β-oxidation, inherited in an autosomal recessive manner.466 Mutations in specific genes result in defective enzymes, such as acylcarnitine translocase, carnitine palmitoyltransferase, and medium-chain acyl-CoA dehydrogenase.467 Among these, medium-chain acyl-CoA dehydrogenase deficiency, due to mutations in the ACADM gene, is the most prevalent form of fatty acid oxidation disorder.468 Clinically, the disorder is characterized by metabolic symptoms, including non-ketotic hypoglycemia, vomiting, encephalopathy, and acidosis, as well as muscular symptoms such as rhabdomyolysis and exercise intolerance.466 The critical role of β-oxidation in energy production means that its failure results in an ATP deficit. Beyond energy stress, mitochondrial dysfunction—driven by the lipotoxicity of accumulating fatty acids and carnitine derivatives—plays a central role in disease progression.467,469 Those toxic lipids can lead to respiratory chain complex inhibition, OXPHOS uncoupling, ROS overproduction, and persistent mPTP opening, resulting in ultimate cell death.467,469
Leigh syndrome
Leigh syndrome, also known as subacute necrotizing encephalopathy, is the most common mitochondrial disorder in childhood. It typically manifests before the age of 2 years and presents with a range of symptoms including hypotonia, epilepsy, respiratory distress, neurodevelopmental delay, ataxia, ophthalmological abnormalities, and lactic acidosis.470 Over 75 genes in both nDNA and mtDNA have been identified as causes of this condition,471 most of which are involved in OXPHOS or other energy production processes.470
Complex I deficiency is the leading cause of Leigh syndrome,472 with mutations in the nuclear gene NDUFS4 being the most common cause of complex I-associated Leigh syndrome. These mutations directly impair complex I function.470 Recent research has shown that NDUFS4 mutations also disrupt direct neuronal reprogramming of proliferating astrocytes through mechanisms involving endoplasmic reticulum stress, mt-UPR, and mt-ISR.473 Besides NDUFS4, other nuclear genes contribute to Leigh syndrome by causing deficiencies in assembly factors, cofactors, or biosynthetic pathways essential for energy metabolism.474 For instance, pyruvate dehydrogenase complex deficiency, often caused by mutations in PDHA1 (which encodes the E1 alpha subunit of pyruvate dehydrogenase),475,476 is another significant contributor to the disease.477 This deficiency disrupts the TCA cycle by depleting substrates needed for OXPHOS, leading to a collapse of energy production. Mutations in nDNA that cause mtDNA depletion are also implicated in Leigh syndrome, particularly those affecting the SUCLA2 or SUCLG1 genes, which encode subunits of succinyl-CoA ligase.470 Defective succinyl-CoA ligase not only hinders the TCA cycle by blocking the conversion of succinyl-CoA but also interferes with mitochondrial nucleotide salvage pathways by disrupting interactions with mitochondrial nucleotide diphosphate kinase, resulting in both metabolic dysfunction and mtDNA depletion.478,479 Primary mtDNA mutations can also lead to Leigh syndrome, with mutations in MT-ND5 (encoding a complex I subunit) and MT-ATP6 (encoding a complex V subunit) being common culprits.476,480 These mutations universally cause a breakdown in mitochondrial energy metabolism, leading to energy failure. MQC mechanisms are also implicated in the pathogenesis of Leigh syndrome-related gene mutations.143,473,481–485
Notably, although hypoxia might reduce energy production of OXPHOS, it alleviates Leigh syndrome in disease models by reducing mtROS generation and activating hypoxia-inducible factor pathway,12 indicating the downstream pathways of genetic mitochondrial dysfunction are promising therapeutic targets.
Mitochondrial neurogastrointestinal encephalomyopathy
Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE), an autosomal recessive disorder first identified in 1978,486 is characterized by a range of clinical symptoms including cachexia, gastrointestinal dysmotility, peripheral neuropathy, ophthalmoparesis, and leukoencephalopathy.487 Mutations in the TYMP gene, which result in the loss of thymidine phosphorylase (TP) activity, are the primary cause of MNGIE.39 TP is responsible for catalyzing the reversible phosphorylation of deoxythymidine and deoxyuridine into thymine, uracil, and 2-deoxyribose 1-phosphate.39,488 The deficiency in TP activity leads to the accumulation of deoxythymidine and deoxyuridine, which disrupts the balance of mitochondrial nucleotide pools, thereby impairing mtDNA replication.489 This imbalance results in the accumulation of point mutations, multiple deletions, and depletion of mtDNA,490,491 ultimately leading to a failure in OXPHOS. The defective TP caused by TYMP mutations leads to the excessive accumulation of nucleosides in both mitochondria and lysosomes.492 The buildup of nucleosides, which possess weak alkaline properties, can alter the acidic environment of lysosomes, potentially suppressing lysosomal activity.492 Consequently, defective mitochondria are not adequately degraded and recycled, undermining the mitochondria’s ability to cope with stress, which may exacerbate mitochondrial dysfunction.
In addition to TYMP mutations, MNGIE-like phenotypes can also arise from mutations in other genes. For example, mutations in RRM2B can cause MNGIE-like symptoms by disrupting the docking interface of the ribonucleoside reductase small subunit homodimer, thereby impairing ribonucleoside reductase activity and damaging the mitochondrial nucleotide pool, which leads to mtDNA depletion.493 Mutations in LIG3, the only mtDNA ligase essential for mtDNA replication and repair, also affect mtDNA maintenance and have been reported in patients with MNGIE, leading to mtDNA depletion.494
Current treatment options for MNGIE include hemodialysis and peritoneal dialysis, enzyme replacement therapy, orthotopic liver transplantation, hematopoietic stem cell transplantation (HSCT), celiac plexus neurolysis, and splenic nerve blockage.495 However, while HSCT can restore biochemical homeostasis, it often fails to alleviate gastrointestinal symptoms.496 Encouragingly, gene therapy has shown efficacy in MNGIE murine models,497,498 and its potential effectiveness in human patients is highly anticipated.
Myopathy, lactic acidosis and sideroblastic anemia
Myopathy, lactic acidosis, and sideroblastic anemia (MLASA) is an autosomal recessive mitochondrial disorder affecting skeletal muscle and bone marrow, characterized by mitochondrial myopathy, lactic acidosis, and sideroblastic anemia.499,500 The condition is primarily caused by mutations in the PUS1 gene, which encodes pseudouridine synthase 1.500 PUS1 mutations result in a defective PUS1p, the catalytic center of pseudouridine synthase 1, which is responsible for pseudouridylating mt-tRNAs.500 In patients with MLASA, the loss of pseudouridylation at tRNA sites typically modified by PUS1p has been observed.501 Recent research using MLASA patient-specific iPSCs and mouse models has shown that PUS1p defects lead to a reduction in mt-tRNA due to the loss of pseudouridylation, which subsequently causes abnormal mitochondrial translation.502,503 Moreover, the pseudouridylation of mt-tRNA-derived fragments is also affected by PUS1 mutations, further contributing to defective mitochondrial protein synthesis.504 This mitochondrial dysfunction, stemming from abnormal mitochondrial proteins, disrupts erythropoiesis, leading to anemia.502,503 Another mutation associated with MLASA involves the YARS2 gene, which encodes mitochondrial tyrosyl-tRNA synthetase, an enzyme that catalyzes the covalent linkage of tyrosine to its corresponding tRNA.505 Mutations in YARS2 reduce the aminoacylation activity of this enzyme, leading to faulty translation of OXPHOS subunits, particularly complexes I and IV.506 Both PUS1 and YARS2 mutations ultimately result in the collapse of OXPHOS.
Sengers syndrome
Sengers syndrome is another autosomal recessive mitochondrial disease, characterized by hypertrophic cardiomyopathy, myopathy, lactic acidosis, and congenital cataracts.507 Mutations in the AGK gene, located in nDNA, have been identified as the cause of this syndrome.508 The AGK gene encodes mitochondrial acylglycerol kinase (AGK), a multi-substrate lipid kinase that phosphorylates monoacylglycerol and diacylglycerol to produce lysophosphatidic acid and phosphatidic acid, thus playing a role in phospholipid synthesis and various signaling pathways.508 AGK is also involved in the synthesis of IMM-specific cardiolipin.509 Beyond lipid metabolism, AGK acts as a subunit of the TIM22 complex, promoting the import of mitochondrial carrier proteins independently of its kinase activity.510,511 AGK is also thought to interact with complex I of the respiratory chain, a function that appears to be disrupted by AGK deficiency rather than mutation, leading to complex I dysfunction.512 This interaction requires further validation. The pathogenesis of Sengers syndrome likely involves disruptions in mitochondrial membrane phospholipid metabolism and the protein import machinery, with further studies needed to elucidate the detailed molecular mechanisms.
Perrault Syndrome
Perrault syndrome is an autosomal recessive disorder characterized by sensorineural hearing loss and ovarian dysgenesis, though other neurological symptoms may also be present.513 The syndrome is associated with mutations in several genes, including CLPP, ERAL1, HARS2, LARS2, and C10orf2.514 The CLPP protein is an endopeptidase component of a mitochondrial ATP-dependent proteolytic complex involved in the degradation of defective proteins through mt-UPR.515 The functional CLPP tetradecamer interacts with the hexameric caseinolytic peptidase X to proteolyze specific protein substrates.516 Mutations in CLPP that reduce its proteolytic activity or disrupt its interaction with caseinolytic peptidase X lead to a breakdown in mitochondrial protein quality control, resulting in mitochondrial dysfunction.517 CLPP plays a critical role in the turnover of complex I, providing protection against mtROS overproduction and related damage.518 This protective function of CLPP may serve as an important supplementary mechanism to mitophagy in clearing ROS-damaged mitochondria.519 However, the role of CLPP in mt-UPR in mammals is debated.520 Some studies suggest that CLPP loss may even alleviate mitochondrial diseases caused by defective DARS2, the mitochondrial aspartyl tRNA synthase.520,521 CLPP also participates in other aspects of mitochondrial metabolism, though its precise role in Perrault syndrome remains unclear.522
HARS2 and LARS2 encode mitochondrial histidyl tRNA synthetase and leucyl tRNA synthetase, respectively, which are responsible for catalyzing the covalent attachment of histidine and leucine to their corresponding tRNAs. The activity of these aminoacyl tRNA synthetases is essential for mtDNA translation.523,524 Mutations in HARS2 or LARS2 reduce the aminoacylation activity of these tRNA synthetases, resulting in impaired mitochondrial translation and subsequent mitochondrial dysfunction.523,524 ERAL1 binds to mitochondrial 12S rRNA as a chaperone and is essential for the assembly of the small 28S subunit of the mitochondrial ribosome.525 Mutations in ERAL1 impair RNA processing and mitochondrial translation.526 In other words, the collapse of mitochondrial protein homeostasis appears to be the predominant mechanism in Perrault syndrome.514
Symptomatic treatments for Perrault syndrome include cochlear implantation for hearing loss and estrogen replacement therapy. For women desiring pregnancy, options such as in vitro fertilization and oocyte cryopreservation may be considered.514
Age-related diseases
Beyond mitochondrial diseases directly caused by hereditary mtDNA and nDNA mutations, aging significantly contributes to the gradual decline in mitochondrial function and the efficiency of oxidative phosphorylation, resulting in decreased ATP production and elevated ROS generation.527 This mitochondrial dysfunction is intricately linked to the onset and progression of various age-related diseases, particularly neurodegenerative disorders such as Alzheimer’s disease (AD) and Parkinson’s disease (PD).528,529 Pathogenic factors, including mitochondrial genome defects, increased oxidative stress, disrupted MQC, impaired mitochondrial proteostasis, and neuroinflammation, are central to the development and progression of these neurodegenerative disorders.529,530 Targeted mitochondrial therapies show promise as potential treatments for these conditions.529 Furthermore, MDPs and mtDNA single nucleotide polymorphism within coding regions are strongly associated with age-related diseases.78,531 For example, humanin has been found to prevent synaptic loss and reduce inflammation, offering therapeutic potential in AD.78 Similarly, SHLP2 and its variants can mitigate mitochondrial dysfunction and protect dopaminergic neurons, thereby lowering the risk of PD.531 Additionally, dysregulation of mitochondrial microRNAs is implicated in mitochondrial dysfunction and is associated with neurodegenerative diseases.532
Damage to mitochondrial antioxidant enzymes, combined with excessive ROS, triggers oxidative stress that leads to mtDNA point mutations and deletions accumulating over time. These genetic alterations disturb MQC, and ultimately result in energy depletion, oxidative damage, and apoptosis.527,529,532 Moreover, ROS accelerates the accumulation of oxidative byproducts through mitochondrial proteases and the mt-UPR.527 Released mtDNA, fragmented mitochondria, and other substances from dying neurons can initiate inflammatory responses, further driving the progression of neurodegenerative diseases.209 Elevated ROS levels, along with mtDNA polymerase mutations that cause replication errors, contribute to mtDNA mutations, multiple deletions, and reduced copy numbers, all of which accumulate with age, leading to mitochondrial dysfunction and cell death.209,530 Notably, AD is associated with mtDNA mutations and reduced copy numbers, while PD is closely linked to mtDNA deletions.530,533
In AD, neuronal damage and dysfunction are closely associated with increased oxidative stress due to amyloid-β accumulation and tau aggregation, as well as impaired mitochondrial bioenergetics and MQC networks.532,533 Mitochondrial and autophagic dysfunctions also contribute to microglial activation and neuroinflammation, further advancing AD pathogenesis.534 Similarly, PD is characterized by increased oxidative stress, abnormal mitochondrial dynamics, impaired biogenesis, and autophagy defects.535,536 These dysfunctions are primarily associated with mutations in the genes of certain proteins, such as α-synuclein, Parkin, and PINK1.3,529 Accumulation of α-synuclein in mitochondria can lead to the formation of oligomers that interact with mitochondrial membranes, inhibiting complex I activity and causing excessive mtROS production, which induces neuronal apoptosis.3,529 Mutations in PINK1 or Parkin disrupt PINK1-Parkin-dependent mitophagy, leading to the accumulation of defective mitochondria.533,536 Moreover, PD-related neurotoxins and mutations can induce mitochondrial fission, exacerbating neuroinflammation.537 The examples of AD and PD illustrate the multifaceted pathological roles that mitochondria play in age-related diseases.
In addition to genetic factors and aging, several other elements contribute to the progression of neurodegenerative diseases, including lifestyle choices and environmental exposures. Genomic instability, telomere attrition, and epigenetic modifications may all increase disease susceptibility.528 Moreover, factors such as dysregulated nutrient sensing, stem cell exhaustion, altered intercellular communication, and immune dysfunction also play significant roles in the development of these diseases.528 Future research should focus on further exploring these factors and their interactions to enhance understanding of the underlying pathogenic mechanisms.
It is evident that age-related diseases are intricately linked to mitochondrial dysfunction associated with aging. Their pathogenesis typically results from the long-term accumulation of factors such as pathological protein aggregation and disrupted MQC. In contrast, inherited mitochondrial diseases arise from specific mutations in mtDNA or nDNA that directly impair mitochondrial function, often leading to significant clinical symptoms in infancy or adolescence. Although both types of diseases share mitochondrial dysfunction as a common feature, they differ in their pathogenesis, age of onset, and clinical phenotypes.
Overall, mitochondrial diseases are increasingly recognized as pathway-based disorders rather than merely energy-related conditions.7 Numerous biological processes, such as autophagy, are energy-dependent, and severe energy stress can suppress these processes.538 Consequently, a genetic deficit in ATP can induce cellular dysfunction. ATP deficit can also serve as a signal to trigger downstream pathways such as AMPK activation, resulting in alterations of mitochondrial function.330 The pathological consequences of the downstream pathways activated by ATP shortage may be of importance for disease development, despite the currently incomplete understanding. As previously discussed, the overproduction of mtROS plays a critical signaling role in triggering MQC, apoptosis, and inflammation. Moreover, elevated mtROS levels can exacerbate the maintenance of mtDNA.539 Oxidative stress may be a common contributing factor in mitochondrial diseases. Additionally, the maintenance of a normal Δψm is essential for ATP synthesis, mitochondrial protein and ion transport, and mito-nuclear communication signaling.83 For example, reduction in Δψm disturbs mitochondrial calcium homeostasis, which may be a pathomechanism in mitochondrial diseases.11 Certain MQC processes, such as IMM fusion, also rely on a healthy Δψm.300 A reduction in Δψm is a key initiator of mt-ISR and mitophagy; however, chronic activation of mt-ISR or mitophagy may accelerate disease progression.540 Thus, the collapse of Δψm can further deteriorate cellular functions. Changes in MQC are observed in mitochondrial diseases. Although these processes are intended to help affected cells resist stress, pathological and chronic activation of them can aggravate mitochondrial dysfunction and promote disease progression.125,248 Despite the significant attention given to MQC in mitochondrial diseases, the underlying molecular mechanisms behind these changes remain largely unknown. The roles of mitochondrial apoptosis and inflammation in these diseases are also not fully understood now. Oxidative stress has been emphasized in the fields.10 Given their established influence in age-related diseases, the potential contributions of mitochondrial apoptosis and inflammation to primary mitochondrial diseases warrant close investigation.
Under OXPHOS defects or mitochondrial stress, the mito-nuclear communication pathways like mt-ISR will be activated. These activations could be significant in those mitochondrial diseases caused by mutations in mt-tRNAs due to defects of mitochondrial protein synthesis.9 Notably, research by Burr et al. has demonstrated cell lineage-specific mitochondrial resilience to mutations during mammalian organ development.541 The specific modes of mito-nuclear communication in response to genetic mitochondrial dysfunction, which determine tissue-specific vulnerability to mtDNA defects, are independent of mt-UPR and mt-ISR.541 Different triggers and degrees of mt-ISR activation in response to identical mutational loads in various cell types may also contribute to this specificity,540 potentially in the later phases.541 A deeper exploration of mito-nuclear communication is essential for understanding the pathogenesis and heterogeneity underlying mitochondrial diseases. In addition to the roles of mt-UPR and mt-ISR in facilitating mito-nuclear communication to manage cellular stress,542 the role of epigenetics, an integral component of mito-nuclear communication, is gaining increasing attention.543 Future research should investigate the pathological role of epigenetic modifications, such as lactylation, particularly given that elevated lactate levels are a common characteristic of mitochondrial diseases.544
While various phenotypic syndromes and their associated gene mutations have been discussed, the genetic and phenotypic heterogeneity of mitochondrial diseases is notably complex. The relationship between common pathogenic variants and the phenotypic syndromes of mitochondrial diseases is summarized in Table 2.
Table 2.
The common mutant genes of mitochondrial phenotypic syndrome
| Phenotype | Mutant gene | Protein | Function/role | Reference |
|---|---|---|---|---|
| Alpers-Huttenlocher syndrome | POLG | MtDNA polymerase gamma | MtDNA replication maintenance | 390,392 |
| ADOA | OPA1 | OPA1 | Mitochondrial dynamics and homeostasis | 903,904 |
| Barth syndrome | TAZ | Phospholipid transacylase | Mitochondrial membrane phospholipid maintenance | 432 |
| Friedreich’s ataxia | Frataxin | Frataxin | Mitochondrial iron-sulphur cluster synthesis | 446,448 |
| Fatty acid oxidation disorders | nDNA, for example, ACADM | Enzymes involved in fatty acid oxidation | Mitochondrial fatty acid oxidation | 468 |
| IOSCA | C10orf2 | Twinkle or Twinky | MtDNA replication maintenance | 428 |
| Kearns-Sayre Syndrome | Single large-scale mtDNA deletions | OXPHOS subunits, mitochondrial tRNA and rRNA | Oxidative phosphorylation and mitochondrial translation maintenance | 323 |
| Leigh syndrome | nDNA and mtDNA such as, NDUFS4, MT-ATP6, and PDHA1 | Subunits of complex I ~ V, pyruvate dehydrogenase, and other enzymes in energy metabolism | Oxidative phosphorylation and the broader process of energy production | 470,474 |
| LHON | mtDNA, for example, m.3460 G > A, m.11778 G > A, m.14484 T > C | Subunits of complex I | Oxidative phosphorylation | 238 |
| MELAS | MT-TL1 | tRNALeu(UUR) | Mitochondrial translation maintenance | 255,256 |
| MERRF | MT‑TK | tRNALys(UUR) | Mitochondrial translation maintenance | 278 |
| MIDD | MT-TL1 | tRNALeu(UUR) | Mitochondrial translation maintenance | 267 |
| MLASA | YARS2 | Mitochondrial tyrosyl-tRNA synthetase | Mitochondrial translation maintenance | 505 |
| PUS1 | Pseudouridine synthase 1 | Mitochondrial translation maintenance | 500 | |
| MIRAS | POLG | MtDNA polymerase gamma | MtDNA replication maintenance | 410 |
| MNGIE | TYMP | Thymidine phosphorylase | Mitochondrial nucleotide pools maintenance | 39 |
| MCHS | POLG | MtDNA polymerase gamma | MtDNA replication maintenance | 905 |
| MEMSA | POLG | MtDNA polymerase gamma | MtDNA replication maintenance | 905 |
| NARP | MT-ATP6 | Subunits of complex V | Oxidative phosphorylation | 291,292 |
| PEO | Single large-scale mtDNA deletions | OXPHOS subunits, mitochondrial tRNA and rRNA | Oxidative phosphorylation and mitochondrial translation maintenance | 303 |
| nDNA, for example, POLG, PLOG2, C10orf2 | Proteins involved in mtDNA maintenance | MtDNA replication maintenance, mitochondrial nucleotide pools maintenance, mitochondrial dynamics | 302 | |
| Pearson syndrome | Single large-scale mtDNA deletions | OXPHOS subunits, mitochondrial tRNA and rRNA | Oxidative phosphorylation and mitochondrial translation maintenance | 345 |
| Perrault syndrome | ERAL | ERAL | Mitochondrial RNA processing and translation maintenance | 526 |
| HARS2, LARS2 | Aminoacyl tRNA synthetase | Mitochondrial translation maintenance | 523,524 | |
| CLPP | Caseinolytic protease P | Mitochondrial protein quality control | 516 | |
| Sengers syndrome | AGK | Mitochondrial acylglycerol kinase | Mitochondrial membrane phospholipids metabolism and protein import machinery | 508 |
| SANDO | POLG | MtDNA polymerase gamma | MtDNA replication maintenance | 408 |
| C10orf2 | Twinkle | MtDNA replication maintenance | 412 |
mtDNA mitochondrial DNA, nDNA nuclear DNA, PEO progressive external ophthalmoplegia, MNGIE mitochondrial neurogastrointestinal encephalomyopathy, SANDO sensory ataxia, neuropathy, dysarthria, and ophthalmoplegia, MIRAS mitochondrial recessive ataxia syndrome, MEMSA myoclonic epilepsy myopathy sensory ataxia, MCHS myocerebrohepatopathy spectrum, IOSCA infantile-onset spinocerebellar ataxia, NARP neurogenic muscle weakness, ataxia and retinitis pigmentosa, LHON Leber hereditary optic neuropathy, ADOA autosomal dominant optic atrophy, MELAS mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes, MERRF myoclonic epilepsy with ragged red fibers, MIDD maternally inherited diabetes and deafness, MLASA myopathy, lactic acidosis and sideroblastic anemia
Diagnostic methodology of mitochondrial diseases
Given the genetic and phenotypic heterogeneity of mitochondrial genetic disorders, accurately diagnosing these diseases remains a significant challenge.545 Beyond careful and detailed clinical observation, comprehensive testing is essential, including biochemical analyses of body fluids, neuroimaging, DNA and RNA sequencing, as well as biochemical or pathological testing of tissues (Fig. 6). When a mitochondrial disease is suspected, initial biochemical testing should be performed on blood, urine, and cerebrospinal fluid.545 For instance, a blood count can reveal anemia or neutropenia, which are critical indicators in certain mitochondrial genetic disorders, such as Pearson syndrome.343
Fig. 6.
Diagnostic Methodology for Mitochondrial Diseases. Diagnostic strategies have evolved from a biopsy-first approach to a genetic-first approach. Initial screenings should utilize blood, urine, and cerebrospinal fluid samples. Biomarker testing is essential in this process. For suspected mitochondrial diseases, mtDNA sequencing and analysis are the preferred methods, while nDNA sequencing should be considered in cases of mtDNA multiple deletions, depletion, or early-onset symptoms. RNA sequencing (transcriptomics) and respirometry also contribute to accurate diagnosis. Biopsy specimens, typically obtained from muscle or skin, remain valuable for confirming tissue-specific mtDNA mutations that may not be detected in blood or urine samples. Histopathological examination and respiratory chain enzyme analysis can be applied to these tissue samples, revealing abnormal mitochondrial structure, morphology, and function. Thus, biopsy retains significant diagnostic value. mtDNA mitochondrial DNA; nDNA nuclear DNA; ccf-mtDNA circulating cell-free mitochondrial DNA; Δψm mitochondrial membrane potential; SDH succinate dehydrogenase; COX cytochrome c oxidase; NADH-TR nicotinamide adenine dinucleotide tetrazolium reductase
Biomarkers
Certain biomarkers for mitochondrial diseases are garnering increasing attention due to their potential diagnostic value. The rise in anaerobic glycolysis, a compensatory response to energy shortages caused by OXPHOS dysfunction, leads to lactate accumulation, making lactic acidemia or lactic acidosis a common feature of these diseases.544 Since lactate is produced from pyruvate by lactate dehydrogenase, elevations in lactate are typically paralleled by increases in pyruvate.546 While lactate elevation exhibits high specificity (83–100%) among patients with mitochondrial diseases, its sensitivity is relatively low (34–62%), compared to pyruvate elevation, which shows a sensitivity of 75% and a specificity of 87.2%.547,548 The balance between lactate and pyruvate is regulated by the cytosolic NAD+/NADH ratio.549 Impaired oxidation of NADH, a function of respiratory complex I, increases NADH levels, driving the equilibrium toward lactate accumulation, which differs from the proportional increase in lactate and pyruvate seen in pyruvate dehydrogenase deficiency.549 Therefore, the lactate/pyruvate ratio offers excellent diagnostic accuracy for distinguishing pyruvate dehydrogenase deficiency from other mitochondrial diseases.550 Additionally, transaminase testing is essential as hepatopathy can be an early sign of mitochondrial diseases like Alpers-Huttenlocher syndrome.394
OXPHOS function measured in blood cells is emerging as a potential biomarker, with an estimated sensitivity of 68.3%, directly reflecting OXPHOS defects.546 The dysfunction of OXPHOS also leads to long-term alterations in Δψm as respiratory chain complexes fail to transfer protons.83 Notably, significant changes in Δψm were consistently observed in blood cell tests, suggesting that Δψm quantification could be a superior diagnostic method.546 Although less sensitive, biomarkers like creatine, creatine kinase, free carnitine, and acylcarnitine in the blood can aid in diagnosing mitochondrial diseases caused by fatty acid β-oxidation disorders.546,547 Additionally, amino acid analysis of both plasma and cerebrospinal fluid, along with organic acid analysis of urine, is valuable for diagnosing specific mitochondrial metabolic disorders, although patient selection prior to testing is recommended.551 For instance, an elevated monolysocardiolipins/tetralinoleoyl-cardiolipin ratio in blood and increased 3-methylglutaconic acid in urine (a precursor to cardiolipin) are strong biochemical indicators of Barth syndrome.438 Notably, two cytokines involved in the mt-ISR, FGF-21 and GDF-15, have shown exceptional diagnostic value for mitochondrial myopathy and tRNA gene mutation-related mitochondrial diseases, and they may also serve as biomarkers for monitoring therapeutic efficacy.548,552,553
The significance of extracellular mitochondrial content has been highlighted by Miliotis et al.554. Recent studies suggest that mitoEVs are involved in encephalopathy associated with mitochondrial disorders, potentially serving as indicators of mitochondrial encephalopathy.555 MitoEVs containing mitochondria or mtDNA are emerging as promising biomarkers,554,556 as their release is part of the process that recycles or eliminates nonfunctional mitochondrial fragments.184 Changes in ncRNAs, which regulate mitochondrial protein expression and mitochondrial function-related signaling pathways, can reflect metabolic and functional alterations in mitochondria.557 MicroRNAs, a specific type of ncRNA, play a role in the pathogenesis of mitochondrial diseases and could aid in diagnosis. For example, oxidative stress-induced microRNA-9/9* has been implicated in the MELAS phenotype, while downregulation of microRNA-181a/b, which promotes MQC through the activation of mitochondrial biogenesis and mitophagy, has shown protective effects on RGCs in an LHON mouse model.558 Further studies on microRNA-181a/b downregulation in the treatment of inherited retinal diseases underscore the importance of microRNAs.559 Additionally, microRNA-27b-3p has demonstrated significant diagnostic value in patients with MELAS.560 However, these biomarker changes are not exclusive to mitochondrial diseases and can also be observed in other conditions,554,561 necessitating further research into their sensitivity and specificity.
Furthermore, under conditions of stress, apoptosis, or necrosis, damaged mtDNA can be released from cells as cell-free circulating mtDNA (ccf-mtDNA).562 The accumulation of damaged mtDNA contributes to the increase of ccf-mtDNA in plasma, making it a potential biomarker for mtDNA genetic disorders, particularly MELAS, with a sensitivity of 44% and specificity of 94%.554
Given the profound impact of metabolic and functional disorders on genetic mitochondrial diseases, the fields of proteomics, lipidomics, and metabolomics are being explored for their potential applications.563–567 While their diagnostic value is not yet fully established, these approaches hold great promise for the future.
DNA and RNA sequencing
Given that mtDNA mutations are the primary cause of genetic mitochondrial diseases, mtDNA testing should be prioritized in the diagnostic process. The advent of NGS has significantly shifted the diagnostic approach from a biopsy-first strategy to a genetic-first strategy.568,569 With the development and widespread application of NGS, the genetic diagnostic yield for these disorders has increased from 10–20% in the pre-NGS era to 40–60% today.41,568 However, the diagnostic yield of NGS in suspected mitochondrial disease cases varies, with reports indicating a range of 7% to 70%.227 Recently, the United Kingdom introduced new practice guidelines for the genetic diagnosis of mitochondrial diseases, aiming to standardize and guide the use of DNA sequencing.569
Comprehensive mtDNA testing should include sequencing the entire mtDNA genome, analyzing mtDNA deletions, assessing mtDNA copy number, and determining the heteroplasmy levels of mtDNA mutations.7,547,551 NGS has become the first-line methodology for mtDNA testing.547 Various laboratory techniques, such as polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), allele-specific oligonucleotide polymerase chain reaction (ASO-PCR), single-strand conformation polymorphism (SSCP), long-range PCR, and Southern blot, have traditionally been used to screen for point mutations and deletions in mtDNA.551 For analyzing mtDNA copy number, which often reflects mtDNA depletion, real-time quantitative PCR (RT-qPCR) is commonly used.551 Assessing the heteroplasmy level of mutant mtDNA is also necessary, as it may correlate with disease severity and progression; this is typically done using methods like pyrosequencing or PCR,546 though these approaches are increasingly being replaced by NGS.570 A promising approach combines long-range PCR-based enrichment with NGS, enabling both quantitative and qualitative detection of every base in the entire mitochondrial genome.570 This method can detect heteroplasmy levels as low as 1–10%, with improved sensitivity due to specific mtDNA enrichment strategies and NGS platforms.7 However, it is essential to distinguish these findings from normal tissue states, which may carry low-level heteroplasmy.228 Although blood is the most commonly used sample for mtDNA testing, it is also advisable to assess mtDNA in urine and other affected tissues due to the possibility of tissue-specific mutations.547
As discussed, multiple mtDNA deletion and depletion are often caused by nDNA mutations. Consequently, 75–90% of pediatric mitochondrial diseases are attributed to nDNA mutations, making nDNA sequencing particularly important for diagnosing childhood-onset mitochondrial diseases.551 NGS of nuclear genes involved in mtDNA maintenance, mitochondrial function, and metabolism using targeted gene panels and whole exome sequencing is a preferred approach.571 Other sequencing methods also play vital roles; for example, Sanger sequencing remains a valuable first-line test for identifying common mutant genes in certain populations.569
Emerging RNA sequencing, or transcriptomics, has recently shown promise for diagnosing mitochondrial diseases.571 This technique serves as an essential complement to genome sequencing and can help diagnose suspected mitochondrial diseases that remain genetically undetermined after genome sequencing.572 RNA sequencing methods can be divided into short-read cDNA sequencing, long-read cDNA sequencing, and direct RNA sequencing.573 RNA sequencing provides insights into aberrant splicing and altered transcript levels due to abnormal gene expression or mono-allelic expression.571 For instance, RNA sequencing has detected a splicing variant in the CLPP gene associated with Perrault syndrome and mono-allelic expressed variants in the ALDH18A1 gene, which encodes an enzyme involved in mitochondrial proline metabolism, linked to cutis laxa III.572 RNA sequencing also has identified splice site mutations in mitochondrial diseases.574,575 Furthermore, tRNA sequencing can uncover defective tRNAs and reduced tRNA levels, which are implicated in many mitochondrial diseases.571 For example, defective N1-methyladenosine (m1A) modification in mt-tRNALys and mtDNA mutation-caused tRNAAla reduction, discovered through tRNA sequencing, have provided valuable insights into the pathogenesis of mitochondrial diseases.69,280
Biopsy
A tissue biopsy is widely regarded as the diagnostic gold standard for mitochondrial diseases, particularly in cases where genetic testing has not provided a definitive diagnosis.547 Detecting OXPHOS or mtDNA defects in biopsy specimens provides compelling evidence for the presence of mitochondrial disease. Muscle tissue is often preferred for biopsy due to its high energy demand, making it particularly susceptible to OXPHOS dysfunction.549 Skin biopsies, which allow for the analysis of fibroblasts, are also a viable alternative.576 Genetic, histopathological, and biochemical evaluations of biopsy specimens are crucial in this diagnostic process.
Testing mtDNA in biopsy specimens offers greater sensitivity in detecting low-level mtDNA heteroplasmy and assessing mtDNA copy number compared to blood samples.547 Histological stains such as Haematoxylin and eosin (H&E) and modified Gomori trichrome are used to examine mitochondrial structure and morphology. The modified Gomori trichrome stain, in particular, can reveal ragged-red fibers, which indicate abnormal mitochondrial proliferation in response to energy failure, and can also highlight cytochrome c oxidase (COX, respiratory complex IV) deficiency.577,578 Sequential staining with succinate dehydrogenase (SDH, respiratory chain complex II) and COX provides additional insights into the activity of these complexes.549 Since SDH is entirely encoded by nuclear DNA, while mtDNA encodes three subunits of COX, COX activity is a more direct reflection of mtDNA maintenance.579 In normal tissue, SDH staining appears as ragged-blue fibers.580,581 Due to the uneven distribution of mtDNA mutations along muscle fibers, COX staining often shows a mosaic pattern of COX-negative and COX-positive fibers in cross-sections.579 In combined COX and SDH staining, the COX activity is typically indicated by a brown stain that overshadows the blue SDH stain. However, reduced COX activity allows the SDH stain to become more visible, presenting as blue fibers.579 This staining combination provides two key insights: a mosaic pattern of SDH-positive but COX-negative fibers indicates an mtDNA defect involving COX, while a mosaic pattern of both SDH-positive and COX-positive fibers suggests an mtDNA defect involving NADH dehydrogenase (respiratory complex I) or cytochrome c reductase (respiratory complex III).579 To specifically detect NADH dehydrogenase activity, nicotinamide adenine dinucleotide tetrazolium reductase (NADH-TR) staining should be employed.582 A reduction in NADH-dehydrogenase activity results in decreased or absent blue-purple color in NADH-TR staining, with an increase in stained ragged blue fibers in the subsarcolemmal region.549
In addition to assessing OXPHOS function in blood, as discussed earlier, the biochemical analysis of mitochondrial respiratory chain enzymes, isolated from muscle tissue or cultured fibroblasts, can directly reflect OXPHOS defects.583,584 CoQ10 assessment is also important, given its critical role in electron transport within OXPHOS.585 Spectrophotometric or colorimetric assays are commonly used to measure enzymatic activity.584,586 Successful enzymological analysis requires the use of internal controls, typically normalized to SDH activity or mitochondrial citrate synthase levels.547,586
Electron microscopy offers detailed visualization of mitochondrial quantity, inclusions, and ultrastructural abnormalities, demonstrating its diagnostic potential for mitochondrial nephropathy, cardiomyopathy, and hepatopathy.587–589
Respirometry
High-resolution respirometry is a valuable methodology for assessing mitochondrial respiration, specifically OXPHOS function.590 This technique can be applied to various subjects, including isolated mitochondria, intact cells, and three-dimensional systems such as tissue slices, and it is typically measured using two primary setups: chamber-based platinum electrodes and microplate-based fluorescent readings.591 The substrate-uncoupler-inhibitor titration (SUIT) protocol is particularly effective in this context, as it allows for the measurement of oxygen consumption (flux) by specific respiratory complexes, thereby reflecting enzyme activity.590,592,593 By calculating flux control ratios—ratios of oxygen flux under different respiratory control conditions—the SUIT protocol enables internal normalization, facilitating comparisons across different studies.593 This method has shown promise in the diagnosis of mitochondrial diseases in several studies.594–596
However, there are no perfect diagnostic methodologies. Diagnosing atypical and novel genetic mitochondrial disorders requires a thoughtful combination of these various diagnostic approaches. Moreover, the importance of integrating clinical observations with laboratory examinations cannot be overstated, as this connection is crucial for accurate diagnosis.
Potential therapeutic strategies of mitochondria in genetic disorders
Effective treatments for most mitochondrial diseases remain elusive. However, mitochondrial replacement therapy (MRT) and gene therapy represent promising foundational approaches for treating mitochondrial genetic disorders and preventing their transmission to future generations. Additionally, ongoing research is exploring the restoration of post-transcriptional tRNA modifications in mitochondria and the development of targeted therapies for specific conditions. Below, we provide a detailed summary of potential therapeutic strategies and discuss their possible clinical applications. (Fig. 7, Fig. 8, Table 3).
Fig. 7.
Procedure for Mitochondrial Replacement Therapy. a The procedure for pronuclear transfer (PNT) involves extracting the pronucleus from a zygote of a healthy donor with wild-type mtDNA and from a patient with defective mtDNA. The cytoplast from the patient and the pronucleus from the healthy donor are then removed. Finally, the pronucleus from the patient and the cytoplast containing wild-type mtDNA from the healthy donor are fused, reconstructing a zygote with the patient’s pronucleus and wild-type mtDNA. b The procedure for spindle-chromosome complex transfer (ST) involves extracting the spindle-chromosome complex from a metaphase II oocyte of a healthy donor with wild-type mtDNA and from a patient with defective mtDNA. The cytoplast from the patient and the spindle-chromosome complex from the healthy donor are removed. Finally, the spindle-chromosome complex from the patient and the cytoplast containing wild-type mtDNA from the healthy donor are fused, reconstructing an oocyte with the patient’s spindle-chromosome complex and wild-type mtDNA. c The first polar body transfer (PB1T) procedure involves extracting the spindle-chromosome complex from a metaphase II oocyte of a healthy donor with wild-type mtDNA and the first polar body from a metaphase II oocyte of a patient with defective mtDNA. The cytoplast from the patient and the spindle-chromosome complex from the healthy donor are then removed. Finally, the first polar body from the patient and the cytoplast containing wild-type mtDNA from the healthy donor are fused, reconstructing an oocyte with the patient’s first polar body and wild-type mtDNA. d The second polar body transfer (PB2T) procedure involves extracting the female pronucleus from a zygote of a healthy donor with wild-type mtDNA and the second polar body from a zygote of a patient with defective mtDNA. The zygote from the patient and the female pronucleus from the healthy donor are then removed. Finally, the second polar body from the patient and the zygote containing wild-type mtDNA and the male pronucleus from the healthy donor are fused, reconstructing a zygote with the patient’s second polar body and wild-type mtDNA
Fig. 8.
Gene Therapy and Post-Transcriptional Modification Strategies for Mitochondrial Genetic Disorders. a MitoTALENs consist of TALE fused with FokI nucleases, while mtZFNs are composed of ZFP linked to FokI nucleases. MTS guides mtZFNs and mitoTALENs to the mitochondria. ZFP and TALE selectively bind to predetermined defective mtDNA target sequences, after which FokI dimerizes and cleaves the mtDNA adjacent to these binding sites, causing double-strand breaks that lead to the elimination of defective mtDNA. The remaining wild-type mtDNA can then replicate, altering the heteroplasmy ratio. b The targeted gene sequence, along with transcriptional regulatory elements and MTS, is packaged into an AAV vector, which is then delivered to the nucleus of the recipient cell. Inside the nucleus, the AAV uncoats and releases single-stranded DNA, which replicates to form double-stranded DNA. RNA polymerase then transcribes this DNA into mRNA. The mRNA exits the nucleus and is translated into the corresponding protein at the ribosomes in the cytoplasm. MTS guides these proteins to the mitochondria, where they undergo further processing and perform their respective functions. c DdCBEs are engineered by fusing MTS, split-DddAtox halves, UGIs, and either ZFPs or TALEs. DddAtox catalyzes the deamination of cytosine to uracil, while UGIs prevent uracil-DNA glycosylase from excising uracil, resulting in C-to-T editing during replication. Additionally, by linking MTS, split-DddAtox halves, TadA8e (an engineered adenine deaminase), and TALEs, TadA8e catalyzes the deamination of adenine to inosine, which pairs with cytosine during replication, thereby achieving targeted A-to-G editing. d Modifying the tRNA binding domain of nuclear-encoded aminoacyl tRNA synthetase or overexpressing aminoacyl tRNA synthetase can enhance aminoacylation efficiency and stabilize translation products. The expression of post-transcriptional negative regulators like microRNAs can inhibit the expression of mitochondrial RNA-modifying enzymes, thereby affecting mt-tRNA modifications. Using microRNA antagonists could potentially reverse disease phenotypes. Furthermore, overexpressing mt-tRNA-modifying enzymes can correct anticodon first nucleotide modification defects in mt-tRNA, improving ribosomal translation within mitochondria. Overexpression of mitochondrial translation elongation factors EFTu and EFG2 can also partially suppress amino acid misincorporation caused by mtDNA mutations during the translation elongation process. mtDNA mitochondrial DNA; mitoTALENs mitochondria-targeted transcription activator-like effector nucleases; mtZFNs mitochondria-targeted zinc-finger nucleases; MTS mitochondrial targeting sequence; NES nuclear export signal; DdCBEs DddAtox-derived cytosine base editors; ZFP zinc finger proteins; TALE transcription activator-like effector; UGIs uracil glycosylase inhibitors; TadA8e deoxyadenosine deaminase; AAV adeno-associated virus
Table 3.
Clinical trials for mitochondrial diseases
| Conditions | Interventions | Phases | Study Status | Times | Countries | ID |
|---|---|---|---|---|---|---|
| Mitochondrial diseases | KH176 | I | Completed | 2015-2015 | Belgium | NCT02544217 |
| KH176 | II | Completed | 2016-2017 | Netherlands | NCT02909400 | |
| KH176 | II | Completed | 2019-2022 | Denmark, Germany, Netherlands, United Kingdom | NCT04165239 | |
| KH176 | II | Completed | 2021-2023 | Netherlands, Denmark, United Kingdom, Germany | NCT04604548 | |
| KH176 | II | Not_recruiting | 2021-2025 (estimated) | Netherlands | NCT04846036 | |
| EPI-743 | I/II | Completed | 2012-2019 | United States | NCT01642056 | |
| EPI-743 | II/III | Completed | 2020-2023 | United States, Canada, France, Italy, Japan, Poland, Spain, Sweden, United Kingdom | NCT04378075 | |
| EPI-743 | III | Enrolling_by_invitation | 2022-2025 (estimated) | United States, France, Italy, Japan, Poland, Spain, United Kingdom | NCT05218655 | |
| OMT-28 | II | Recruiting | 2023-2025 (estimated) | Germany, Italy, Netherlands | NCT05972954 | |
| KL1333 | I | Completed | 2019-2021 | United Kingdom | NCT03888716 | |
| KL1333 | I | Completed | 2020-2020 | United Kingdom | NCT04643249 | |
| KL1333 | II | Not_recruiting | 2022-2025 (estimated) | United States, Belgium, Denmark, France, Spain, United Kingdom | NCT05650229 | |
| Elamipretide | II | Completed | 2016-2017 | United States | NCT02805790 | |
| Elamipretide | III | Not_recruiting | 2022-2024 (estimated) | United States, Australia, Germany, Hungary, Italy, Netherlands, New Zealand, Norway, Spain, United Kingdom | NCT05162768 | |
| Coenzyme Q10 | III | Completed | 2007-2013 | United States, Canada | NCT00432744 | |
| Sodium succinate | II/III | Completed | 2014-2019 | Japan | JPRN-UMIN000013512 | |
| N-acetylcysteine | I | Recruiting | 2023-2024 (estimated) | United States | NCT05241262 | |
| Cysteamine bitartrate | II | Completed | 2014-2016 | United States | NCT02023866 | |
| Leber hereditary optic neuropathy | NR082 | I/II | Not_recruiting | 2023-2029 (estimated) | United States | NCT05293626 |
| NR082 | II/III | Recruiting | 2021-2028 (estimated) | China | NCT04912843 | |
| NFS-02 | I/II | Recruiting | 2023-2029 (estimated) | United States, China | NCT05820152 | |
| Curcumin | III | Completed | 2005-2007 | Thailand | NCT00528151 | |
| Idebenone | I/II | Completed | 2013-2017 | Japan | JPRN-UMIN000017939 | |
| Idebenone | II | Completed | 2007-2010 | Canada, Germany, United Kingdom | NCT00747487 | |
| Idebenone | III | Not_recruiting | 2022-2025 (estimated) | China | ChiCTR2200059044 | |
| Idebenone | IV | Completed | 2016-2021 | United States, Austria, Belgium, Bulgaria, Germany, Italy, Poland, Portugal, Spain, United Kingdom | NCT02774005 | |
| Idebenone | IV | Completed | 2018-2018 | China | ChiCTR-IPR-17013821 | |
| Bezafibrate | II/III | Completed | 2019-2023 | France | NCT04561466 | |
| Elamipretide | II | Completed | 2016-2019 | United States | NCT02693119 | |
| rAAV2-ND1 | 0 | Completed | 2021-2022 | China | ChiCTR2000041574 | |
| rAAV2-ND4 | 0 | Completed | 2020-2021 | China | ChiCTR2000038570 | |
| rAAV2-ND4 | I/II | Completed | 2014-2020 | France | NCT02064569 | |
| rAAV2-ND4 | II/III | Not_recruiting | 2017-2025 (estimated) | China | NCT03153293 | |
| rAAV2-ND4 | III | Completed | 2016-2018 | United States, France, Germany, Italy, United Kingdom | NCT02652780 | |
| rAAV2-ND4 | III | Completed | 2016-2019 | United States, France, Germany, Italy, United Kingdom | NCT02652767 | |
| rAAV2-ND4 | III | Completed | 2018-2022 | United States, France, Germany, Italy, United Kingdom | NCT03406104 | |
| rAAV2-ND4 | III | Not_recruiting | 2018-2024 (estimated) | United States,Belgium, France, Italy, Spain, Taiwan, United Kingdom | NCT03293524 | |
| scAAV2-P1ND4v2 | I | Not_recruiting | 2014-2024 (estimated) | United States | NCT02161380 | |
| Skin electrical stimulation | I/II | Completed | 2018-2019 | Japan | JPRN-jRCTs052180066 | |
| Friedreich’s ataxia | A0001 | II | Completed | 2009-2011 | United States | NCT01035671 |
| RT001 | I/II | Completed | 2015-2016 | United States | NCT02445794 | |
| RT001 | III | Completed | 2019-2021 | United States | NCT04102501 | |
| DT-216 | I | Completed | 2022-2022 | United States | NCT05285540 | |
| DT-216 | I | Completed | 2022-2023 | United States | NCT05573698 | |
| EPI-743 | II | Completed | 2012-2016 | United States | NCT01728064 | |
| EPI-743 | II | Completed | 2013-2016 | United States | NCT01962363 | |
| EPI-743 | II | Not_recruiting | 2022-2024 (estimated) | United States | NCT05485987 | |
| EPI-743 | II/III | Completed | 2020-2023 | United States, Australia, Brazil, Canada, France, Germany, Italy, New Zealand, Spain | NCT04577352 | |
| EPI-743 | III | Enrolling_by_invitation | 2022-2027 (estimated) | United States, Australia, Brazil, Canada, France, Germany, Italy, New Zealand, Spain | NCT05515536 | |
| EGb 761 | II | Completed | 2008-2011 | France | NCT00824512 | |
| TAK-831 | II | Completed | 2017-2018 | United States | NCT03214588 | |
| MIN-102 | II | Completed | 2019-2020 | Belgium, France, Germany, Spain | NCT03917225 | |
| MIB-626 | II | Completed | 2021-2022 | United States | NCT04817111 | |
| CTI-1601 | I | Completed | 2019-2020 | United States | NCT04176991 | |
| CTI-1601 | I | Completed | 2020-2021 | United States | NCT04519567 | |
| CTI-1601 | II | Completed | 2022-2023 | United States | NCT05579691 | |
| CTI-1601 | II | Enrolling_by_invitation | 2024-2027 (estimated) | United States | NCT06447025 | |
| VP 20629 | I | Completed | 2013-2015 | United States | NCT01898884 | |
| Calcitriol | IV | Completed | 2021-2023 | Spain | NCT04801303 | |
| Etravirine | II | Completed | 2020-2023 | Italy | NCT04273165 | |
| Artesunate | I/II | Recruiting | 2022-2023 (estimated) | France | NCT04921930 | |
| Idebenone | I | Completed | 2001-2006 | United States | NCT00015808 | |
| Idebenone | I | Completed | 2004-2006 | United States | NCT00078481 | |
| Idebenone | II | Completed | 2005-2007 | United States | NCT00229632 | |
| Idebenone | III | Completed | 2006-2010 | Austria, Belgium, France, Germany, Netherlands, United Kingdom | NCT00905268 | |
| Idebenone | III | Completed | 2007-2009 | United States | NCT00537680 | |
| Idebenone | III | Completed | 2007-2012 | Austria, Belgium, France, Germany, Netherlands | NCT00993967 | |
| Idebenone | III | Completed | 2008-2010 | United States | NCT00697073 | |
| Idebenone | III | Completed | 2011-2012 | Austria, Germany, Netherlands, United Kingdom | NCT01303406 | |
| Pioglitazone | III | Completed | 2008-2013 | France | NCT00811681 | |
| Deferiprone | I/II | Completed | 2008-2009 | Australia, Belgium, Canada, France, Italy, Spain | NCT00530127 | |
| Deferiprone | II | Completed | 2009-2011 | Belgium, France, Italy, Spain | NCT00897221 | |
| Resveratrol | I/II | Completed | 2011-2012 | Australia | NCT01339884 | |
| Resveratrol | II | Completed | 2019-2024 | Australia | NCT03933163 | |
| Epoetin alfa | II | Completed | 2008-2009 | Italy | NCT00631202 | |
| Epoetin alfa | II | Completed | 2013-2015 | Italy | NCT01493973 | |
| Rosuvastatin | Early_I | Completed | 2016-2017 | United States | NCT02705547 | |
| Iron chelating | I/II | Completed | 2005-2008 | France | NCT00224640 | |
| (+)-Epicatechin | II | Completed | 2016-2018 | United States | NCT02660112 | |
| Interferon γ-1b | II | Completed | 2013-2014 | United States | NCT01965327 | |
| Interferon γ-1b | II | Completed | 2013-2014 | Italy | NCT02035020 | |
| Interferon γ-1b | II | Completed | 2016-2017 | No location data | NCT03888664 | |
| Interferon γ-1b | III | Completed | 2015-2016 | United States | NCT02415127 | |
| Interferon γ-1b | III | Completed | 2015-2017 | United States | NCT02593773 | |
| Interferon γ-1b | III | Completed | 2016-2017 | United States | NCT02797080 | |
| Omaveloxolone | I | Not_yet_recruiting | 2023-2024 (estimated) | United States | NCT06054893 | |
| Omaveloxolone | II | Not_recruiting | 2015-2024 (estimated) | United States, Australia, Austria, Italy, United Kingdom | NCT02255435 | |
| AAVrh.10hFXN | I/II | Recruiting | 2022-2029 (estimated) | United States, Canada | NCT05445323 | |
| AAVrh.10hFXN | I | Recruiting | 2022-2029 (estimated) | United States | NCT05302271 | |
| Methylprednisolone | Early_I | Completed | 2015-2018 | United States | NCT02424435 | |
| Carbamylated erythropoietin | II | Completed | 2009-2011 | Austria, Germany, Italy | NCT01016366 | |
| Bupropion and citalopram | IV | Completed | 2012-2013 | No location data | NCT01716221 | |
| Mitochondrial myopathy | REN001 | I | Completed | 2022-2022 | England, United Kingdom | ISRCTN57533271 |
| REN001 | II | Completed | 2021-2023 | United States, Australia, Belgium, Canada, Czechia, Denmark, France, Germany, Hungary, Italy, Netherlands, New Zealand, Norway, Spain, United Kingdom | NCT04535609 | |
| Bocidelpar | II/III | Not_recruiting | 2021-2025 (estimated) | United States | NCT04641962 | |
| L-arginine | III | Completed | 2008-2012 | Japan | JPRN-jRCT2091220023 | |
| Bezafibrate | II | Completed | 2015-2017 | United Kingdom | NCT02398201 | |
| Elamipretide | I/II | Completed | 2015-2016 | United States | NCT02367014 | |
| Omaveloxolone | II | Completed | 2015-2017 | United States, Denmark | NCT02255422 | |
| Periodic acceleration | I | Recruiting | 2023-2024 (estimated) | United States | NCT05569122 | |
| Nicotinamide riboside | II | Recruiting | 2023-2025 (estimated) | United States | NCT05590468 | |
| Autologous mesoangioblasts | I | Completed | 2020-2022 | Netherlands | NCT05063721 | |
| Autologous mesoangioblasts | II | Recruiting | 2023-2025 (estimated) | Netherlands | NCT05962333 | |
| Mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes | Taurine | III | Completed | 2017-2019 | Japan | JPRN-jRCTs061180015 |
| KL1333 | I | Completed | 2017-2018 | Korea | NCT03056209 | |
| Idebenone | II | Completed | 2009-2012 | United States | NCT00887562 | |
| L-arginine | II | Completed | 2012-2013 | Canada | NCT01603446 | |
| L-citrulline | I | Recruiting | 2021-2024 (estimated) | United States | NCT03952234 | |
| Zagociguat | II | Not_yet_recruiting | 2024-2024 (estimated) | United States, Australia, Canada, Germany, Italy, United Kingdom | NCT06402123 | |
| Arginine and citrulline | Early_I | Completed | 2009-2016 | United States | NCT01339494 | |
| Medium-chain triglycerides | Early_I | Completed | 2010-2011 | United States | NCT01252979 | |
| Leigh syndrome | EPI-743 | II | Completed | 2012-2015 | United States | NCT01721733 |
| EPI-743 | II | Completed | 2014-2023 | United States | NCT02352896 | |
| EPI-743 | III | Completed | 2014-2021 | Japan | JPRN-jRCT2080222577 | |
| Pearson syndrome | MNV-201 | I | Recruiting | 2023-2027 (estimated) | Israel | NCT06017869 |
| MNV-BM-BLD | I/II | Completed | 2019-2021 | Israel | NCT03384420 | |
| Pyruvate dehydrogenase complex deficiency | Triheptanoin | I | Recruiting | 2024-2027 (estimated) | United States | NCT06340685 |
| Dichloroacetate | III | Not_recruiting | 2020-2025 (estimated) | United States | NCT02616484 | |
| Sodium phenylbutyrate | II | Completed | 2018-2020 | Italy | NCT03734263 | |
| Barth syndrome | Elamipretide | II/III | Completed | 2017-2021 | United States | NCT03098797 |
| Mitochondrial depletion syndrome | Deoxycytidine and deoxythymidine | II | Recruiting | 2021-2026 (estimated) | Canada | NCT04802707 |
Data from ClinicalTrials.gov and International Clinical Trials Registry Platform
Mitochondrial replacement therapy
Pronuclear transfer
Pronuclear transfer (PNT) is a technique that replaces the mitochondrial genome by transferring the parental pronucleus from zygotes with mutated mtDNA into enucleated zygotes containing healthy mitochondria.597 Initial studies show that less than 2% of the donor’s mtDNA persists in early embryos, and it becomes undetectable after development to the blastocyst stage in vitro.598 In mouse models with large-scale mtDNA deletions, PNT has successfully corrected mtDNA-related phenotypes in offspring,599 demonstrating its potential in treating mitochondrial genetic diseases. In 2015, the United Kingdom Parliament approved regulations allowing the use of PNT and ST, with other countries also exploring these techniques.600 However, the long-term effects on live-born offspring remain uncertain. Technical challenges, such as cytoplasmic leakage or incomplete separation of the nucleoplasm, may result in the transfer of a portion of the donor’s cytoplasm during PNT.601 Over time, the proportion of donor mtDNA may increase,599,601 potentially affecting the efficacy of MRT due to factors like enrichment, genetic drift, and mitochondrial bottleneck effects.602–605 This highlights the importance of minimizing or even eliminating mtDNA carryover during embryo transfer procedures.605 Vitrification of patient oocytes could help reduce mtDNA carryover and provide the option for oocyte storage.601,606 Additionally, female pronuclei, being smaller and containing fewer mitochondria than those used in PNT and ST, are easier to isolate and position, reducing the need for cytoskeletal inhibitors and avoiding premature oocyte activation.607 In a notable advancement, researchers isolated the female pronucleus from the second polar body and transferred it to another zygote, resulting in the live birth of four healthy cynomolgus monkeys.608 Moreover, techniques that enforce mitochondrial autophagy in reconstructed embryos produced by PNT have shown promise in reducing or eliminating mtDNA heterogeneity, enhancing safety.605
Despite these advancements, several potential issues require careful consideration. PNT results in the loss of approximately half of the embryos.609 While some ethical frameworks define pronuclear fusion as the beginning of embryonic life, PNT does not involve such fusion, which may reduce ethical concerns.605,610 Nevertheless, the loss of embryos remains a significant consideration.605,610 Furthermore, interactions between nDNA and mtDNA are extensive, with normal mitochondrial function relying on the coevolution of these two genomes.611,612 Disruption of these interactions post-PNT could potentially alter gene expression and phenotypic traits in offspring.600 This functional incompatibility between nuclear and mitochondrial genomes could even lead to reproductive isolation in mammals.613 To mitigate the risks associated with mito-nuclear interactions after mitochondrial replacement, it is recommended to match mitochondrial genotypes between donors and recipients and to determine acceptable variation levels between donor and patient mtDNA haplotypes.600 Until the implications of these interactions are better understood, combining mitochondrial replacement therapy with prenatal screening is advised.601 Ongoing follow-up and research will be essential to fully understand and address these issues.
Spindle-chromosome complex transfer
A study comparing mtDNA carryover and embryo development outcomes using germinal vesicle nuclear transfer, ST, and PNT in a mouse model614 found that germinal vesicle nuclear transfer did not result in blastocyst formation, while PNT and ST performed comparably well.614 ST involves replacing mutated mtDNA in patient oocytes by isolating the spindle-chromosome complex from the patient’s unfertilized mature MII oocytes and transferring it into the cytoplasm of enucleated healthy oocytes.597 Notably, Tachibana et al. successfully employed ST to produce healthy rhesus monkey offspring, which contained nDNA from the spindle donor and nearly homogenous mtDNA from the cytoplasm donor.44 In human studies, ST performed on unfertilized mature MII oocytes resulted in zygotes and derived embryonic stem cell lines with normal euploid karyotypes and donor-only mtDNA.615 Furthermore, the transfer of vitrified spindles into fresh cytoplasm led to the birth of four healthy rhesus monkey infants, with no significant changes in mtDNA heterogeneity.615 The use of cryopreserved human oocytes for ST has also been shown to eliminate mtDNA heteroplasmy while maintaining normal mitochondrial activity.616 Cryopreservation primarily affects the oocyte cytoplasm rather than the spindle, making it suitable for storing patient oocytes.615
There has been a reported case of a female patient with Leigh syndrome, carrying the m.8993 T > G mutation, giving birth to a healthy child using ST technology.46 However, the live birth following ST involved electrofusion,46 a technique that can prematurely activate the oocyte to enter late anaphase II, potentially leading to incomplete meiotic recovery post-fertilization and increasing the risk of abnormal pronuclear formation and aneuploidy.615 To mitigate the risk of premature oocyte activation, researchers have explored alternative approaches such as using chemical or mechanical methods,617 lowering temperatures to induce partial depolymerization of the spindle-chromosome complex in mature MII oocytes,616 and adjusting the sequence of intracytoplasmic sperm injection and ST.618 Despite the potential of ST, the procedure can result in donor mtDNA carryover.44,609 Although low-level mtDNA heteroplasmy often diminishes during MRT, genetic drift can sometimes cause mtDNA to revert to its original genotype, compromising the effectiveness of MRT.602,619 This reversion may be linked to the preferential replication of specific D-loop conservative sequence box II region polymorphisms.619 To address this, ongoing research is being conducted to reduce mitochondrial carryover and enhance the efficiency and safety of MRT.620 Strategies such as lowering cytochalasin B concentration before ST and PNT,621 generating aggregated chromosomes using the phosphodiesterase inhibitor 3-isobutyl 1-methylxanthine followed by transfer,622 and implementing ST with maximal residue removal in MII oocytes618 have shown promise in significantly reducing residual mtDNA levels. While ST is considered safe and effective44,618 and ethically acceptable,623 the optimal methodology has yet to be determined, and the risks of genetic drift necessitate further research.623
Polar body transfer
Wang et al. demonstrated that germline genomes derived from ST, PNT, and polar body transfer (PBT) in mice could lead to embryos capable of normal fertilization and producing viable offspring.609 The F1 generation from PBT exhibited minimal donor mtDNA carryover, which remained stable into the F2 generation.609 Notably, blastocysts resulting from first polar body transfer (PB1T) displayed lower average levels of mtDNA carryover compared to those from second polar body transfer (PB2T).624 Embryonic stem cells derived from PB1T blastocysts maintained low and stable mtDNA carryover during extended proliferation and differentiation, both in vitro and in vivo.624 Due to the mitochondrial inheritance bias during meiosis, polar bodies harbor minimal to undetectable mitochondria.609 PBT proceeds without the need for cytoskeletal disruptors,624 thereby further reducing donor mtDNA carryover. Additionally, the membrane encasing polar bodies protects the genome and facilitates their isolation.609,624 Combining PBT with ST or PNT in individual donor oocytes could potentially halve the number of donor oocytes needed, thereby improving MRT efficiency.609 Given that polar bodies and oocytes share the same genome, they might be more suitable as nuclear transfer donors than pronucleus or spindle-chromosome complexes.609 Wang et al. also demonstrated that PB1T could effectively substitute ST, yielding healthy macaque monkeys with stable mtDNA heteroplasmy below 5% and no mtDNA drift.625 However, due to the higher mitochondrial concentration in dense clusters within the first polar body in intact oocytes—compared to the uniform mitochondrial distribution in the ooplasm in ST—and the slightly larger volume of the first polar body relative to ST, the isolated first polar body contained more mtDNA than the isolated spindle complex,625 contrary to findings in mice.609 Nonetheless, polar bodies are distinct byproducts of meiosis, tasked with extruding the surplus genome, which might affect their quality.626 Further refinements in PBT techniques and additional preclinical studies on human oocytes and zygotes are necessary to assess the safety, efficacy, and feasibility of PBT.620
Gene therapy
Allotopic expression
Mitochondrial proteins, encoded by nuclear genes, are synthesized in the cytoplasm before being imported into mitochondria.627 This process has propelled advances in treating mitochondrial genetic diseases through the allotopic expression approach. This method re-encodes defective mtDNA gene sequences into nuclear-compatible sequences, aligned with the “universal” genetic code.628,629 These sequences are combined with appropriate transcriptional regulatory elements and mitochondrial targeting peptides, then delivered to the cell nucleus via suitable vectors.628,629 The resultant mRNA is translated in the cytosol,628,629 after which the mitochondrial targeting peptides facilitate the import of these proteins into mitochondria, achieving the allotopic expression of functional mtDNA genes to compensate for genetic defects.628,629 Specifically, re-engineered and stabilized allotopic expression of MT-ATP6 and MT-ATP8 allows their processing, import, and integration into complex V,630–632 while MT-ND3,633 MT-ND4,628 and MT-ND6634 are successfully imported into mitochondria and incorporated into complex I, rescuing mitochondrial dysfunction caused by corresponding mtDNA mutations. These mutations can lead to the loss of mitochondrial proteins, defects in respiratory chain complexes, and impaired oxidative phosphorylation.
Although human tRNA is typically not imported into mitochondria,635 Kolesnikova et al. demonstrated partial mitochondrial import and proper aminoacylation of nDNA-encoded yeast tRNALys derivatives in human cells, partially restoring mitochondrial function in MERRF cells.636 Furthermore, the Leishmania RNA import complex, entering human cells via a caveolin-1-dependent pathway, can induce the import of endogenous cytosolic tRNA, encoded by the nucleus, into mitochondria, rescuing dysfunction caused by mutated mt-tRNA genes.637
In LHON mice or rats, intravitreal injection of adeno-associated virus (AAV) vectors carrying nuclear-encoded human MT-ND4 genes resulted in the allotopic expression of the wild-type MT-ND4 gene, with subsequent accumulation of mRNA and protein in RGCs and optic nerve axons, successfully importing them into mitochondria.629,638 The ND4 protein assembles with three complex I subunits, integrating into the respiratory chain complex I without disrupting its activity.629 This integration preserves complex I function, preventing vision loss, RGC apoptosis, and degeneration, as well as optic nerve atrophy induced by mtDNA mutations.629,639 AAV-mediated allotopic expression of the nuclear-encoded MT-ND4 gene appears to be a feasible and safe treatment for LHON with mutated mtDNA, with clinical trials underway.47 However, the timing of gene therapy is crucial, as oxidative damage, RGC apoptosis, and axonal loss may be partly irreversible.639 Therefore, administering gene therapy after vision loss but before optic nerve atrophy, or targeting the asymptomatic contralateral eye in patients with acute unilateral vision loss, may offer benefits.628,639
Despite the promise of allotopic expression, challenges remain. Only a small fraction of allotopically expressed mitochondrial proteins localize to mitochondria632,640 and functionally integrate,641 likely due to the high hydrophobicity of these proteins, which hinders efficient mitochondrial import.640 This necessitates further refinement in processing and import efficiency.632,642 Combining recoded mtDNA sequences with optimized 3’ untranslated regions and 5’ mitochondrial targeting sequences (MTS) can enhance protein localization and import.629,642 The importability of hydrophobic peptides may also be improved by enhancing the expression of molecular chaperones.640 Additionally, codon optimization of mitochondrial genes can improve the efficiency and stability of recoded mtDNA gene expression in the nucleus.632 LHON is characterized by a 100% m.11778 G > A mutation. Although LHON mammalian models exhibit symptoms of acute vision loss similar to human LHON, the LHON model retains a normal MT-ND4 gene.638 Continued research is needed to explore the therapeutic potential of allotopic expression for other mitochondrial diseases.
Gene replacement therapy
Currently, the application of gene replacement therapy for various mitochondrial genetic diseases is the subject of extensive research and development. Normal genes can be delivered via AAV transduction to replace defective nuclear genes in mitochondrial genetic disorders, achieving therapeutic effects. Frataxin plays a key role in Fe-S cluster biosynthesis.643 In Friedreich’s ataxia, Frataxin deficiency leads to primary Fe-S cluster defects, reducing enzyme activity associated with these clusters, resulting in mitochondrial iron accumulation, dysfunction, and cellular damage.644 An AAV vector containing the Frataxin gene facilitates the expression of human Frataxin in Friedreich’s ataxia mouse models, restoring Fe-S cluster-associated protein levels and enzyme activity in cardiomyocytes.644,645 This restoration normalizes Fe-S biosynthesis, corrects iron accumulation, improves mitochondrial ultrastructure and abnormal cardiac myofibrils, and thus prevents and reverses the cardiomyopathy phenotype.644,645 However, therapeutic outcomes are highly dependent on the cardiac biodistribution of the vector.646 Furthermore, the therapeutic window for AAV-mediated Frataxin gene therapy is narrow; overexpression of Frataxin may induce oxidative stress and significantly increase labile iron pool levels, leading to hepatotoxicity and cardiotoxicity.647
A single intrathecal administration of AAV9/hSURF1 partially restores complex IV levels and activity, showing potential for treating SURF1-related Leigh syndrome.648 Additionally, intravenous and intracerebroventricular injections of AAV2/9-hNDUFS4 enhance complex I activity, improving weight, motor function, and lifespan in NDUFS4 knockout Leigh syndrome mouse models.649 However, neither intravenous nor intracerebroventricular administration alone fully improved the clinical phenotype, indicating limitations in standard AAV vector transduction.649 In contrast, intravenous injection of the brain-penetrating AAV.PHP.B-NDUFS4 vector restored mitochondrial complex I activity and function, improved behavior, corrected brain, retina, and heart pathologies, restored weight, and extended lifespan in mice.650,651 However, the AAV.PHP.B vector’s efficacy is limited to certain mouse strains and is not applicable to primates.652,653 The use of self-complementary AAV9 vectors, effective across mammals and enhancing the transcription rate of recombinant human NDUFS4, restored complex I activity and assembly in Leigh syndrome mice, significantly extending their lifespan.654
Delivering CRISPR/Cas9 and TYMP cDNA via lipid nanoparticles, polymeric nanoparticles, or AAV2/8 viral vector can efficiently integrate TYMP into the TYMP and Alb loci of hepatocytes in the MNGIE mouse model.655 This approach increases TP activity in plasma, reduces nucleoside levels, and shows promise for treating MNGIE.655 AAV-TYMP has been shown to elevate hepatic TP activity in MNGIE mouse models, normalize nucleoside and mitochondrial nucleotide metabolism, enhance mtDNA replication, correct mitochondrial dysfunction, and alleviate functional phenotypes.656 The alpha-1-antitrypsin promoter, demonstrating optimal efficacy,497 could also minimize the dosage required for clinical effectiveness.656
Moreover, the AAV2/8 vector can mediate the expression of the human ETHE1 gene in the liver of ethylmalonic encephalopathy mouse models, effectively clearing circulating hydrogen sulfide, correcting plasma thiosulfate levels, restoring sulfur dioxygenase activity, significantly improving disease phenotypes, and extending lifespan—highlighting its potential for future clinical applications in treating ethylmalonic encephalopathy.657
In Barth syndrome cells, AAV-TAZ transduction increases mtDNA copy number and enhances mitochondrial structure and function.658 Similarly, AAV9-TAZ ameliorates mitochondrial structural defects in a Barth syndrome mouse model, improving cardiac and skeletal muscle function659,660 and rescuing neonatal mice from mortality, cardiac dysfunction, and fibrosis. However, its efficacy and duration depend on the number of cardiomyocytes transduced.660 Notably, administration of AAV9 following low-intensity aerobic exercise can enhance AAV transduction efficiency in the heart and skeletal muscles.661
RNA-based therapy
Delivering double-stranded RNA, single-stranded silencing RNA, or antisense oligonucleotides (ASOs) to Friedreich’s ataxia cells can specifically target or excise the intronic GAA trinucleotide repeat sequence, thereby reducing R-loop formation between the expanded repeat RNA and complementary genomic DNA.662–664 This intervention reverses the transcriptional silencing of Frataxin, decreases the production of aberrant early-terminated Frataxin transcripts, and increases both Frataxin mRNA and protein levels, presenting a promising therapeutic strategy for Friedreich’s ataxia.662–664 Gapmer oligonucleotides complementary to the adenine-adenine-guanine repeat sequence within the Frataxin gene have shown a higher efficacy in activating Frataxin RNA and protein expression.665 Research by Li et al. indicates that co-delivering oligonucleotides targeting the 5’ or 3’ untranslated regions of Frataxin can extend the mRNA half-life, leading to increased steady-state levels of Frataxin mRNA and protein, suggesting a novel approach to upregulating mRNA levels in any transcriptionally downregulated disorder.666 Despite the successful delivery of ASOs in Frataxin mouse models, the anticipated increase in Frataxin expression was not observed, possibly due to the limited potency of the compounds, differences in the regulatory mechanisms of the Frataxin gene,667 or off-target effects.668 Additionally, two phosphorothioate-based ASOs with G-rich motifs were identified668 that indirectly activate Frataxin expression in Friedreich’s ataxia cells but similarly failed to induce Frataxin expression in Friedreich’s ataxia mouse models.668 Thus, determining the efficacy of double-stranded RNA, single-stranded silencing RNA, and ASOs in animal models is essential, necessitating further investigation.
Peptide nucleic acid oligomers
PNAs were the pioneering tool for achieving mitochondrial heteroplasmy shift.669 These molecules can selectively bind to complementary DNA or RNA sequences, effectively inhibiting replication and translation.670 Researchers have synthesized PNAs that are complementary to human mtDNA templates containing deletion breakpoints or single-base mutations, which specifically inhibit the replication of mutant human mtDNA templates in vitro.42 To address the challenge of delivering PNAs to mitochondria within cells, Muratovska et al. conjugated an 11-mer PNA to a lipophilic phosphonium cation.670 This phosphonium-PNA conjugate is non-cytotoxic, remains stable within cells, and can selectively inhibit the in vitro replication of mtDNA carrying the human m.8344 A > G mutation associated with MERRF.670 Despite the promising results PNAs have shown in inhibiting mutant mtDNA replication, their broader application is constrained. A PNA must be at least 7-mer in length to target a unique site within mtDNA.669 Additionally, during mtDNA replication, nucleic acid derivatives may fail to bind to their complementary sequences, and it remains uncertain whether single-stranded mtDNA at the replication fork can effectively interact with PNAs.669,670 Consequently, the widespread use of PNAs remains limited.
Mitochondria-targeted restriction endonucleases
Pathogenic mtDNA variations can introduce unique restriction endonuclease cleavage sites.671 MTS directs the restriction endonucleases into mitochondria, which cleaves pathogenic mtDNA at specific recognition sites, resulting in double-strand breaks (DSBs) that lead to the elimination of defective mtDNA.671–673 This process allows the remaining wild-type mtDNA to replicate, thereby altering the degree of heteroplasmy and offering a potential treatment for mitochondrial genetic disorders.671–673 Early research in 2002 demonstrated that mitochondria-targeted SmaI restriction endonuclease could selectively eliminate mutated mtDNA, allowing the replication of wild-type mtDNA and restoring normal cellular ATP levels and Δψm.674 Subsequent in vivo and in vitro studies have shown that mitochondria-targeted restriction endonucleases, such as PstI,675 ApaLI,676 Scal,677 and R.XmaI,678 can effectively eliminate mutant mtDNA at corresponding sites, thereby facilitating a shift in heteroplasmy. These interventions resulted in notable improvements in ATP synthase function and alleviated mitochondrial dysfunction, demonstrating the potential of mitochondria-targeted restriction endonucleases (mtREs) to prevent disease onset or reverse clinical symptoms in patients with specific pathogenic heteroplasmic mtDNA mutations. Additionally, they may inhibit the transgenerational transmission of human mitochondrial diseases.673,676,678–680 Compared to other gene-editing technologies, mtREs offer a significantly higher specificity, minimizing off-target activity and preventing mtDNA copy number depletion.681 However, two major challenges may limit the application of this method. First, the cellular delivery of mtREs could raise safety concerns.680 Second, the utility of mtREs is restricted to targeting specific heterogeneous mtDNA mutations.680 Many mtDNA mutations do not create new restriction enzyme sites, rendering mtREs ineffective against these mutations,42 which hinders their clinical application.669 To extend this approach to other pathogenic mtDNA mutations, the development of nucleases with novel specificities is essential.679
Mitochondria-targeted zinc-finger nucleases
Zinc finger nucleases (ZFNs) consist of zinc finger proteins (ZFPs) linked to FokI nucleases, with ZFPs further connected to MTS and nuclear export signals (NES). These modifications guide mitochondria-targeted zinc-finger nucleases (mtZFNs) to the mitochondria, where ZFPs can selectively bind to specific DNA target sequences. Upon binding, FokI nucleases dimerize and cleave the DNA adjacent to the ZFP binding sites.669,682 While traditional ZFNs can target mtDNA, identifying suitable ZFN pairs for certain mutations poses a challenge. To address this, researchers developed heterodimeric ZFNs that bind both mutant and adjacent wild-type mtDNA sequences. However, this approach often results in the degradation of wild-type mtDNA and rapid mtDNA depletion. To overcome this, researchers created single-chain ZFNs by conjugating two FokI nuclease catalytic domains to ZFPs. These single-chain ZFNs demonstrated greater selectivity for pathogenic point mutations in mtDNA682 and proved more effective than their single-domain counterparts.682 Despite this improvement, single-chain ZFNs are ineffective against large mtDNA deletions and present potential safety concerns.683 To address these limitations, researchers redesigned conventional dimeric mtZFNs, ensuring that monomers did not affect mtDNA.683 The improved mtZFNs effectively eliminated point mutations and large-scale mtDNA deletions, reducing the mutant mtDNA haplotype load below the pathogenic threshold, thereby restoring OXPHOS function and improving mitochondrial respiration.683 Both in vitro and in vivo experiments with mtZFN-AAV targeting the m.5024 C > T tRNAAla mutation demonstrated a partial shift in heteroplasmy, leading to improved steady-state levels of mt-tRNAAla, as well as enhanced mitochondrial respiration and metabolic function.672 Additionally, Gammage et al. achieved near-complete correction of mtDNA mutations and rescued mitochondrial respiratory function and metabolic defects through either consecutive short-term mtZFN treatments or finely controlled, optimized mtZFN expression.681 This approach minimized off-target effects and unwanted depletion of mtDNA copy numbers, proving more efficient than mtREs and mitochondria-targeted transcription activator-like effector nucleases (mitoTALENs).681 Despite these advancements, ZFN expression is associated with cytotoxicity due to off-target site cleavage. This issue might be mitigated by equipping mtZFNs with tightly regulated expression systems or by optimizing NES and ZFP sequences to reduce cytotoxicity.682,684
Mitochondria-targeted transcription activator-like effector nucleases
MitoTALENs are composed of targeted transcription activator-like effectors (TALEs) that bind to specific DNA sequences, coupled with FokI nucleases that dimerize to cleave mtDNA. Numerous in vitro and in vivo studies have demonstrated that mitoTALENs can reduce the load of pathogenic mtDNA and rescue associated functional phenotypes.685,686 Additionally, mitoTALENs have shown effectiveness in reducing the human m.3243 A > G mtDNA mutation in porcine oocytes,685 as well as the NZB mtDNA in MII oocytes of the NZB/BALB heterozygous mouse model.680 Targeting human m.14459 G > A and m.9176 T > C mutant mtDNA using mitoTALENs has led to the specific elimination of these mutant mitochondrial genomes.680 These results highlight the potential of mitoTALENs for selectively eliminating mutant mtDNAs and preventing their germline transmission. Furthermore, mitoTALEN nickases, which are derivatives of mitoTALEN with an inactive FokI domain on one monomer, can induce single-strand breaks at specific sites in human mtDNA. This process leads to mtDNA deletions687,688 and facilitates the creation of new animal models for studying single large-scale mtDNA deletion diseases.689
Although mitoTALENs may not be as potent as mtREs in preventing the spread of germline mitochondrial diseases,680 they offer greater design flexibility compared to mtREs and mtZFNs.690 However, mitoTALENs come with several limitations. Unlike more precise gene-editing technologies, ZFN and TALEN are incapable of performing precise single-base editing, making them unsuitable for correcting homogenous mtDNA mutations.691 Additionally, most mitoTALENs require a thymidine base at position 0 of the target DNA binding site,692 and the mutation’s sequence context, along with methylation or other epigenetic modifications of the mtDNA target sequence, can impact the TALE’s efficiency.693,694 The large size of mitoTALENs also complicates their encapsulation in many vector systems.695,696 Some researchers have attempted to overcome this by fusing a monomeric nuclease domain derived from the I-TevI homologous endonuclease to the TALE DNA-binding domain697 or by designing shorter, more specific mitoTALENs.698 However, these approaches often limit DNA sequence recognition.698 Another significant challenge is the potential for non-specific cleavage by mitoTALENs, which can lead to substantial depletion of mtDNA copies and induce cytotoxicity,314,692 necessitating precise dose control of the constructs.681 Moreover, while mitoTALENs exhibit minimal cleavage activity against wild-type mtDNA,699 further development is needed to create more effective, safer, and easier-to-deliver mitoTALENs in the future.
CRISPR/Cas9
The design of single-guide RNA (sgRNA) for targeting mtDNA mutations involves adding MTS upstream and downstream of the Cas9 gene and the 3’ untranslated region of the target gene.700,701 This configuration enables mitochondria-targeted Cas9 (mito-Cas9) to cleave mtDNA at specific sites dictated by the sgRNA.700,701 It has been shown that the mito-Cas9 system can be successfully translocated into mitochondria, where it can introduce exogenous single-stranded DNA oligonucleotides into mtDNA, thereby facilitating the creation of cellular models of disease-causing mtDNA mutations.701 However, the efficiency of mtDNA editing using mito-Cas9 systems can be limited by several factors, including the sequence characteristics of the target region and the variable targeting efficiencies of different sgRNAs.701 Optimizing the mito-Cas9 system might involve enhancing mitochondrial RNA transport702 or employing engineered Cas proteins with higher editing efficiency.703 Nevertheless, the lack of an RNA transporter system within the double-membrane structure of mammalian mitochondria31,704 results in inefficient or defective nucleic acid import into mitochondria.691,700 Additionally, the absence of homologous recombination and non-homologous end-joining pathways for repairing DSBs in mitochondria further complicates the manipulation of mtDNA.689,705 In summary, any attempt to import synthetic RNA molecules into mitochondria based on naturally occurring mechanisms in human cells is likely to be sporadic and inefficient.681 Consequently, the application of CRISPR/Cas9 technology to mtDNA manipulation remains highly challenging and will require further extensive research to overcome these obstacles.
Mitochondria-targeted meganucleases
Mitochondria-targeted meganucleases (mitoARCUS), derived from the naturally occurring I-CreI nucleic acid endonucleases, are small, highly specific single-component proteins.706 These nucleases possess the ability to recognize DNA sequences with single-base pair differences, generating DSBs. They can be efficiently packaged into individual viral vectors, requiring only minimal AAV titers, and do not show signs of vector-associated or transient mtDNA depletion toxicity.707,708 In heterozygous mice with the m.5024 C > T mutation, AAV9-mitoARCUS significantly altered heteroplasmy and restored mt-tRNAAla levels.708,709 Additionally, AAV9-mitoARCUS modulated heterogeneity and enhanced mitochondrial-encoded protein homeostasis and respiratory function in m.3243 A > G cell lines and mouse models.707 However, the need to redesign target-specific mitoARCUS for each mutation and the complexity involved in reengineering I-CreI to recognize new targets remain substantial challenges that must be addressed.705,707,710
DddA-derived cytosine base editors
DddA-derived cytosine base editors (DdCBEs) are engineered by fusing MTS, split-DddAtox halves, TALE, and uracil glycosylase inhibitors.710 The DddAtox enzyme catalyzes the conversion of cytosine to uracil,710 while the uracil glycosylase inhibitors prevent uracil-DNA glycosylase from removing uracil, leading to C-to-T mutations during subsequent DNA replication without introducing DSBs. This mechanism ensures high specificity and product purity in targeted edits 715. DdCBEs have facilitated mitochondrial base editing in human embryos711,712 and enabled precise, heritable C-to-T base editing at specific mtDNA sites in zebrafish, mice, and rats. These models replicate phenotypes akin to human mitochondrial diseases, thereby advancing the precise modeling of these conditions.710,713–716 Additionally, Mok et al. developed monomeric DdCBEs derived from non-toxic, full-length DddAtox variants, achieving C-to-T editing with reduced off-target effects.717 Zinc finger deaminases, which combine zinc finger DNA-binding proteins, DddAtox, and uracil glycosylase inhibitors, also catalyze targeted C-to-T transitions in mtDNA.718 Optimizing the architecture of zinc finger DdCBEs has improved editing efficiency, reduced off-target effects, facilitated packaging in AAV vectors, and potentially decreased immunogenicity.719 These advances enable the correction of pathogenic mtDNA mutations and the modeling of mitochondrial diseases.
Ongoing research focuses on enhancing the base editing efficiency of DdCBEs and expanding their applications. For example, co-injection of mitoTALEN has been shown to enhance DdCBE-NES-mediated mtDNA editing.720 Variants of DddA with increased base editing efficiency and a broader target range have been developed using phage-assisted continuous evolution and phage-assisted discontinuous evolution.720,721 The DddA11 variant, in particular, has expanded HC (H = A, C, or T) sequence compatibility, although it remains less effective for GC targets.722 CRISPR-mediated nuclear and TALE-based mitochondrial DdCBEs, utilizing dsDNA deaminase derived from Roseburia intestinalis interbacterial toxin (riDddAtox), have successfully achieved C-to-T editing of HC and GC targets.723 Fusion of transactivators to DddAtox or riDddAtox has significantly increased editing efficiency in both nDNA and mtDNA.723 Moreover, combining the DddA11 variant with activation-inducible cytidine deaminase has further improved C-to-T editing efficiencies across various targets.724 In another approach, TALE-linked deaminases designed by Cho et al. catalyze the hydrolytic deamination of adenine to produce inosine, which pairs with cytosine during replication, enabling targeted A-to-G editing in mitochondria with high efficiency.725 The A-to-G editing efficiency was further enhanced by fusing the DddA6 variant with TALE-linked deoxyadenosine deaminase.724
However, DdCBEs may induce off-target activities in mtDNA,712,726 likely due to non-specific interactions between TALE and DNA or spontaneous assembly of split DddAtox deaminases.727 To mitigate these issues, Lee et al. developed high-fidelity DdCBEs that are both efficient and precise, avoiding off-target mutations.727 Adding NES sequences also helps reduce off-target editing while improving on-target efficiency.724 DdCBEs offer precise mtDNA base editing both in vitro and in vivo, with the ability for germline transmission.728 This method is particularly valuable for creating mitochondrial disease-associated cell lines and animal models, deepening our understanding of mitochondrial disorders and providing potential avenues for correcting both homoplasmic and heteroplasmic pathogenic variants.
Post-transcriptional modifications
Post-transcriptional modifications of mitochondrial RNA play a critical role in finely regulating the synthesis and stability of the 13 mitochondrial proteins.280 These modifications stabilize tRNA and introduce wobble modifications, which can ameliorate mitochondrial translation defects and present potential therapeutic avenues for mitochondrial genetic diseases.729 In vitro studies have demonstrated that the m.3290 T > C mutation can restore the hypomodified 5-taurinomethyluridine in mt-tRNALeu(UUR) with the m.3243 A > G mutation, thereby improving mitochondrial translation in MELAS, facilitating respiratory chain complex formation, and enhancing oxygen consumption rates.730 Additionally, acquiring wobble modifications in mt-tRNALeu(CUN) with the m.12300 G > A mutation can also alleviate respiratory defects associated with the m.3243 A > G mutation.731 Introducing wobble modifications in other isoacceptor tRNAs may also yield similar benefits.731
In addition, the defects in mt-tRNA modification and subsequent mitochondrial protein translation can be restored by regulating the expression of mt-tRNA-modifying enzymes. Mitochondrial translation optimization 1 (MTO1) and GTP-binding protein 3 (GTPBP3) are responsible for catalyzing the biosynthesis of 5-taurinomethyluridine.732 High-dose oral taurine has been shown to increase MTO1 expression,733 effectively preventing stroke-like episodes in MELAS by correcting the first anticodon nucleotide modification defect in mt-tRNALeu(UUR).734 Moreover, MTO1 overexpression restores 5-taurinomethyluridine in mutant mt-tRNALeu(UUR) in MELAS and the 2-thiouridine derivative in mutant mt-tRNALys in MERRF, enabling efficient decoding of homologous codons independently of taurine supplementation.729 MTO1 may also function as an RNA chaperone, stabilizing pathogenic mt-tRNA mutations, enhancing tRNA aminoacylation efficiency, and supporting mitochondrial protein synthesis.280,729 Furthermore, ketogenic diet can improve OXPHOS defects independently of MTO1-mediated tRNA modifications, suggesting an alternative therapeutic approach.735 Overexpression of TRMT61B can restore the N1-methyladenosine modification at position 58 in mt-tRNALys with the m.8344 A > G mutation in MERRF, thereby finely regulating mitochondrial protein synthesis and stability.280 Cysteine is essential for the 2-thiomodification of mt-tRNA,736 and L-cysteine has been shown to partially rescue mitochondrial translation defects in cells with m.3243 A > G and m.8344 A > G mutations.737 Additionally, N-acetyl-cysteine has demonstrated benefits for mitochondrial translation in cells deficient in tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase (TRMU) and MTO1,736 indicating that restoring specific tRNA modifications could mitigate mitochondrial disease symptoms.
Further research by Meseguer et al. has revealed that retrograde signals from mitochondria to the nucleus, such as ROS or Ca2+, can increase microRNA expression in cells with various mtDNA mutations. These microRNAs, acting as post-transcriptional negative regulators, also influence mt-tRNA modifications by modulating the expression of mt-tRNA-modifying enzymes, thereby exacerbating disease phenotypes. Notably, microRNA-335/335* regulates GTPBP3 and MTO1 expression.738 High ROS levels induce microRNA-9/9* expression via the NF-κB pathway, directly targeting and reducing TRMU, GTPBP3, and MTO1 mRNA and protein levels, impairing non-mutant mt-tRNA modifications and worsening the MELAS phenotype.264 Consequently, microRNA antagonists may offer potential strategies to counteract these deleterious effects.264,738
Additional approaches to improve mitochondrial protein synthesis function include the use of the mTOR inhibitor rapamycin, which can enhance erythroid differentiation by inhibiting mTOR signaling and protein synthesis, effectively alleviating anemia symptoms in MLASA.502,739 Modifying the tRNA-binding domain of nuclear-encoded human mitochondrial phenylalanyl-tRNA synthetase can increase the aminoacylation efficiency of mt-tRNAPhe with the G34A mutation in MERRF.740 Overexpressing human mitochondrial leucyl-tRNA synthetase can stabilize tRNALeu(UUR) and mitochondrial translation products, thereby rescuing respiratory chain defects in cells with the m.3243 A > G mutation.741 Furthermore, overexpressing mitochondrial translation elongation factors EFTu and EFG2 can improve quality control during translational elongation, partially suppressing amino acid misincorporation in complex III, complex II, and ATP6 caused by the m.3243 A > G mutation, thus ameliorating respiratory chain assembly defects.742
Drug therapy
Advancements in pharmaceutical technology have led to the development of numerous drugs that are now being applied to treat mitochondrial disorders, with many having entered the clinical trial stage. These include CoQ10, idebenone, EPI-743, N-acetylcysteine, elamipretide, RT001, KH176, omaveloxolone, bezafibrate, pioglitazone, deferiprone, sodium dichloroacetate, L-arginine, interferon-gamma (IFN-γ), recombinant human erythropoietin, deoxynucleotide monophosphate and deoxynucleoside, among others. Each of these drugs operates through distinct mechanisms, targeting various aspects of mitochondrial dysfunction to potentially alleviate the symptoms or slow the progression of these disorders.
Antioxidants
CoQ10 plays a critical role in the mitochondrial respiratory chain,743 enhancing electron transport and ATP production, regulating redox signaling, stabilizing the mPTP, and preventing autophagy and apoptosis.744 Consequently, CoQ10 supplementation can reverse pathophysiological alterations and significantly improve clinical outcomes in conditions such as Leigh syndrome and MELAS.263,745,746 Furthermore, combining Idebenone with CoQ10 therapy may enhance therapeutic efficacy.746 CoQ10 offers tangible benefits for patients with mitochondrial diseases.
Idebenone, a CoQ10 analog, is the first drug approved in Europe for the treatment of LHON.45 It is reduced by cytosolic NAD(P)H oxidoreductase I (NQO1)747 and bypasses LHON-associated complex I dysfunction, restoring mitochondrial function by shuttling electrons directly from the cytoplasm to complex III. This mechanism maintains cellular energy production, restores ATP levels, and reduces ROS production.747,748 Both in vitro and in vivo studies have demonstrated Idebenone’s protective effects on RGCs and retinal integrity, preserving visual function.749,750 Idebenone has been shown to restore or maintain visual function in LHON, prevent color vision loss, and improve extraocular nerve dysfunction.751,752 Its long-term efficacy for LHON has been documented,753 although the therapeutic effect varies depending on the disease stage and specific pathogenic mtDNA mutation.754 Innovative delivery methods, such as PCL intravitreal implants loaded with Idebenone755 and biodegradable poly microspheres,756 offer controlled and prolonged intraocular administration, providing a new strategy for sustained LHON treatment. Idebenone is also used in the treatment of MELAS,757 Leigh syndrome,758 and Friedreich’s ataxia.759 However, Idebenone’s effect on mitochondrial respiratory chain function is dose-dependent; it can inhibit complex I activity while increasing complex II activity,760 potentially transforming from an antioxidant to a prooxidant and inducing mitochondrial dysfunction depending on its concentration and NQO1 expression levels.761,762 Due to its narrow therapeutic range, high doses of Idebenone can be cytotoxic, particularly in the ganglion cell layer.761 Genetic variations in NQO1 protein levels significantly influence Idebenone’s efficacy and toxicity, especially in NQO1-deficient cell lines,747,761 necessitating consideration of the patient’s NQO1 genotype and mtDNA mutation before treatment.747 Additionally, Idebenone is ineffective in correcting mitochondrial energy metabolism defects in conditions of CoQ deficiency.763
EPI-743, a novel p-benzoquinone therapeutic agent, enhances endogenous glutathione biosynthesis and improves oxidative status by modulating oxidoreductase enzyme activity, leading to clinical improvements in some hereditary mitochondrial disorders.764,765 EPI-743 has been shown to delay disease progression in Leigh syndrome765,766 and positively impact the recovery of visual function in LHON.767 Some evidence suggests that EPI-743 may be more potent than Idebenone.768
N-acetylcysteine, a precursor to glutathione, plays a critical role in restoring glutathione balance, improving mitochondrial complex IV dysfunction, reducing cellular oxidative damage, and ameliorating neuromuscular dysfunctions in Leigh syndrome models.769 When combined with cysteamine bitartrate, these benefits are further enhanced. Additionally, a combination therapy involving glucose, niacin, and N-acetylcysteine has been shown to synergistically improve respiratory chain complex I dysfunction, reduce mitochondrial stress, and boost metabolic and glutathione levels, thereby increasing resilience and preventing acute neurological and biochemical decompensation.770
Elamipretide, a mitochondria-targeted aromatic-cationic tetrapeptide, efficiently penetrates the OMM and rapidly localizes to the inner membrane, where it binds to cardiolipin or monolysocardiolipin.771 By improving the function of specific proteins involved in mitochondrial dynamics and mitophagy, Elamipretide can restore mitochondrial morphology and function,772 thereby enhancing skeletal muscle and cardiovascular performance in Barth syndrome and alleviating related clinical symptoms and disease progression.773,774 Additionally, Elamipretide may improve visual function in LHON775 and increase exercise capacity in patients with primary mitochondrial myopathy.776
RT001, a deuterated ethyl linoleate, is an orally bioavailable synthetic deuterated polyunsaturated fatty acid designed to inhibit lipid autoxidation and protect cells from oxidative stress.777 It has shown potential therapeutic effects in Friedreich’s ataxia, where it has been demonstrated to improve peak work capacity and oxygen consumption.778 However, clinical trials have yielded mixed results, with one trial indicating that RT001 may not be beneficial for treating Friedreich’s ataxia.779
KH176 exhibits dual antioxidant and redox-modulating properties.780 As a ROS-redox modulator, KH176 preserves the microstructure of the NDUFS4 mouse brain, reduces lipid peroxidation, and mitigates RGC degeneration, leading to improved rotational and gait performance in NDUFS4 mice.781 By targeting the thioredoxin/peroxiredoxin system, KH176 effectively lowers ROS levels and offers protection to cells with OXPHOS deficiencies.780 It also ameliorates neuronal network dysfunction and transcriptomic changes linked to m.3243 A > G heteroplasmy in neurons derived from human iPSCs.782 However, clinical studies by Janssen et al. suggest that KH176 may not significantly enhance clinical outcomes in patients with mitochondrial m.3243 A > G spectrum disorders.783
Metabolic modifiers
Omaveloxolone, an Nrf2 activator and NF-kB inhibitor, targets inflammatory and metabolic pathways.784,785 It enhances substrate availability and complex I activity, reduces endogenous lipid peroxidation and mitochondrial ROS levels, and elevates glutathione levels, thereby protecting cells from oxidative stress, maintaining Δψm, promoting mitochondrial respiration, and preventing cell death.786 Omaveloxolone has shown significant improvements in neurological function in Friedreich’s ataxia and can markedly slow disease progression with a favorable safety and tolerability profile.787 It is the first drug approved in the United States and Europe for treating Friedreich’s ataxia in patients aged 16 and older.48 However, given the multi-system nature of Friedreich’s ataxia, a cure may ultimately require combination therapy. Omaveloxolone may also enhance mitochondrial function and submaximal exercise tolerance, reducing heart rate and lactate production during exercise, which could benefit those with mitochondrial myopathy.788 Other compounds, such as (+)-Epicatechin,789 A0001,790 and Nomlacofusp,791 have also shown potential in treating Friedreich’s ataxia, though further research is necessary.
Bezafibrate, a PPAR activator, upregulates downstream PPAR target genes, increases mitochondrial biogenesis, and prevents cardiac dysfunction in mouse models of Barth syndrome792,793 and exercise intolerance.793 It also improves disturbances in the antioxidant system, mitochondrial quality control proteins, and mitochondrial function, offering potential treatment for Barth syndrome and dilated cardiomyopathy with ataxia syndrome.794 Additionally, bezafibrate enhances metabolic programming in Leigh syndrome neural progenitor cells by promoting SURF1 gene expression and inducing PGC-1α, thereby restoring neuronal morphogenesis.795 It increases survival rates and mitigates disease progression in Leigh syndrome mouse models.796 In carriers of the m.3243 A > G mutation, bezafibrate induces mitochondrial biogenesis, improves cardiac function, and alters metabolomic profiles while increasing mitochondrial disease biomarkers in serum.797
Pioglitazone, a PPAR-γ agonist, increases levels of the human insulin-degrading enzyme and PITRM1 protein through PPAR-γ activation.798 This mechanism restores mitochondrial targeting sequence pre-processing and alleviates feedback inhibition of mitochondrial processing peptidase activity in fibroblasts from PITRM1-deficient patients, improving Frataxin maturation and mitochondrial function.798 In combination with deoxynucleosides, pioglitazone may increase mtDNA copy number and mitochondrial mass, reduce mtDNA-encoded transcripts, and improve mitochondrial respiration in MELAS cells with the m.3243 A > G mutation.799 Similarly, rosiglitazone, another PPAR-γ agonist, enhances Frataxin levels in Frataxin-deficient dorsal root ganglia neurons, boosting mitochondrial biogenesis, function, calcium homeostasis, and cell survival.800 Rosiglitazone also improves energy metabolism by increasing fatty acid β-oxidation in Frataxin-deficient cardiomyocytes and enhances motor function in Friedreich’s ataxia mouse models,800 indicating its potential as a treatment for this condition both in vitro and in vivo.800
Sodium dichloroacetate inhibits accelerated lactate production in mtDNA deletion mice, ameliorates chronic lactic acidosis, improves mitochondrial biogenesis, restores respiratory function, and extends lifespan.801 It may offer certain benefits for patients with mitochondrial diseases.802 Oral sodium dichloroacetate stimulates cellular energy metabolism by activating residual enzyme activity, effectively reducing blood and cerebrospinal fluid lactate levels in children with congenital lactic acidosis due to pyruvate dehydrogenase complex mutations.803
L-arginine, a precursor for nitric oxide synthesis, enhances nitric oxide formation in patients with impaired endothelial function.804 It has been shown to improve aerobic capacity and muscle metabolism in MELAS,805 and reverse endothelial dysfunction.806 L-arginine can also reduce the frequency and severity of stroke-like episodes and slow the progression of MELAS.807 Long-term L-arginine supplementation appears promising for MELAS therapy,808 though the existing evidence is of poor methodological quality, and both intravenous and oral L-arginine have shown limited clinical benefits in the acute or preventive treatment of MELAS.809 L-arginine may primarily serve to prevent the onset of stroke-like episodes.810 Citrulline, which increases de novo arginine synthesis and enhances nitric oxide production, could potentially offer better therapeutic effects than arginine.811 More rigorous trials are needed to fully evaluate the efficacy and safety of L-arginine and citrulline therapies.
Chelator
Deferiprone is known to redistribute iron and stimulate the expression of Frataxin, a mitochondrial iron chaperone.459,812 This action facilitates the chelation of mitochondrial labile iron, which plays a role in oxidative stress, and leads to the reactivation of iron-deficient aconitase, reduction of iron accumulation, and mitigation of iron-induced ROS synthesis and mitochondrial stress in Friedreich’s ataxia cardiomyocytes.459,812 As a result, deferiprone helps restore respiratory chain protein levels, salvages mitochondrial function, inhibits TSFR gene expression, and improves calcium handling dynamics, thereby enhancing cardiac function.459,812 However, the use of high doses of deferiprone can negatively impact cellular Fe-S enzyme activity and reduce Frataxin levels.459,813 Additionally, Lim et al. identified a novel lipophilic iron chelator, 2-pyridylcarboxaldehyde 2-thiophenecarboxyl hydrazone, which rapidly penetrates cells to induce iron efflux and protect Friedreich’s ataxia fibroblasts from hydrogen peroxide-induced cytotoxicity, showing potential as a treatment for iron overload.814
Others
IFN-γ has emerged as a potential treatment for Friedreich’s ataxia. Both in vitro and in vivo studies have demonstrated that IFN-γ can upregulate Frataxin levels by modulating Frataxin gene transcription, which helps improve sensory and motor deficits in Friedreich’s ataxia mouse models.815 IFN-γ enhances the expression of Nrf2 and manganese-dependent superoxide dismutase in Friedreich’s ataxia cells, activating the non-canonical Nrf2 pathway via p21. This activation reduces the cells’ sensitivity to hydrogen peroxide-induced cell death, thereby offering protective effects.816 IFN-γ treatment may also reduce cardiomyocyte damage and improve cardiac function in Friedreich’s ataxia cardiomyopathy.817
A single high dose of erythropoietin has been shown to sustainably elevate Frataxin levels in Friedreich’s ataxia, reduce oxidative stress markers, and improve clinical symptoms.818 However, this treatment can lead to an increase in hematocrit levels.818 In vitro studies have demonstrated that carbamylated erythropoietin, an erythropoietin derivative, can increase Frataxin levels independently of erythropoietin receptor activity and without inducing erythropoiesis.819 Despite these promising results, a Phase II clinical trial indicated that carbamylated erythropoietin might not have significant therapeutic effects on Friedreich’s ataxia820
In TK2 deficiency, deoxythymidine triphosphate levels are significantly diminished, resulting in an imbalance in the mitochondrial deoxyribonucleoside triphosphate pool.821 Administering oral TK2 products such as deoxycytidine and deoxythymidine monophosphates, or rapidly degradable deoxypyrimidine monophosphate products like deoxythymidine and deoxycytidine, can effectively increase deoxythymidine triphosphate concentrations in TK2-deficient mouse models.822–824 This treatment has been shown to restore mtDNA copy numbers and improve the activity and levels of mitochondrial respiratory chain enzymes, thereby delaying disease onset, alleviating symptoms, and extending lifespan in these models.822–824 Clinical studies have further demonstrated that deoxynucleotide monophosphate and deoxynucleoside therapies significantly enhance survival, swallowing, respiratory, and motor functions in patients with TK2-deficient myopathy, showcasing both efficacy and a favorable safety profile.825,826 However, because deoxythymidine and deoxycytidine therapy alone cannot completely halt or reverse the progression of TK2 deficiency, a combined therapeutic approach using AAV-TK2 gene therapy alongside deoxynucleosides may provide a more comprehensive and effective treatment for TK2 deficiency.827
Cell therapy
Unlike skeletal muscles, satellite cells—dormant myoblasts that can be activated to re-enter the cell cycle and fuse with existing muscle fibers—rarely or never harbor mutant mtDNA.828,829 This unique characteristic enables muscle fiber regeneration through satellite cells, which can be stimulated by resistance training or injury-induced muscle regeneration,828–830 promoting hypertrophy and the incorporation of satellite cells into muscle fibers.828–830 This process facilitates the transfer of wild-type mtDNA from satellite cells to mature muscle fibers, altering the heteroplasmy and ultimately enhancing muscle strength and oxidative capacity.828–830 Although satellite cells and myoblasts are primarily suited for local muscle administration,831 mesoangioblasts—stem cells capable of fusing with damaged muscles—can either directly repair muscle tissue or augment the satellite cell pool to promote muscle regeneration.832,833 Mesoangioblasts can adhere to and traverse vascular endothelial cells, allowing for systemic arterial administration.834 In vitro studies have shown that the fusion of wild-type mesoangioblasts with myotubes carrying the m.3271 T > C or m.3291 T > C mutations can reduce the mtDNA mutation load and improve mitochondrial function.831 Moreover, mesoangioblasts from patients with mtDNA mutations or large-scale deletions show negligible corresponding mtDNA mutations or deletions and demonstrate mitochondrial function, proliferation, and myogenic differentiation abilities comparable to those of wild-type mesoangioblasts.835 Thus, mesoangioblasts present a viable option for autologous myogenic cell therapy, enabling the regeneration of new muscle fibers without mtDNA mutations or deletions following muscle injury.835 Additionally, eccentric exercise can induce skeletal muscle inflammation, prompting monocyte migration, which may enhance the efficacy of such treatments.835,836
In MNGIE, the infusion of healthy donor platelets or the use of hemodialysis and peritoneal dialysis can transiently restore circulating TP and temporarily reduce plasma levels of thymidine and deoxyuridine.837,838 However, these effects are short-lived and do not address neurological functions.837 HSCT offers a more definitive solution by permanently restoring TP function and thereby curing MNGIE.839 Both in vitro and in vivo studies have shown that HSCT can restore TP activity, correct the imbalance in the mitochondrial deoxyribonucleoside triphosphate pool in the liver, and safely halt disease progression.840,841 HSCT has been shown to correct the biochemical abnormalities and clinical manifestations of MNGIE,489,842 with a standardized protocol now in place.843 Recent research has also explored haploidentical transplantation as a potential treatment option.844 However, the timing of treatment, the preconditioning regimen, and donor selection are critical factors in determining therapeutic outcomes.839 HSCT is recommended for patients with the best donor match, younger age, and milder symptoms.489 Due to the increased risk of morbidity and mortality associated with advanced disease progression and complications, HSCT is not advised for patients with advanced MNGIE.489,845
Enzyme replacement therapy
The high mortality risk associated with HSCT and the scarcity of matched donors limit the accessibility of this treatment for many patients.844,845 An alternative approach involves lentiviral transduction of hematopoietic stem and erythroid cell lines to produce reticulocytes containing active TP.846 Erythrocyte-encapsulated thymidine phosphorylase (EE-TP) can catalyze the metabolism of thymidine and deoxyuridine, which freely diffuse across the red blood cell membrane, converting them into normal products.847 This process reduces plasma nucleosides, improves mitochondrial dysfunction, alleviates clinical symptoms, and has shown good safety and tolerability, making EE-TP a potential enzyme replacement therapy for MNGIE.848 However, the therapeutic effects are temporary, as metabolite levels return to abnormal values once treatment is discontinued.848 Regular intravenous injections of EE-TP can help manage intracellular nucleotide imbalance in patients with MNGIE,847 positioning EE-TP as a viable treatment option for those without suitable HSCT donors or for patients with irreversible end-stage disease.848 Nonetheless, preclinical toxicity assessments have indicated that EE-TP might pose severe toxicity risks in MNGIE, necessitating careful management of immune reactions.847 To address these concerns, Vocht et al. developed active TP-encapsulating nanoreactors, which, due to their stability and lack of cytotoxicity and inflammatory response, could offer a more effective and safer enzyme replacement therapy option.849
Organ transplantation therapy
Liver transplantation has emerged as a novel therapeutic option for mitochondrial genetic diseases involving the liver. The liver is a major source of TP,850 and in MNGIE, liver transplantation has been shown to restore TP activity, rapidly normalize nucleoside levels, and maintain them at stable low levels, leading to the improvement and stabilization of various clinical symptoms.851,852 While liver transplantation may not achieve complete clinical recovery, it offers a potentially safer alternative to allogeneic HSCT, especially in patients with underlying liver disease.851 Additionally, liver transplantation has been explored as a treatment for ethylmalonic encephalopathy.853 However, in an 18-month-old patient with ethylmalonic encephalopathy, liver transplantation only partially improved symptoms and did not result in a complete cure,854 suggesting that the procedure is most effective when performed before irreversible neurological damage occurs. In Wolcott-Rallison syndrome, which is caused by EIF2AK3 mutations, single or combined transplantation of the liver, pancreas, and kidneys has been associated with improved overall health, although further clinical follow-up is necessary to confirm these outcomes.855,856 The decision to perform liver transplantation in patients with mtDNA depletion syndromes and deoxyguanosine kinase deficiency remains controversial, as post-transplant outcomes for these conditions are generally poor.857
Exercise therapy
Aerobic training
Exercise intolerance is a prevalent symptom in patients with mitochondrial disease, directly correlating with the severity of impaired muscle OXPHOS.858,859 Reduced physical activity significantly contributes to decreased exercise capacity in individuals with mitochondrial myopathies.860 Several studies have shown that moderate-intensity aerobic training can enhance OXPHOS in skeletal muscle, promote mitochondrial proliferation, and increase the levels of respiratory chain enzymes.861 However, the impact of exercise on muscle mutational load remains unclear, with some studies suggesting that mutational load may increase with training,861 while others report no significant change in muscle mtDNA amounts or mutational load levels.859,860 Consequently, long-term studies are necessary to evaluate the safety and efficacy of exercise as a treatment for patients with mitochondrial myopathy. Exercise limitations in mitochondrial myopathy may also be compounded by chronic conditions such as cytochrome oxidase deficiency, in addition to primary mitochondrial dysfunction.862 Combining aerobic training with oral sodium dichloroacetate therapy could potentially improve aerobic capacity and motor function.862 Moreover, low-intensity aerobic exercise has been shown to facilitate targeted transgene delivery to specific organs, potentially enhancing the safety of gene therapy in human patients.661 Further research is needed to optimize exercise training regimens to maximize their benefits for patients with mitochondrial disease, ensuring both safety and therapeutic efficacy.
Resistance training
Endurance exercise promotes mitochondrial turnover, biogenesis, and angiogenesis by activating PGC-1α expression in skeletal muscle, thereby enhancing mitochondrial function.863,864 It also improves mitochondrial morphology and boosts antioxidant capacity.864 Training interventions that combine aerobic and resistance exercise have been shown to enhance mTOR-activated signaling pathways, PGC-1α signaling related to muscle mitochondrial biogenesis and anabolism, as well as OXPHOS complex activity and redox balance in muscle tissues.865 These combined improvements lead to increased aerobic fitness and muscle strength, as demonstrated in mice.865
Induced pluripotent stem cells and organoid
Recent advancements in generating patient-specific human iPSCs and their derived cells and organoids have established robust models for investigating the pathophysiological mechanisms of various mitochondrial genetic diseases, including MELAS,866 MERRF,283 Leigh syndrome,795 Friedreich’s ataxia,867 and LHON868 These iPSCs, along with their derived cells and organoids, address the challenges of sample acquisition while preserving patient-specific genetic backgrounds and accurately replicating disease characteristics.869 Notably, these technologies have become powerful tools for screening potential therapeutic drugs and developing novel treatment strategies. For example, Guo et al. reprogrammed skin fibroblasts from a patient with DGUOK-mutated mtDNA depletion syndrome into iPSCs, which were then differentiated into hepatocyte-like cells and liver organoids.869 This study not only uncovered the mechanism linking iron overload to hepatocyte death but also identified N-acetylcysteine as a potential therapeutic intervention to inhibit ferroptosis in patients with mtDNA depletion syndrome.869 Similarly, iPSCs derived from patients with Alpers’ syndrome, as well as their differentiated neural stem cells and cortical organoids, revealed that nicotinamide riboside could ameliorate mitochondrial defects and exert neuroprotective effects.870 Furthermore, researchers are also utilizing iPSCs derived from patients with mitochondrial diseases to investigate the role and efficacy of gene-editing tools.685 Extensive research highlights the potential of iPSCs and their derived cells and organoids in modeling mitochondrial genetic diseases and exploring therapeutic strategies, offering renewed hope to patients. However, mtDNA alterations may occur during iPSC reprogramming, potentially affecting the reprogramming and differentiation processes.871 These changes could lead to the emergence of immunogenic neo-epitopes.872 Therefore, monitoring and dynamic analysis of mtDNA integrity should be integral to the quality control processes in iPSC production to ensure both safety and efficacy.871
Overall, interventions such as MRT, gene therapy, pharmacological treatments, and cell therapy have demonstrated significant therapeutic potential in clinical trials for mitochondrial genetic disorders. These approaches primarily function through mechanisms including the replacement of defective mtDNA/nDNA, antioxidation, metabolic regulation, and mitochondrial protection. Although the widespread clinical application of these technologies continues to face numerous challenges, efforts can be made to overcome these issues. For example, reducing the possibility of genetic drift in MRT might be achievable through gene editing technologies or the development of new biomaterials, such as the generation of in vitro oocytes.873 Additionally, the timing of gene therapy, tailored approaches for different mtDNA/nDNA defects, and more efficient delivery systems will further enhance the efficacy and application of gene therapy. Research on personalized therapeutic strategies and the synergistic effects of multiple drugs could mitigate the variability in drug efficacy caused by factors such as administration methods, individual differences, and varying disease stages. Numerous clinically non-translational but potentially effective therapeutic approaches remain. Mesenchymal stem cells can transfer functionally normal mitochondria to cells with hereditary mtDNA defects via TNT and mitoEVs, thereby increasing the proportion of normal mtDNA, improving and sustaining mitochondrial function in the long term.874 Further in vivo studies on the long-term effects and mechanisms of mitochondrial transfer are essential to ensure the safety and efficacy of these therapies, ultimately optimizing cell therapy for clinical application.874 In mitochondrial diseases, oxidative stress due to the disruption of oxidative phosphorylation leads to an imbalance between oxygen delivery and utilization, resulting in oxygen toxicity.12 Hypoxia can reduce the production of free radicals or abnormal signaling substrates and activate the vHL-PHD-HIF hypoxic transcriptional program, which in turn activates glycolysis to generate ATP and reduces ROS production caused by impaired electron transfer in ETC, thereby rescuing the ETC-inhibited cellular phenotype.12 Reducing oxygen delivery and consumption may offer therapeutic or preventive benefits for mitochondrial diseases.12,875 Further research is required to optimize and investigate the long-term effects of hypoxia, as well as its applicability in humans.875 The advent of induced pluripotent stem cells and organoids also provides powerful models for exploring the pathogenesis and therapeutic strategies of mitochondrial genetic disorders, potentially leading to the discovery of more effective treatments.
Conclusion and Future perspectives
Since the first description of LHON in 1871,19 and the subsequent discoveries of the roles of mtDNA and nDNA in mitochondrial genetic disorders in 198832,33 and 1995,37 the field has transitioned from the pre-molecular era to the molecular era. This deeper understanding of mitochondrial biology has significantly advanced our ability to elucidate pathogenesis and explore effective therapies.
Significant progress has been made in understanding the pathogenesis, improving diagnosis, and developing treatments for mitochondrial diseases. Novel mechanisms of MQC, such as migrasome-mediated exocytosis,190 mitolysosome exocytosis,188 and mitopherogenesis,194 have been identified, highlighting the role of a robust MQC network in enhancing mitochondrial resilience to stress and damage. Beyond OXPHOS disturbances, the broader implications of mitochondrial dysfunction and its downstream effects are receiving increasing attention. Abnormalities induced by mtDNA or nDNA mutations—such as impaired mito-nuclear communication, mitochondrial dynamics, and mitophagy—are now recognized as key factors in mitochondrial diseases.126,227 Notably, mutation-specific and cell-specific mito-nuclear communication has been shown to significantly influence the tissue heterogeneity observed in these disorders.541 Advances in NGS and other technologies have markedly improved diagnostic accuracy. Promising therapeutic strategies are also advancing, with MRT showing significant potential in treating and preventing mitochondrial genetic diseases and their germline transmission.609 The success of ST technology in particular is encouraging.46 Gene therapy, especially AAV-mediated allogeneic expression and gene replacement, is another highly promising approach, with several therapies now in clinical trials.47 Additional interventions such as exercise,876 pharmacological treatments,48,754,826 cell therapy,843 enzyme replacement therapy,848 and organ transplantation877 have demonstrated beneficial effects on mitochondrial diseases.
However, several urgent challenges remain. The genotype-phenotype correlation in mitochondrial diseases is complex and not fully understood.4 Despite advancements in iPSC and organoid technology, as well as gene editing, developing accurate disease models continues to be a significant challenge, limiting research progress. Existing cell (cybrid) and animal models are insufficient, particularly due to issues such as tissue specificity and heteroplasmy shift.226 Additionally, not all mutations can be modeled, as some homozygous mutations result in embryonic lethality.369 In terms of diagnosis, DNA sequencing remains the primary method for detecting mitochondrial diseases, but it faces significant limitations. Secondary mtDNA mutations caused by nDNA mutations, tissue heteroplasmy, and overlapping phenotypes complicate sequencing efforts, leading to a low overall diagnostic yield.549,571 This often necessitates repeated and combined testing methods, increasing the economic burden. Regarding treatment, a complete cure for mitochondrial diseases remains elusive. The application of MRT faces potential limitations due to functional incompatibilities between nuclear and mitochondrial genomes,613 residual donor mtDNA,609 and ethical concerns.878 Current mtDNA editing approaches are limited to correcting a few specific point mutations,879 and concerns about off-target effects and safety undermine confidence in gene therapy.680,719 Additionally, allogeneically expressed mitochondrial proteins are highly hydrophobic, and only a small portion successfully localizes to mitochondria, limiting the effectiveness of allogeneic expression therapy.880 Significant challenges remain in overcoming these obstacles.
Future research should increasingly focus on the relationship between genetic defects and mitochondrial dysfunction, along with the subsequent consequences. Novel MQC mechanisms offer promising avenues for investigation in mitochondrial diseases. Delving into tissue-specific and cell-specific mito-nuclear communication pathways is crucial to understanding these conditions better. Epigenetics, particularly lactylation, may play a significant role in mitochondrial diseases and warrant further exploration. A deeper understanding of mito-nuclear communication could also provide solutions to mito-nuclear incompatibility issues encountered after MRT. Given the challenges of unstable diagnostic yield and variants of uncertain significance in genome sequencing, integrating multi-omics approaches as complementary diagnostic tools could enhance diagnostic accuracy. To address the delivery limitations of gene therapy, future efforts should concentrate on developing methods to efficiently introduce nuclear-encoded products into mitochondria to ensure they function correctly. Prioritizing the development of gene therapy tools that are more precise, efficient, minimally off-target, safer, and easier to deliver is essential. Additionally, the combination of gene therapy with MRT could yield unexpected therapeutic benefits. Therapies such as drugs and exercise also require clinical evidence-based guidance for their application. Notably, combining different therapeutic approaches may offer synergistic benefits. The use of iPSCs and organoid technologies should be expanded to create models for specific mutations associated with mitochondrial genetic disorders. These models would facilitate the study of disease phenotypes and post-treatment changes, advancing our understanding of disease mechanisms and aiding in the development of new treatment strategies. Exploring the differentiation of pluripotent stem cells into oocytes for MRT could reduce the need for egg retrieval from healthy donors and minimize the waste of oocytes. The application of iPSCs and organoids as models for gene therapy also holds significant potential.
This review systematically summarizes the physiological aspects of mitochondrial metabolism, the intracellular and intercellular MQC network, and mitochondrial inflammation and apoptosis. It highlights potential molecular mechanisms, recent diagnostic advancements, and therapeutic developments in mitochondrial diseases, contributing to a deeper understanding of these conditions and guiding future research design and clinical translation of precise diagnostics and effective therapies. The future directions for research, diagnosis, and treatment outlined here aim to inspire and inform subsequent studies.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82271110), the Undergraduate Training Program for Innovation and Entrepreneurship of Hunan Province (No. S202410533161), and the New Technology Incubation Funds in Ophthalmology. All figures in this manuscript are created by Adobe Illustrator 2022 (Adobe systems, USA).
Author contributions
Y.Z. conceptualized and designed the manuscript. H.W. and H.D. wrote and drafted the manuscript. H.W., H.D., B.L., J.C., and J.Z. contributed to the literature collection. Y.Z., X.Z., and S.Y. reviewed and revised the manuscript. All authors have read and approved the final version of the manuscript.
Competing interests
The authors declare no competing interest.
Footnotes
These authors contributed equally: Haipeng Wen, Hui Deng
References
- 1.Mitchell, P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature191, 144–148 (1961). [DOI] [PubMed] [Google Scholar]
- 2.Gorman, G. S. et al. Mitochondrial diseases. Nat. Rev. Dis. Prim.2, 16080 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Schapira, A. H. Mitochondrial diseases. Lancet379, 1825–1834 (2012). [DOI] [PubMed] [Google Scholar]
- 4.La Morgia, C., Maresca, A., Caporali, L., Valentino, M. L. & Carelli, V. Mitochondrial diseases in adults. J. Intern Med.287, 592–608 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Gropman, A. L. Diagnosis and treatment of childhood mitochondrial diseases. Curr. Neurol. Neurosci. Rep.1, 185–194 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Klopstock, T. et al. Mitochondrial Disorders. Dtsch. Arztebl. Int.118, 741–748 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCormick, E., Place, E. & Falk, M. J. Molecular genetic testing for mitochondrial disease: from one generation to the next. Neurotherapeutics10, 251–261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schon, K. R., Ratnaike, T., van den Ameele, J., Horvath, R. & Chinnery, P. F. Mitochondrial Diseases: A Diagnostic Revolution. Trends Genet36, 702–717 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Suomalainen, A. & Battersby, B. J. Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat. Rev. Mol. Cell Biol.19, 77–92 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Balsa, E. et al. Defective NADPH production in mitochondrial disease complex I causes inflammation and cell death. Nat. Commun.11, 2714 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Kleist-Retzow, J. C. et al. Impaired mitochondrial Ca2+ homeostasis in respiratory chain-deficient cells but efficient compensation of energetic disadvantage by enhanced anaerobic glycolysis due to low ATP steady state levels. Exp. Cell Res313, 3076–3089 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Jain, I. H. et al. Hypoxia as a therapy for mitochondrial disease. Science352, 54–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu, H. et al. Mitochondrial Unfolded Protein Response and Integrated Stress Response as Promising Therapeutic Targets for Mitochondrial Diseases. Cells12, 20 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eldeeb, M. A., Thomas, R. A., Ragheb, M. A., Fallahi, A. & Fon, E. A. Mitochondrial quality control in health and in Parkinson’s disease. Physiol. Rev.102, 1721–1755 (2022). [DOI] [PubMed] [Google Scholar]
- 15.Liu, L., Li, Y., Chen, G. & Chen, Q. Crosstalk between mitochondrial biogenesis and mitophagy to maintain mitochondrial homeostasis. J. Biomed. Sci.30, 86 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borcherding, N. & Brestoff, J. R. The power and potential of mitochondria transfer. Nature623, 283–291 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchi, S., Guilbaud, E., Tait, S. W. G., Yamazaki, T. & Galluzzi, L. Mitochondrial control of inflammation. Nat. Rev. Immunol.23, 159–173 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vafai, S. B. & Mootha, V. K. Mitochondrial disorders as windows into an ancient organelle. Nature491, 374–383 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Piotrowska, A., Korwin, M., Bartnik, E. & Tońska, K. Leber hereditary optic neuropathy - historical report in comparison with the current knowledge. Gene555, 41–49 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Luft, R., Ikkos, D., Palmieri, G., Ernster, L. & Afzelius, B. A case of severe hypermetabolism of nonthyroid origin with a defect in the maintenance of mitochondrial respiratory control: a correlated clinical, biochemical, and morphological study. J. Clin. Invest41, 1776–1804 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimauro, S. A history of mitochondrial diseases. J. Inherit. Metab. Dis.34, 261–276 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Shy, G. M., Gonatas, N. K. & Perez, M. Two childhood myopathies with abnormal mitochondria. I. Megaconial myopathy. II. Pleoconial myopathy. Brain89, 133–158 (1966). [DOI] [PubMed] [Google Scholar]
- 23.Shy, G. M. & Gonatas, N. K. HUMAN MYOPATHY WITH GIANT ABNORMAL MITOCHONDRIA. Science145, 493–496 (1964). [DOI] [PubMed] [Google Scholar]
- 24.Gonatas, N. K. & Shy, G. M. in Proceedings of the Vth International Congress of Neuropathology Vol. 100 606-612 (Amsterdam: Excerpta Medica International Congress Series, 1965).
- 25.Engel, W. K. & Cunningham, G. G. RAPID EXAMINATION OF MUSCLE TISSUE. AN IMPROVED TRICHROME METHOD FOR FRESH-FROZEN BIOPSY SECTIONS. Neurology13, 919–923 (1963). [DOI] [PubMed] [Google Scholar]
- 26.Blass, J. P., Avigan, J. & Uhlendorf, B. W. A defect in pyruvate decarboxylase in a child with an intermittent movement disorder. J. Clin. Invest49, 423–432 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engel, A. G. & Angelini, C. Carnitine deficiency of human skeletal muscle with associated lipid storage myopathy: a new syndrome. Science179, 899–902 (1973). [DOI] [PubMed] [Google Scholar]
- 28.DiMauro, S. & DiMauro, P. M. Muscle carnitine palmityltransferase deficiency and myoglobinuria. Science182, 929–931 (1973). [DOI] [PubMed] [Google Scholar]
- 29.Willems, J. L. et al. Leigh’s encephalomyelopathy in a patient with cytochrome c oxidase deficiency in muscle tissue. Pediatrics60, 850–857 (1977). [PubMed] [Google Scholar]
- 30.Shapira, Y., Harel, S. & Russell, A. Mitochondrial encephalomyopathies: a group of neuromuscular disorders with defects in oxidative metabolism. Isr. J. Med Sci.13, 161–164 (1977). [PubMed] [Google Scholar]
- 31.Anderson, S. et al. Sequence and organization of the human mitochondrial genome. Nature290, 457–465 (1981). [DOI] [PubMed] [Google Scholar]
- 32.Holt, I. J., Harding, A. E. & Morgan-Hughes, J. A. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature331, 717–719 (1988). [DOI] [PubMed] [Google Scholar]
- 33.Wallace, D. C. et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science242, 1427–1430 (1988). [DOI] [PubMed] [Google Scholar]
- 34.DiMauro, S. & Garone, C. Historical perspective on mitochondrial medicine. Dev. Disabil. Res Rev.16, 106–113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeviani, M. et al. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature339, 309–311 (1989). [DOI] [PubMed] [Google Scholar]
- 36.Moraes, C. T. et al. mtDNA depletion with variable tissue expression: a novel genetic abnormality in mitochondrial diseases. Am. J. Hum. Genet48, 492–501 (1991). [PMC free article] [PubMed] [Google Scholar]
- 37.Bourgeron, T. et al. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat. Genet11, 144–149 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Kogelnik, A. M., Lott, M. T., Brown, M. D., Navathe, S. B. & Wallace, D. C. MITOMAP: a human mitochondrial genome database. Nucleic Acids Res24, 177–179 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishino, I., Spinazzola, A. & Hirano, M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science283, 689–692 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Vasta, V., Ng, S. B., Turner, E. H., Shendure, J. & Hahn, S. H. Next generation sequence analysis for mitochondrial disorders. Genome Med1, 100 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahman, J. & Rahman, S. Mitochondrial medicine in the omics era. Lancet391, 2560–2574 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Taylor, R. W., Chinnery, P. F., Turnbull, D. M. & Lightowlers, R. N. Selective inhibition of mutant human mitochondrial DNA replication in vitro by peptide nucleic acids. Nat. Genet15, 212–215 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Spees, J. L., Olson, S. D., Whitney, M. J. & Prockop, D. J. Mitochondrial transfer between cells can rescue aerobic respiration. Proc. Natl. Acad. Sci. USA103, 1283–1288 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tachibana, M. et al. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature461, 367–372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amore, G. et al. Therapeutic Options in Hereditary Optic Neuropathies. Drugs81, 57–86 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, J. et al. Live birth derived from oocyte spindle transfer to prevent mitochondrial disease. Reprod. Biomed. Online34, 361–368 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Vignal, C. et al. Safety of rAAV2/2-ND4 Gene Therapy for Leber Hereditary Optic Neuropathy. Ophthalmology125, 945–947 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Pilotto, F., Chellapandi, D. M. & Puccio, H. Omaveloxolone: a groundbreaking milestone as the first FDA-approved drug for Friedreich ataxia. Trends Mol. Med30, 117–125 (2024). [DOI] [PubMed] [Google Scholar]
- 49.Thorburn, D. R. Mitochondrial disorders: prevalence, myths and advances. J. Inherit. Metab. Dis.27, 349–362 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Chinnery, P. F. et al. The epidemiology of pathogenic mitochondrial DNA mutations. Ann. Neurol.48, 188–193 (2000). [PubMed] [Google Scholar]
- 51.Hong, S., Kim, S., Kim, K. & Lee, H. Clinical Approaches for Mitochondrial Diseases. Cells12, 2494 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart, J. B. & Chinnery, P. F. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet16, 530–542 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Chinnery, P. F. & Hudson, G. Mitochondrial genetics. Br. Med Bull.106, 135–159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russell, O. M., Gorman, G. S., Lightowlers, R. N. & Turnbull, D. M. Mitochondrial Diseases: Hope for the Future. Cell181, 168–188 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Elliott, H. R., Samuels, D. C., Eden, J. A., Relton, C. L. & Chinnery, P. F. Pathogenic mitochondrial DNA mutations are common in the general population. Am. J. Hum. Genet83, 254–260 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahuja, A. S. Understanding mitochondrial myopathies: a review. PeerJ6, e4790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gray, M. W. The pre-endosymbiont hypothesis: a new perspective on the origin and evolution of mitochondria. Cold Spring Harb. Perspect. Biol.6, a016097 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Supinski, G. S., Schroder, E. A. & Callahan, L. A. Mitochondria and Critical Illness. Chest157, 310–322 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mourier, T., Hansen, A. J., Willerslev, E. & Arctander, P. The Human Genome Project reveals a continuous transfer of large mitochondrial fragments to the nucleus. Mol. Biol. Evol.18, 1833–1837 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Mercer, T. R. et al. The human mitochondrial transcriptome. Cell146, 645–658 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crews, S., Ojala, D., Posakony, J., Nishiguchi, J. & Attardi, G. Nucleotide sequence of a region of human mitochondrial DNA containing the precisely identified origin of replication. Nature277, 192–198 (1979). [DOI] [PubMed] [Google Scholar]
- 62.Ott, M., Amunts, A. & Brown, A. Organization and Regulation of Mitochondrial Protein Synthesis. Annu Rev. Biochem85, 77–101 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Sun, W. et al. Mitochondrial Non-Coding RNAs Are Potential Mediators of Mitochondrial Homeostasis. Biomolecules12, 1863 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vendramin, R., Marine, J. C. & Leucci, E. Non-coding RNAs: the dark side of nuclear-mitochondrial communication. Embo j.36, 1123–1133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, Z. et al. Mitochondrial Genome-Derived circRNA mc-COX2 Functions as an Oncogene in Chronic Lymphocytic Leukemia. Mol. Ther. Nucleic Acids20, 801–811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu, X. et al. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China Life Sci.63, 1429–1449 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Rackham, O. et al. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. Rna17, 2085–2093 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tomar, A. et al. Epigenetic inheritance of diet-induced and sperm-borne mitochondrial RNAs. Nature630, 720–727 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meseguer, S. et al. The MELAS mutation m.3243A>G alters the expression of mitochondrial tRNA fragments. Biochim Biophys. Acta Mol. Cell Res1866, 1433–1449 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Pagliarini, D. J. et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell134, 112–123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baker, Z. N., Forny, P. & Pagliarini, D. J. Mitochondrial proteome research: the road ahead. Nat. Rev. Mol. Cell Biol.25, 65–82 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morgenstern, M. et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep.19, 2836–2852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wachoski-Dark, E., Zhao, T., Khan, A., Shutt, T. E. & Greenway, S. C. Mitochondrial Protein Homeostasis and Cardiomyopathy. Int. J. Mol. Sci.23, 3353 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benayoun, B. A. & Lee, C. MOTS-c: A Mitochondrial-Encoded Regulator of the Nucleus. Bioessays41, e1900046 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li, Y. et al. Mitochondrial-derived peptides in cardiovascular disease: Novel insights and therapeutic opportunities. J. Adv. Res64, 99–115 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu, Z. et al. A novel protein CYTB-187AA encoded by the mitochondrial gene CYTB modulates mammalian early development. Cell Metab.36, 1586–1597.e1587 (2024). [DOI] [PubMed] [Google Scholar]
- 77.Sreekumar, P. G. & Kannan, R. Mechanisms of protection of retinal pigment epithelial cells from oxidant injury by humanin and other mitochondrial-derived peptides: Implications for age-related macular degeneration. Redox Biol.37, 101663 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller, B., Kim, S. J., Kumagai, H., Yen, K. & Cohen, P. Mitochondria-derived peptides in aging and healthspan. J. Clin. Invest132, e158449 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim, K. H., Son, J. M., Benayoun, B. A. & Lee, C. The Mitochondrial-Encoded Peptide MOTS-c Translocates to the Nucleus to Regulate Nuclear Gene Expression in Response to Metabolic Stress. Cell Metab.28, 516–524.e517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller, B. et al. Mitochondrial DNA variation in Alzheimer’s disease reveals a unique microprotein called SHMOOSE. Mol. Psychiatry28, 1813–1826 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Protasoni, M. & Zeviani, M. Mitochondrial Structure and Bioenergetics in Normal and Disease Conditions. Int. J. Mol. Sci.22, 586 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen, L. et al. Mitochondrial heterogeneity in diseases. Signal. Transduct. Target Ther.8, 311 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zorova, L. D. et al. Mitochondrial membrane potential. Anal. Biochem552, 50–59 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rottenberg, H. The Reduction in the Mitochondrial Membrane Potential in Aging: The Role of the Mitochondrial Permeability Transition Pore. Int. J. Mol. Sci.24, 12295 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilson, D. F. Oxidative phosphorylation: regulation and role in cellular and tissue metabolism. J. Physiol.595, 7023–7038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Frey, T. G. & Mannella, C. A. The internal structure of mitochondria. Trends Biochem. Sci.25, 319–324 (2000). [DOI] [PubMed] [Google Scholar]
- 87.Spinelli, J. B. & Haigis, M. C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol.20, 745–754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nesci, S. et al. Molecular and Supramolecular Structure of the Mitochondrial Oxidative Phosphorylation System: Implications for Pathology. Life (Basel)11, 242 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tang, J. X., Thompson, K., Taylor, R. W. & Oláhová, M. Mitochondrial OXPHOS Biogenesis: Co-Regulation of Protein Synthesis, Import, and Assembly Pathways. Int. J. Mol. Sci.21, 3820 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mergenthaler, P., Lindauer, U., Dienel, G. A. & Meisel, A. Sugar for the brain: the role of glucose in physiological and pathological brain function. Trends Neurosci.36, 587–597 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vuoristo, K. S., Mars, A. E., Sanders, J. P. M., Eggink, G. & Weusthuis, R. A. Metabolic Engineering of TCA Cycle for Production of Chemicals. Trends Biotechnol.34, 191–197 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Srere, P. A. The Molecular Physiology of Citrate. Nature205, 766–770 (1965). [Google Scholar]
- 93.Srere, P. A. The citrate cleavage enzyme. I. Distribution and purification. J. Biol. Chem.234, 2544–2547 (1959). [PubMed] [Google Scholar]
- 94.Shi, L. & Tu, B. P. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr. Opin. Cell Biol.33, 125–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rui, L. Energy metabolism in the liver. Compr. Physiol.4, 177–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fu, Z. et al. Dyslipidemia in retinal metabolic disorders. EMBO Mol. Med11, e10473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Adeva-Andany, M. M., Carneiro-Freire, N., Seco-Filgueira, M., Fernández-Fernández, C. & Mouriño-Bayolo, D. Mitochondrial β-oxidation of saturated fatty acids in humans. Mitochondrion46, 73–90 (2019). [DOI] [PubMed] [Google Scholar]
- 98.Nguyen, P. et al. Liver lipid metabolism. J. Anim. Physiol. Anim. Nutr. (Berl.)92, 272–283 (2008). [DOI] [PubMed] [Google Scholar]
- 99.Chandel, N. S. Lipid Metabolism. Cold Spring Harb. Perspect. Biol.13, a040576 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Staerck, C. et al. Microbial antioxidant defense enzymes. Micro Pathog.110, 56–65 (2017). [DOI] [PubMed] [Google Scholar]
- 101.Turrens, J. F. Mitochondrial formation of reactive oxygen species. J. Physiol.552, 335–344 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zorov, D. B., Juhaszova, M. & Sollott, S. J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev.94, 909–950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bhatti, J. S., Bhatti, G. K. & Reddy, P. H. Mitochondrial dysfunction and oxidative stress in metabolic disorders - A step towards mitochondria based therapeutic strategies. Biochim Biophys. Acta Mol. Basis Dis.1863, 1066–1077 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Murphy, M. P. How mitochondria produce reactive oxygen species. Biochem J.417, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang, B. et al. Role of mitochondrial reactive oxygen species in homeostasis regulation. Redox Rep.27, 45–52 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brown, G. C. & Borutaite, V. There is no evidence that mitochondria are the main source of reactive oxygen species in mammalian cells. Mitochondrion12, 1–4 (2012). [DOI] [PubMed] [Google Scholar]
- 107.Sies, H. & Jones, D. P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol.21, 363–383 (2020). [DOI] [PubMed] [Google Scholar]
- 108.Ray, P. D., Huang, B. W. & Tsuji, Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal24, 981–990 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Le Belle, J. E. et al. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell8, 59–71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jang, Y. Y. & Sharkis, S. J. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood110, 3056–3063 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hayashi, G. & Cortopassi, G. Oxidative stress in inherited mitochondrial diseases. Free Radic. Biol. Med88, 10–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bertero, E. & Maack, C. Calcium Signaling and Reactive Oxygen Species in Mitochondria. Circ. Res122, 1460–1478 (2018). [DOI] [PubMed] [Google Scholar]
- 113.Shadel, G. S. & Horvath, T. L. Mitochondrial ROS signaling in organismal homeostasis. Cell163, 560–569 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cross, C. E. et al. Oxygen radicals and human disease. Ann. Intern Med107, 526–545 (1987). [DOI] [PubMed] [Google Scholar]
- 115.Zorov, D. B., Filburn, C. R., Klotz, L. O., Zweier, J. L. & Sollott, S. J. Reactive oxygen species (ROS)-induced ROS release: a new phenomenon accompanying induction of the mitochondrial permeability transition in cardiac myocytes. J. Exp. Med192, 1001–1014 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brenner, C. & Grimm, S. The permeability transition pore complex in cancer cell death. Oncogene25, 4744–4756 (2006). [DOI] [PubMed] [Google Scholar]
- 117.Murakami, Y. et al. Photoreceptor cell death and rescue in retinal detachment and degenerations. Prog. Retin Eye Res37, 114–140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chang, X. et al. Zishen Tongyang Huoxue decoction (TYHX) alleviates sinoatrial node cell ischemia/reperfusion injury by directing mitochondrial quality control via the VDAC1-β-tubulin signaling axis. J. Ethnopharmacol.320, 117371 (2024). [DOI] [PubMed] [Google Scholar]
- 119.De Nicolo, B., Cataldi-Stagetti, E., Diquigiovanni, C. & Bonora, E. Calcium and Reactive Oxygen Species Signaling Interplays in Cardiac Physiology and Pathologies. Antioxid. (Basel)12, 353 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chang, X. et al. Zishenhuoxue decoction-induced myocardial protection against ischemic injury through TMBIM6-VDAC1-mediated regulation of calcium homeostasis and mitochondrial quality surveillance. Phytomedicine132, 155331 (2023). [DOI] [PubMed] [Google Scholar]
- 121.Li, Y., Yu, J., Li, R., Zhou, H. & Chang, X. New insights into the role of mitochondrial metabolic dysregulation and immune infiltration in septic cardiomyopathy by integrated bioinformatics analysis and experimental validation. Cell Mol. Biol. Lett.29, 21 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chang, X. et al. Molecular Mechanisms of Mitochondrial Quality Control in Ischemic Cardiomyopathy. Int. J. Biol. Sci.19, 426–448 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu, B. H. et al. Mitochondrial quality control in human health and disease. Mil. Med Res11, 32 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chang, X. et al. Quercetin inhibits necroptosis in cardiomyocytes after ischemia-reperfusion via DNA-PKcs-SIRT5-orchestrated mitochondrial quality control. Phytother. Res38, 2496–2517 (2024). [DOI] [PubMed] [Google Scholar]
- 125.Khan, N. A. et al. mTORC1 Regulates Mitochondrial Integrated Stress Response and Mitochondrial Myopathy Progression. Cell Metab.26, 419–428.e415 (2017). [DOI] [PubMed] [Google Scholar]
- 126.Liu, S., Liu, S. & Jiang, H. Multifaceted roles of mitochondrial stress responses under ETC dysfunction - repair, destruction and pathogenesis. Febs j.289, 6994–7013 (2022). [DOI] [PubMed] [Google Scholar]
- 127.Shpilka, T. & Haynes, C. M. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol.19, 109–120 (2018). [DOI] [PubMed] [Google Scholar]
- 128.Costa-Mattioli, M. & Walter, P. The integrated stress response: From mechanism to disease. Science368, eaat5314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Harding, H. P. et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell6, 1099–1108 (2000). [DOI] [PubMed] [Google Scholar]
- 130.Zhou, D. et al. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J. Biol. Chem.283, 7064–7073 (2008). [DOI] [PubMed] [Google Scholar]
- 131.Jousse, C. et al. Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5’UTR. Nucleic Acids Res29, 4341–4351 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sturm, G. et al. OxPhos defects cause hypermetabolism and reduce lifespan in cells and in patients with mitochondrial diseases. Commun. Biol.6, 22 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jornayvaz, F. R. & Shulman, G. I. Regulation of mitochondrial biogenesis. Essays Biochem.47, 69–84 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Puigserver, P. et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell92, 829–839 (1998). [DOI] [PubMed] [Google Scholar]
- 135.Virbasius, J. V. & Scarpulla, R. C. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl. Acad. Sci. USA91, 1309–1313 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cameron, R. B., Beeson, C. C. & Schnellmann, R. G. Development of Therapeutics That Induce Mitochondrial Biogenesis for the Treatment of Acute and Chronic Degenerative Diseases. J. Med Chem.59, 10411–10434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Collins, T. J., Berridge, M. J., Lipp, P. & Bootman, M. D. Mitochondria are morphologically and functionally heterogeneous within cells. Embo j.21, 1616–1627 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chan, D. C. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev. Genet46, 265–287 (2012). [DOI] [PubMed] [Google Scholar]
- 139.Mishra, P. & Chan, D. C. Metabolic regulation of mitochondrial dynamics. J. Cell Biol.212, 379–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zorov, D. B. et al. Lessons from the Discovery of Mitochondrial Fragmentation (Fission): A Review and Update. Cells8, 175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Twig, G. et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo j.27, 433–446 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang, X., Zhou, H. & Chang, X. Involvement of mitochondrial dynamics and mitophagy in diabetic endothelial dysfunction and cardiac microvascular injury. Arch. Toxicol.97, 3023–3035 (2023). [DOI] [PubMed] [Google Scholar]
- 143.Tokuyama, T. et al. Mitochondrial Dynamics Regulation in Skin Fibroblasts from Mitochondrial Disease Patients. Biomolecules10, 0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Quiles, J. M. & Gustafsson, Å. B. The role of mitochondrial fission in cardiovascular health and disease. Nat. Rev. Cardiol.19, 723–736 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Smirnova, E., Griparic, L., Shurland, D. L. & van der Bliek, A. M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell12, 2245–2256 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.James, D. I., Parone, P. A., Mattenberger, Y. & Martinou, J. C. hFis1, a novel component of the mammalian mitochondrial fission machinery. J. Biol. Chem.278, 36373–36379 (2003). [DOI] [PubMed] [Google Scholar]
- 147.Otera, H. et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol.191, 1141–1158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Palmer, C. S. et al. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep.12, 565–573 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yu, R., Jin, S. B., Lendahl, U., Nistér, M. & Zhao, J. Human Fis1 regulates mitochondrial dynamics through inhibition of the fusion machinery. Embo j.38, e99748 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ishihara, N., Eura, Y. & Mihara, K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell Sci.117, 6535–6546 (2004). [DOI] [PubMed] [Google Scholar]
- 151.Cipolat, S., Martins de Brito, O., Dal Zilio, B. & Scorrano, L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA101, 15927–15932 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ishihara, N., Fujita, Y., Oka, T. & Mihara, K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. Embo j.25, 2966–2977 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ban, T. et al. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat. Cell Biol.19, 856–863 (2017). [DOI] [PubMed] [Google Scholar]
- 154.Mishra, P., Carelli, V., Manfredi, G. & Chan, D. C. Proteolytic cleavage of Opa1 stimulates mitochondrial inner membrane fusion and couples fusion to oxidative phosphorylation. Cell Metab.19, 630–641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ge, Y. et al. Two forms of Opa1 cooperate to complete fusion of the mitochondrial inner-membrane. Elife9, e50973 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wallace, D. C. & Chalkia, D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol.5, a021220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Youle, R. J. & van der Bliek, A. M. Mitochondrial fission, fusion, and stress. Science337, 1062–1065 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Labbé, K., Murley, A. & Nunnari, J. Determinants and functions of mitochondrial behavior. Annu Rev. Cell Dev. Biol.30, 357–391 (2014). [DOI] [PubMed] [Google Scholar]
- 159.Granatiero, V. & Manfredi, G. Mitochondrial Transport and Turnover in the Pathogenesis of Amyotrophic Lateral Sclerosis. Biol. (Basel)8, 36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zheng, Y. R., Zhang, X. N. & Chen, Z. Mitochondrial transport serves as a mitochondrial quality control strategy in axons: Implications for central nervous system disorders. CNS Neurosci. Ther.25, 876–886 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sheng, Z. H. & Cai, Q. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat. Rev. Neurosci.13, 77–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kumar Sharma, R., Chafik, A. & Bertolin, G. Mitochondrial transport, partitioning, and quality control at the heart of cell proliferation and fate acquisition. Am. J. Physiol. Cell Physiol.322, C311–c325 (2022). [DOI] [PubMed] [Google Scholar]
- 163.Lemasters, J. J. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res8, 3–5 (2005). [DOI] [PubMed] [Google Scholar]
- 164.Ploumi, C., Daskalaki, I. & Tavernarakis, N. Mitochondrial biogenesis and clearance: a balancing act. Febs j.284, 183–195 (2017). [DOI] [PubMed] [Google Scholar]
- 165.Gao, K., Chen, Y., Mo, R. & Wang, C. Excessive BNIP3- and BNIP3L-dependent mitophagy underlies the pathogenesis of FBXL4-mutated mitochondrial DNA depletion syndrome. Autophagy20, 460–462 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen, Y. et al. FBXL4 mutations cause excessive mitophagy via BNIP3/BNIP3L accumulation leading to mitochondrial DNA depletion syndrome. Cell Death Differ.30, 2351–2363 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cho, H. M. et al. Drp1-Zip1 Interaction Regulates Mitochondrial Quality Surveillance System. Mol. Cell73, 364–376.e368 (2019). [DOI] [PubMed] [Google Scholar]
- 168.de Vries, R. L., Gilkerson, R. W., Przedborski, S. & Schon, E. A. Mitophagy in cells with mtDNA mutations: being sick is not enough. Autophagy8, 699–700 (2012). [DOI] [PubMed] [Google Scholar]
- 169.Redza-Dutordoir, M. & Averill-Bates, D. A. Interactions between reactive oxygen species and autophagy: Special issue: Death mechanisms in cellular homeostasis. Biochim Biophys. Acta Mol. Cell Res1868, 119041 (2021). [DOI] [PubMed] [Google Scholar]
- 170.Frank, M. et al. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim Biophys. Acta1823, 2297–2310 (2012). [DOI] [PubMed] [Google Scholar]
- 171.Xu, Y., Shen, J. & Ran, Z. Emerging views of mitophagy in immunity and autoimmune diseases. Autophagy16, 3–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lu, Y. et al. Cellular mitophagy: Mechanism, roles in diseases and small molecule pharmacological regulation. Theranostics13, 736–766 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Deas, E. et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum. Mol. Genet20, 867–879 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Matsuda, N. et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol.189, 211–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Koyano, F. et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature510, 162–166 (2014). [DOI] [PubMed] [Google Scholar]
- 176.Harper, J. W., Ordureau, A. & Heo, J. M. Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol.19, 93–108 (2018). [DOI] [PubMed] [Google Scholar]
- 177.Lazarou, M. et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature524, 309–314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Geisler, S. et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol.12, 119–131 (2010). [DOI] [PubMed] [Google Scholar]
- 179.Sarraf, S. A. et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature496, 372–376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chen, Y. & Dorn, G. W. 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science340, 471–475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Heo, J. M., Ordureau, A., Paulo, J. A., Rinehart, J. & Harper, J. W. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol. Cell60, 7–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Palikaras, K., Lionaki, E. & Tavernarakis, N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol.20, 1013–1022 (2018). [DOI] [PubMed] [Google Scholar]
- 183.Shu, L. et al. ATAD3B is a mitophagy receptor mediating clearance of oxidative stress-induced damaged mitochondrial DNA. Embo j.40, e106283 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Liu, Y. et al. Mitochondrial transfer between cell crosstalk - An emerging role in mitochondrial quality control. Ageing Res Rev.91, 102038 (2023). [DOI] [PubMed] [Google Scholar]
- 185.Liu, D. et al. Intercellular mitochondrial transfer as a means of tissue revitalization. Signal Transduct. Target Ther.6, 65 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sugiura, A., McLelland, G. L., Fon, E. A. & McBride, H. M. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. Embo j.33, 2142–2156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Todkar, K. et al. Selective packaging of mitochondrial proteins into extracellular vesicles prevents the release of mitochondrial DAMPs. Nat. Commun.12, 1971 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bao, F. et al. Mitolysosome exocytosis, a mitophagy-independent mitochondrial quality control in flunarizine-induced parkinsonism-like symptoms. Sci. Adv.8, eabk2376 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bao, F., Zhou, L., Xiao, J. & Liu, X. Mitolysosome exocytosis: a novel mitochondrial quality control pathway linked with parkinsonism-like symptoms. Biochem Soc. Trans.50, 1773–1783 (2022). [DOI] [PubMed] [Google Scholar]
- 190.Jiao, H. et al. Mitocytosis, a migrasome-mediated mitochondrial quality-control process. Cell184, 2896–2910.e2813 (2021). [DOI] [PubMed] [Google Scholar]
- 191.Ma, L. et al. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res25, 24–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fan, C. et al. Cell migration orchestrates migrasome formation by shaping retraction fibers. J. Cell Biol.221, e202109168 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Huang, Y. et al. Migrasome formation is mediated by assembly of micron-scale tetraspanin macrodomains. Nat. Cell Biol.21, 991–1002 (2019). [DOI] [PubMed] [Google Scholar]
- 194.Liu, P. et al. Mitopherogenesis, a form of mitochondria-specific ectocytosis, regulates sperm mitochondrial quantity and fertility. Nat. Cell Biol.25, 1625–1636 (2023). [DOI] [PubMed] [Google Scholar]
- 195.Tan, H. W. S. et al. A degradative to secretory autophagy switch mediates mitochondria clearance in the absence of the mATG8-conjugation machinery. Nat. Commun.13, 3720 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Galluzzi, L. et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ.25, 486–541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lenaers, G. et al. Dominant optic atrophy: Culprit mitochondria in the optic nerve. Prog. Retin Eye Res83, 100935 (2021). [DOI] [PubMed] [Google Scholar]
- 198.Chen, B. S., Yu-Wai-Man, P. & Newman, N. J. Developments in the Treatment of Leber Hereditary Optic Neuropathy. Curr. Neurol. Neurosci. Rep.22, 881–892 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Czabotar, P. E. & Garcia-Saez, A. J. Mechanisms of BCL-2 family proteins in mitochondrial apoptosis. Nat. Rev. Mol. Cell Biol.24, 732–748 (2023). [DOI] [PubMed] [Google Scholar]
- 200.Estaquier, J., Vallette, F., Vayssiere, J. L. & Mignotte, B. The mitochondrial pathways of apoptosis. Adv. Exp. Med Biol.942, 157–183 (2012). [DOI] [PubMed] [Google Scholar]
- 201.Subburaj, Y. et al. Bax monomers form dimer units in the membrane that further self-assemble into multiple oligomeric species. Nat. Commun.6, 8042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Vringer, E. & Tait, S. W. G. Mitochondria and cell death-associated inflammation. Cell Death Differ.30, 304–312 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bibo-Verdugo, B. & Salvesen, G. S. Caspase mechanisms in the regulation of inflammation. Mol. Asp. Med88, 101085 (2022). [DOI] [PubMed] [Google Scholar]
- 204.Bock, F. J. & Tait, S. W. G. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol.21, 85–100 (2020). [DOI] [PubMed] [Google Scholar]
- 205.Rodríguez-Nuevo, A. & Zorzano, A. The sensing of mitochondrial DAMPs by non-immune cells. Cell Stress3, 195–207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Krysko, D. V. et al. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol.32, 157–164 (2011). [DOI] [PubMed] [Google Scholar]
- 207.Nakahira, K., Hisata, S. & Choi, A. M. The Roles of Mitochondrial Damage-Associated Molecular Patterns in Diseases. Antioxid. Redox Signal23, 1329–1350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kim, J., Kim, H. S. & Chung, J. H. Molecular mechanisms of mitochondrial DNA release and activation of the cGAS-STING pathway. Exp. Mol. Med55, 510–519 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Shang, D. et al. mtDNA Maintenance and Alterations in the Pathogenesis of Neurodegenerative Diseases. Curr. Neuropharmacol.21, 578–598 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.West, A. P. & Shadel, G. S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol.17, 363–375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kato, K., Omura, H., Ishitani, R. & Nureki, O. Cyclic GMP-AMP as an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Annu Rev. Biochem86, 541–566 (2017). [DOI] [PubMed] [Google Scholar]
- 212.Zhang, C. et al. Structural basis of STING binding with and phosphorylation by TBK1. Nature567, 394–398 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tanaka, Y. & Chen, Z. J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal5, ra20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Fitzgerald, K. A. et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol.4, 491–496 (2003). [DOI] [PubMed] [Google Scholar]
- 215.Brokatzky, D. et al. A non-death function of the mitochondrial apoptosis apparatus in immunity. Embo j.38, e100907 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.McArthur, K. et al. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science359, eaao6047 (2018). [DOI] [PubMed] [Google Scholar]
- 217.Ning, X. et al. Apoptotic Caspases Suppress Type I Interferon Production via the Cleavage of cGAS, MAVS, and IRF3. Mol. Cell74, 19–31.e17 (2019). [DOI] [PubMed] [Google Scholar]
- 218.van Horssen, J., van Schaik, P. & Witte, M. Inflammation and mitochondrial dysfunction: A vicious circle in neurodegenerative disorders? Neurosci. Lett.710, 132931 (2019). [DOI] [PubMed] [Google Scholar]
- 219.Warren, E. B. et al. Inflammatory and interferon gene expression signatures in patients with mitochondrial disease. J. Transl. Med21, 331 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kwon, H. S. & Koh, S. H. Neuroinflammation in neurodegenerative disorders: the roles of microglia and astrocytes. Transl. Neurodegener.9, 42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Butterfield, D. A. & Halliwell, B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat. Rev. Neurosci.20, 148–160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Li, A. L. et al. The role of mitochondria in myocardial damage caused by energy metabolism disorders: From mechanisms to therapeutics. Free Radic. Biol. Med208, 236–251 (2023). [DOI] [PubMed] [Google Scholar]
- 223.Pacheu-Grau, D., Rucktäschel, R. & Deckers, M. Mitochondrial dysfunction and its role in tissue-specific cellular stress. Cell Stress2, 184–199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Cheng, X. T., Huang, N. & Sheng, Z. H. Programming axonal mitochondrial maintenance and bioenergetics in neurodegeneration and regeneration. Neuron110, 1899–1923 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Olsen, R. K., Cornelius, N. & Gregersen, N. Redox signalling and mitochondrial stress responses; lessons from inborn errors of metabolism. J. Inherit. Metab. Dis.38, 703–719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tolle, I., Tiranti, V. & Prigione, A. Modeling mitochondrial DNA diseases: from base editing to pluripotent stem-cell-derived organoids. EMBO Rep.24, e55678 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Stenton, S. L. & Prokisch, H. Genetics of mitochondrial diseases: Identifying mutations to help diagnosis. EBioMedicine56, 102784 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Stewart, J. B. & Chinnery, P. F. Extreme heterogeneity of human mitochondrial DNA from organelles to populations. Nat. Rev. Genet22, 106–118 (2021). [DOI] [PubMed] [Google Scholar]
- 229.Aryaman, J., Johnston, I. G. & Jones, N. S. Mitochondrial Heterogeneity. Front Genet9, 718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.McFarland, R., Taylor, R. W. & Turnbull, D. M. A neurological perspective on mitochondrial disease. Lancet Neurol.9, 829–840 (2010). [DOI] [PubMed] [Google Scholar]
- 231.Yu-Wai-Man, P., Griffiths, P. G., Hudson, G. & Chinnery, P. F. Inherited mitochondrial optic neuropathies. J. Med Genet46, 145–158 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kirkman, M. A. et al. Quality of life in patients with leber hereditary optic neuropathy. Invest Ophthalmol. Vis. Sci.50, 3112–3115 (2009). [DOI] [PubMed] [Google Scholar]
- 233.Yu, D. Y. et al. Retinal ganglion cells: Energetics, compartmentation, axonal transport, cytoskeletons and vulnerability. Prog. Retin Eye Res36, 217–246 (2013). [DOI] [PubMed] [Google Scholar]
- 234.Zeviani, M. & Carelli, V. Mitochondrial Retinopathies. Int. J. Mol. Sci.23, 210 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Fuller, J. T. et al. Coenzyme Q10 trapping in mitochondrial complex I underlies Leber’s hereditary optic neuropathy. Proc. Natl. Acad. Sci. USA120, e2304884120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Bristow, E. A., Griffiths, P. G., Andrews, R. M., Johnson, M. A. & Turnbull, D. M. The distribution of mitochondrial activity in relation to optic nerve structure. Arch. Ophthalmol.120, 791–796 (2002). [DOI] [PubMed] [Google Scholar]
- 237.Osborne, N. N., Núñez-Álvarez, C., Del Olmo-Aguado, S. & Merrayo-Lloves, J. Visual light effects on mitochondria: The potential implications in relation to glaucoma. Mitochondrion36, 29–35 (2017). [DOI] [PubMed] [Google Scholar]
- 238.Mackey, D. A. et al. Primary pathogenic mtDNA mutations in multigeneration pedigrees with Leber hereditary optic neuropathy. Am. J. Hum. Genet59, 481–485 (1996). [PMC free article] [PubMed] [Google Scholar]
- 239.Yang, T. C. et al. Mitochondrial transport mediates survival of retinal ganglion cells in affected LHON patients. Hum. Mol. Genet29, 1454–1464 (2020). [DOI] [PubMed] [Google Scholar]
- 240.Ghelli, A. et al. Leber’s hereditary optic neuropathy (LHON) pathogenic mutations induce mitochondrial-dependent apoptotic death in transmitochondrial cells incubated with galactose medium. J. Biol. Chem.278, 4145–4150 (2003). [DOI] [PubMed] [Google Scholar]
- 241.Zhang, J. et al. Assocation Between Leber’s Hereditary Optic Neuropathy and MT-ND1 3460G>A Mutation-Induced Alterations in Mitochondrial Function, Apoptosis, and Mitophagy. Invest Ophthalmol. Vis. Sci.62, 38 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zanna, C. et al. Caspase-independent death of Leber’s hereditary optic neuropathy cybrids is driven by energetic failure and mediated by AIF and Endonuclease G. Apoptosis10, 997–1007 (2005). [DOI] [PubMed] [Google Scholar]
- 243.Buonfiglio, F., Böhm, E. W., Pfeiffer, N. & Gericke, A. Oxidative Stress: A Suitable Therapeutic Target for Optic Nerve Diseases? Antioxid. (Basel)12, 1465 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Cortopassi, G. et al. Mitochondrial disease activates transcripts of the unfolded protein response and cell cycle and inhibits vesicular secretion and oligodendrocyte-specific transcripts. Mitochondrion6, 161–175 (2006). [DOI] [PubMed] [Google Scholar]
- 245.Silva, J. M., Wong, A., Carelli, V. & Cortopassi, G. A. Inhibition of mitochondrial function induces an integrated stress response in oligodendroglia. Neurobiol. Dis.34, 357–365 (2009). [DOI] [PubMed] [Google Scholar]
- 246.Bao, X. R. et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife5, e10575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wassmer, S. J. et al. XIAP Protects Retinal Ganglion Cells in the Mutant ND4 Mouse Model of Leber Hereditary Optic Neuropathy. Invest Ophthalmol. Vis. Sci.61, 49 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Danese, A. et al. Pathological mitophagy disrupts mitochondrial homeostasis in Leber’s hereditary optic neuropathy. Cell Rep.40, 111124 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Giordano, C. et al. Efficient mitochondrial biogenesis drives incomplete penetrance in Leber’s hereditary optic neuropathy. Brain137, 335–353 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Carelli, V., Ross-Cisneros, F. N. & Sadun, A. A. Optic nerve degeneration and mitochondrial dysfunction: genetic and acquired optic neuropathies. Neurochem. Int.40, 573–584 (2002). [DOI] [PubMed] [Google Scholar]
- 251.Kim, U. S., Jurkute, N. & Yu-Wai-Man, P. Leber Hereditary Optic Neuropathy-Light at the End of the Tunnel? Asia Pac. J. Ophthalmol. (Philos.)7, 242–245 (2018). [DOI] [PubMed] [Google Scholar]
- 252.Pavlakis, S. G., Phillips, P. C., DiMauro, S., De Vivo, D. C. & Rowland, L. P. Mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes: a distinctive clinical syndrome. Ann. Neurol.16, 481–488 (1984). [DOI] [PubMed] [Google Scholar]
- 253.Alves, C. et al. MELAS: Phenotype Classification into Classic-versus-Atypical Presentations. AJNR Am. J. Neuroradiol.44, 602–610 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Wang, Y. X. & Le, W. D. Progress in Diagnosing Mitochondrial Myopathy, Encephalopathy, Lactic Acidosis, and Stroke-like Episodes. Chin. Med J. (Engl.)128, 1820–1825 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Goto, Y., Nonaka, I. & Horai, S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature348, 651–653 (1990). [DOI] [PubMed] [Google Scholar]
- 256.Tarnopolsky, M. A., Maguire, J., Myint, T., Applegarth, D. & Robinson, B. H. Clinical, physiological, and histological features in a kindred with the T3271C melas mutation. Muscle Nerve21, 25–33 (1998). [DOI] [PubMed] [Google Scholar]
- 257.Stenqvist, L., Paetau, A., Valanne, L., Suomalainen, A. & Pihko, H. A juvenile case of MELAS with T3271C mitochondrial DNA mutation. Pediatr. Res58, 258–262 (2005). [DOI] [PubMed] [Google Scholar]
- 258.Chomyn, A., Enriquez, J. A., Micol, V., Fernandez-Silva, P. & Attardi, G. The mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episode syndrome-associated human mitochondrial tRNALeu(UUR) mutation causes aminoacylation deficiency and concomitant reduced association of mRNA with ribosomes. J. Biol. Chem.275, 19198–19209 (2000). [DOI] [PubMed] [Google Scholar]
- 259.Loguercio Polosa, P., Capriglia, F. & Bruni, F. Molecular Investigation of Mitochondrial RNA19 Role in the Pathogenesis of MELAS Disease. Life (Basel)13, 1863 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Pek, N. M. Q. et al. Mitochondrial 3243A > G mutation confers pro-atherogenic and pro-inflammatory properties in MELAS iPS derived endothelial cells. Cell Death Dis.10, 802 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Hämäläinen, R. H. et al. Tissue- and cell-type-specific manifestations of heteroplasmic mtDNA 3243A>G mutation in human induced pluripotent stem cell-derived disease model. Proc. Natl. Acad. Sci. USA110, E3622–E3630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Morán, M. et al. Bulk autophagy, but not mitophagy, is increased in cellular model of mitochondrial disease. Biochim. Biophys. Acta1842, 1059–1070 (2014). [DOI] [PubMed] [Google Scholar]
- 263.Cotán, D. et al. Secondary coenzyme Q10 deficiency triggers mitochondria degradation by mitophagy in MELAS fibroblasts. Faseb j.25, 2669–2687 (2011). [DOI] [PubMed] [Google Scholar]
- 264.Meseguer, S., Martínez-Zamora, A., García-Arumí, E., Andreu, A. L. & Armengod, M. E. The ROS-sensitive microRNA-9/9* controls the expression of mitochondrial tRNA-modifying enzymes and is involved in the molecular mechanism of MELAS syndrome. Hum. Mol. Genet24, 167–184 (2015). [DOI] [PubMed] [Google Scholar]
- 265.El-Hattab, A. W., Adesina, A. M., Jones, J. & Scaglia, F. MELAS syndrome: Clinical manifestations, pathogenesis, and treatment options. Mol. Genet Metab.116, 4–12 (2015). [DOI] [PubMed] [Google Scholar]
- 266.Orsucci, D., Caldarazzo Ienco, E., Montano, V., Siciliano, G. & Mancuso, M. Mitochondrial stroke-like episodes: The search for new therapies. Pharm. Res180, 106228 (2022). [DOI] [PubMed] [Google Scholar]
- 267.van den Ouweland, J. M. et al. Mutation in mitochondrial tRNA(Leu)(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat. Genet1, 368–371 (1992). [DOI] [PubMed] [Google Scholar]
- 268.Kadowaki, T. et al. A subtype of diabetes mellitus associated with a mutation of mitochondrial DNA. N. Engl. J. Med330, 962–968 (1994). [DOI] [PubMed] [Google Scholar]
- 269.Yang, M. et al. The Mutations and Clinical Variability in Maternally Inherited Diabetes and Deafness: An Analysis of 161 Patients. Front Endocrinol. (Lausanne)12, 728043 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.de Wit, H. M., Westeneng, H. J., van Engelen, B. G. & Mudde, A. H. MIDD or MELAS : that’s not the question MIDD evolving into MELAS : a severe phenotype of the m.3243A>G mutation due to paternal co-inheritance of type 2 diabetes and a high heteroplasmy level. Neth. J. Med70, 460–462 (2012). [PubMed] [Google Scholar]
- 271.McMillan, R. P. et al. Quantitative Variation in m.3243A > G Mutation Produce Discrete Changes in Energy Metabolism. Sci. Rep.9, 5752 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Robinson, K. N., Terrazas, S., Giordano-Mooga, S. & Xavier, N. A. THE ROLE OF HETEROPLASMY IN THE DIAGNOSIS AND MANAGEMENT OF MATERNALLY INHERITED DIABETES AND DEAFNESS. Endocr. Pr.26, 241–246 (2020). [DOI] [PubMed] [Google Scholar]
- 273.Tan, W. J. T. & Song, L. Role of mitochondrial dysfunction and oxidative stress in sensorineural hearing loss. Hear Res434, 108783 (2023). [DOI] [PubMed] [Google Scholar]
- 274.Raimundo, N. et al. Mitochondrial stress engages E2F1 apoptotic signaling to cause deafness. Cell148, 716–726 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Pickett, S. J. et al. Phenotypic heterogeneity in m.3243A>G mitochondrial disease: The role of nuclear factors. Ann. Clin. Transl. Neurol.5, 333–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Fukuhara, N., Tokiguchi, S., Shirakawa, K. & Tsubaki, T. Myoclonus epilepsy associated with ragged-red fibres (mitochondrial abnormalities): disease entity or a syndrome? Light-and electron-microscopic studies of two cases and review of literature. J. Neurol. Sci.47, 117–133 (1980). [DOI] [PubMed] [Google Scholar]
- 277.Lorenzoni, P. J. et al. MERRF: Clinical features, muscle biopsy and molecular genetics in Brazilian patients. Mitochondrion11, 528–532 (2011). [DOI] [PubMed] [Google Scholar]
- 278.Shoffner, J. M. et al. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell61, 931–937 (1990). [DOI] [PubMed] [Google Scholar]
- 279.Park, S. Y., Kim, S. H. & Lee, Y. M. Molecular Diagnosis of Myoclonus Epilepsy Associated with Ragged-Red Fibers Syndrome in the Absence of Ragged Red Fibers. Front Neurol.8, 520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Richter, U. et al. RNA modification landscape of the human mitochondrial tRNA(Lys) regulates protein synthesis. Nat. Commun.9, 3966 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Wu, S. B., Ma, Y. S., Wu, Y. T., Chen, Y. C. & Wei, Y. H. Mitochondrial DNA mutation-elicited oxidative stress, oxidative damage, and altered gene expression in cultured cells of patients with MERRF syndrome. Mol. Neurobiol.41, 256–266 (2010). [DOI] [PubMed] [Google Scholar]
- 282.Wu, Y. T. et al. Identification of new variants in MTRNR1 and MTRNR2 genes using whole mitochondrial genome sequencing in a Taiwanese family with MERRF (myoclonic epilepsy with ragged-red fibers) syndrome. Hear Res438, 108876 (2023). [DOI] [PubMed] [Google Scholar]
- 283.Wu, Y. T. et al. Mitochondrial impairment and synaptic dysfunction are associated with neurological defects in iPSCs-derived cortical neurons of MERRF patients. J. Biomed. Sci.30, 70 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Chou, S. J. et al. Impaired ROS Scavenging System in Human Induced Pluripotent Stem Cells Generated from Patients with MERRF Syndrome. Sci. Rep.6, 23661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Villanueva-Paz, M. et al. Pathophysiological characterization of MERRF patient-specific induced neurons generated by direct reprogramming. Biochim Biophys. Acta Mol. Cell Res1866, 861–881 (2019). [DOI] [PubMed] [Google Scholar]
- 286.Villanueva-Paz, M. et al. Parkin-mediated mitophagy and autophagy flux disruption in cellular models of MERRF syndrome. Biochim Biophys. Acta Mol. Basis Dis.1866, 165726 (2020). [DOI] [PubMed] [Google Scholar]
- 287.Mirabella, M., Di Giovanni, S., Silvestri, G., Tonali, P. & Servidei, S. Apoptosis in mitochondrial encephalomyopathies with mitochondrial DNA mutations: a potential pathogenic mechanism. Brain123(Pt 1), 93–104 (2000). [DOI] [PubMed] [Google Scholar]
- 288.Ikezoe, K. et al. Apoptosis is suspended in muscle of mitochondrial encephalomyopathies. Acta Neuropathol.103, 531–540 (2002). [DOI] [PubMed] [Google Scholar]
- 289.Nakase, H. et al. Transcription and translation of deleted mitochondrial genomes in Kearns-Sayre syndrome: implications for pathogenesis. Am. J. Hum. Genet46, 418–427 (1990). [PMC free article] [PubMed] [Google Scholar]
- 290.Sudoyo, H., Marzuki, S., Byrne, E. & Mastaglia, F. Phenotypic expression of mtDNA heteroplasmy in the skeletal muscle of patients with oculomyopathy: defect in mitochondrial protein synthesis. J. Neurol. Sci.117, 83–91 (1993). [DOI] [PubMed] [Google Scholar]
- 291.Holt, I. J., Harding, A. E., Petty, R. K. & Morgan-Hughes, J. A. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am. J. Hum. Genet46, 428–433 (1990). [PMC free article] [PubMed] [Google Scholar]
- 292.Rantamäki, M. T., Soini, H. K., Finnilä, S. M., Majamaa, K. & Udd, B. Adult-onset ataxia and polyneuropathy caused by mitochondrial 8993T–>C mutation. Ann. Neurol.58, 337–340 (2005). [DOI] [PubMed] [Google Scholar]
- 293.Mäkelä-Bengs, P. et al. Correlation between the clinical symptoms and the proportion of mitochondrial DNA carrying the 8993 point mutation in the NARP syndrome. Pediatr. Res37, 634–639 (1995). [DOI] [PubMed] [Google Scholar]
- 294.Tatuch, Y. et al. Heteroplasmic mtDNA mutation (T—G) at 8993 can cause Leigh disease when the percentage of abnormal mtDNA is high. Am. J. Hum. Genet50, 852–858 (1992). [PMC free article] [PubMed] [Google Scholar]
- 295.de Vries, D. D., van Engelen, B. G., Gabreëls, F. J., Ruitenbeek, W. & van Oost, B. A. A second missense mutation in the mitochondrial ATPase 6 gene in Leigh’s syndrome. Ann. Neurol.34, 410–412 (1993). [DOI] [PubMed] [Google Scholar]
- 296.Gelfand, J. M. et al. Heterogeneous patterns of tissue injury in NARP syndrome. J. Neurol.258, 440–448 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Paumard, P. et al. The ATP synthase is involved in generating mitochondrial cristae morphology. Embo j.21, 221–230 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Hayashi, N., Geraghty, M. T. & Green, W. R. Ocular histopathologic study of a patient with the T 8993-G point mutation in Leigh’s syndrome. Ophthalmology107, 1397–1402 (2000). [DOI] [PubMed] [Google Scholar]
- 299.He, B. et al. Mitochondrial cristae architecture protects against mtDNA release and inflammation. Cell Rep.41, 111774 (2022). [DOI] [PubMed] [Google Scholar]
- 300.Sauvanet, C. et al. Mitochondrial DNA mutations provoke dominant inhibition of mitochondrial inner membrane fusion. PLoS One7, e49639 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Chakrabarti, L., Eng, J., Ivanov, N., Garden, G. A. & La Spada, A. R. Autophagy activation and enhanced mitophagy characterize the Purkinje cells of pcd mice prior to neuronal death. Mol. Brain2, 24 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.McClelland, C., Manousakis, G. & Lee, M. S. Progressive External Ophthalmoplegia. Curr. Neurol. Neurosci. Rep.16, 53 (2016). [DOI] [PubMed] [Google Scholar]
- 303.Rodríguez-López, C. et al. Clinical, pathological and genetic spectrum in 89 cases of mitochondrial progressive external ophthalmoplegia. J. Med Genet57, 643–646 (2020). [DOI] [PubMed] [Google Scholar]
- 304.Poulton, J., Deadman, M. E. & Gardiner, R. M. Duplications of mitochondrial DNA in mitochondrial myopathy. Lancet1, 236–240 (1989). [DOI] [PubMed] [Google Scholar]
- 305.Chen, X. et al. Rearranged mitochondrial genomes are present in human oocytes. Am. J. Hum. Genet57, 239–247 (1995). [PMC free article] [PubMed] [Google Scholar]
- 306.Grady, J. P. et al. Disease progression in patients with single, large-scale mitochondrial DNA deletions. Brain137, 323–334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Hirano, M. & Pitceathly, R. D. S. Progressive external ophthalmoplegia. Handb. Clin. Neurol.194, 9–21 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Tyynismaa, H. et al. Thymidine kinase 2 mutations in autosomal recessive progressive external ophthalmoplegia with multiple mitochondrial DNA deletions. Hum. Mol. Genet21, 66–75 (2012). [DOI] [PubMed] [Google Scholar]
- 309.Peter, B. & Falkenberg, M. TWINKLE and Other Human Mitochondrial DNA Helicases: Structure, Function and Disease. Genes (Basel)11, 408 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Van Goethem, G., Dermaut, B., Löfgren, A., Martin, J. J. & Van Broeckhoven, C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat. Genet28, 211–212 (2001). [DOI] [PubMed] [Google Scholar]
- 311.Young, M. J., Humble, M. M., DeBalsi, K. L., Sun, K. Y. & Copeland, W. C. POLG2 disease variants: analyses reveal a dominant negative heterodimer, altered mitochondrial localization and impaired respiratory capacity. Hum. Mol. Genet24, 5184–5197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Longley, M. J. et al. Mutant POLG2 disrupts DNA polymerase gamma subunits and causes progressive external ophthalmoplegia. Am. J. Hum. Genet78, 1026–1034 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Lujan, S. A. et al. Ultrasensitive deletion detection links mitochondrial DNA replication, disease, and aging. Genome Biol.21, 248 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Milenkovic, D. et al. TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum. Mol. Genet22, 1983–1993 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Tyynismaa, H. et al. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc. Natl. Acad. Sci. USA102, 17687–17692 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Oexner, R. R. et al. Extraocular Muscle Reveals Selective Vulnerability of Type IIB Fibers to Respiratory Chain Defects Induced by Mitochondrial DNA Alterations. Invest Ophthalmol. Vis. Sci.61, 14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Sen, A. et al. Mitochondrial membrane proteins and VPS35 orchestrate selective removal of mtDNA. Nat. Commun.13, 6704 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Garone, C. et al. Retrospective natural history of thymidine kinase 2 deficiency. J. Med Genet55, 515–521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Saada, A. et al. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat. Genet29, 342–344 (2001). [DOI] [PubMed] [Google Scholar]
- 320.Kearns, T. P. & Sayre, G. P. Retinitis pigmentosa, external ophthalmophegia, and complete heart block: unusual syndrome with histologic study in one of two cases. AMA Arch. Ophthalmol.60, 280–289 (1958). [PubMed] [Google Scholar]
- 321.Phillips, C. I. & Gosden, C. M. Leber’s hereditary optic neuropathy and Kearns-Sayre syndrome: mitochondrial DNA mutations. Surv. Ophthalmol.35, 463–472 (1991). [DOI] [PubMed] [Google Scholar]
- 322.Katsanos, K. H., Elisaf, M., Bairaktari, E. & Tsianos, E. V. Severe hypomagnesemia and hypoparathyroidism in Kearns-Sayre syndrome. Am. J. Nephrol.21, 150–153 (2001). [DOI] [PubMed] [Google Scholar]
- 323.Zeviani, M. et al. Deletions of mitochondrial DNA in Kearns-Sayre syndrome. Neurology38, 1339–1346 (1988). [DOI] [PubMed] [Google Scholar]
- 324.Yamashita, S., Nishino, I., Nonaka, I. & Goto, Y. I. Genotype and phenotype analyses in 136 patients with single large-scale mitochondrial DNA deletions. J. Hum. Genet53, 598 (2008). [DOI] [PubMed] [Google Scholar]
- 325.Kisilevsky, E., Freund, P. & Margolin, E. Mitochondrial disorders and the eye. Surv. Ophthalmol.65, 294–311 (2020). [DOI] [PubMed] [Google Scholar]
- 326.Pitceathly, R. D., Rahman, S. & Hanna, M. G. Single deletions in mitochondrial DNA–molecular mechanisms and disease phenotypes in clinical practice. Neuromuscul. Disord.22, 577–586 (2012). [DOI] [PubMed] [Google Scholar]
- 327.Russell, O. M. et al. Preferential amplification of a human mitochondrial DNA deletion in vitro and in vivo. Sci. Rep.8, 1799 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Grigalionienė, K., Burnytė, B., Balkelienė, D., Ambrozaitytė, L. & Utkus, A. Kearns-Sayre syndrome case. Novel 5,9 kb mtDNA deletion. Mol. Genet Genom. Med11, e2059 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Wei, Y. H. et al. Increases of mitochondrial mass and mitochondrial genome in association with enhanced oxidative stress in human cells harboring 4,977 BP-deleted mitochondrial DNA. Ann. N. Y Acad. Sci.928, 97–112 (2001). [DOI] [PubMed] [Google Scholar]
- 330.Alemi, M. et al. Mitochondrial DNA deletions inhibit proteasomal activity and stimulate an autophagic transcript. Free Radic. Biol. Med42, 32–43 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Lin, Y. F. et al. Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature533, 416–419 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Salvador, C. L. et al. Increased Sphingomyelin and Free Sialic Acid in Cerebrospinal Fluid of Kearns-Sayre Syndrome: New Findings Using Untargeted Metabolomics. Pediatr. Neurol.143, 68–76 (2023). [DOI] [PubMed] [Google Scholar]
- 333.Quntanilla, R. A. & Tapia-Monsalves, C. The Role of Mitochondrial Impairment in Alzheimer’s Disease Neurodegeneration: The Tau Connection. Curr. Neuropharmacol.18, 1076–1091 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Bresolin, N. et al. Progressive cytochrome c oxidase deficiency in a case of Kearns-Sayre syndrome: morphological, immunological, and biochemical studies in muscle biopsies and autopsy tissues. Ann. Neurol.21, 564–572 (1987). [DOI] [PubMed] [Google Scholar]
- 335.Brown, D. A. & O’Rourke, B. Cardiac mitochondria and arrhythmias. Cardiovasc Res88, 241–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Deng, J. et al. Mitochondrial Dysfunction in Cardiac Arrhythmias. Cells12, 679 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Pitceathly, R. D. et al. Kearns-Sayre syndrome caused by defective R1/p53R2 assembly. J. Med Genet48, 610–617 (2011). [DOI] [PubMed] [Google Scholar]
- 338.Pontarin, G. et al. Ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage. Proc. Natl. Acad. Sci. USA105, 17801–17806 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Imamura, T. et al. The necessity of implantable cardioverter defibrillators in patients with Kearns-Sayre syndrome - systematic review of the articles. Int. J. Cardiol.279, 105–111 (2019). [DOI] [PubMed] [Google Scholar]
- 340.Di Nora, C. et al. Heart Transplantation in Kearns-Sayre Syndrome. Transplantation103, e393–e394 (2019). [DOI] [PubMed] [Google Scholar]
- 341.Liu, Y. et al. Long-term safety of human retinal progenitor cell transplantation in retinitis pigmentosa patients. Stem Cell Res Ther.8, 209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Drag, S., Dotiwala, F. & Upadhyay, A. K. Gene Therapy for Retinal Degenerative Diseases: Progress, Challenges, and Future Directions. Invest Ophthalmol. Vis. Sci.64, 39 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Pearson, H. A. et al. A new syndrome of refractory sideroblastic anemia with vacuolization of marrow precursors and exocrine pancreatic dysfunction. J. Pediatr.95, 976–984 (1979). [DOI] [PubMed] [Google Scholar]
- 344.Rötig, A. et al. Deletion of blood mitochondrial DNA in pancytopenia. Lancet2, 567–568 (1988). [DOI] [PubMed] [Google Scholar]
- 345.Rötig, A., Bourgeron, T., Chretien, D., Rustin, P. & Munnich, A. Spectrum of mitochondrial DNA rearrangements in the Pearson marrow-pancreas syndrome. Hum. Mol. Genet4, 1327–1330 (1995). [DOI] [PubMed] [Google Scholar]
- 346.Rocha, M. C. et al. Pathological mechanisms underlying single large-scale mitochondrial DNA deletions. Ann. Neurol.83, 115–130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Yoshimi, A., Ishikawa, K., Niemeyer, C. & Grünert, S. C. Pearson syndrome: a multisystem mitochondrial disease with bone marrow failure. Orphanet. J. Rare Dis.17, 379 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Nilay, M. & Phadke, S. R. Pearson Syndrome: Spontaneously Recovering Anemia and Hypoparathyroidism. Indian J. Pediatr.87, 1070–1072 (2020). [DOI] [PubMed] [Google Scholar]
- 349.Ying, Y., Liang, Y., Luo, X. & Wei, M. Case Report: Clinical and Genetic Characteristics of Pearson Syndrome in a Chinese Boy and 139 Patients. Front Genet13, 802402 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Reynolds, E., Byrne, M., Ganetzky, R. & Parikh, S. Pediatric single large-scale mtDNA deletion syndromes: The power of patient reported outcomes. Mol. Genet Metab.134, 301–308 (2021). [DOI] [PubMed] [Google Scholar]
- 351.Mancuso, M. et al. Redefining phenotypes associated with mitochondrial DNA single deletion. J. Neurol.262, 1301–1309 (2015). [DOI] [PubMed] [Google Scholar]
- 352.Lee, H. F. et al. The neurological evolution of Pearson syndrome: case report and literature review. Eur. J. Paediatr. Neurol.11, 208–214 (2007). [DOI] [PubMed] [Google Scholar]
- 353.Sabella-Jiménez, V., Otero-Herrera, C., Silvera-Redondo, C. & Garavito-Galofre, P. Mitochondrial DNA deletion and duplication in Kearns-Sayre Syndrome (KSS) with initial presentation as Pearson Marrow-Pancreas Syndrome (PMPS): Two case reports in Barranquilla, Colombia. Mol. Genet Genom. Med8, e1509 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Larsson, N. G., Holme, E., Kristiansson, B., Oldfors, A. & Tulinius, M. Progressive increase of the mutated mitochondrial DNA fraction in Kearns-Sayre syndrome. Pediatr. Res28, 131–136 (1990). [DOI] [PubMed] [Google Scholar]
- 355.McShane, M. A. et al. Pearson syndrome and mitochondrial encephalomyopathy in a patient with a deletion of mtDNA. Am. J. Hum. Genet48, 39–42 (1991). [PMC free article] [PubMed] [Google Scholar]
- 356.Muraki, K. et al. The association between haematological manifestation and mtDNA deletions in Pearson syndrome. J. Inherit. Metab. Dis.20, 697–703 (1997). [DOI] [PubMed] [Google Scholar]
- 357.Yanagihara, I. et al. Fluorescence in situ hybridization analysis of peripheral blood cells in Pearson marrow-pancreas syndrome. J. Pediatr.139, 452–455 (2001). [DOI] [PubMed] [Google Scholar]
- 358.Katada, S., Mito, T., Ogasawara, E., Hayashi, J. & Nakada, K. Mitochondrial DNA with a large-scale deletion causes two distinct mitochondrial disease phenotypes in mice. G3 (Bethesda)3, 1545–1552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Ishikawa, K. et al. Pearson syndrome-like anemia induced by accumulation of mutant mtDNA and anemia with imbalanced white blood cell lineages induced by Drp1 deletion in a murine model. Pharm. Res185, 106467 (2022). [DOI] [PubMed] [Google Scholar]
- 360.Hinge, A. et al. Asymmetrically Segregated Mitochondria Provide Cellular Memory of Hematopoietic Stem Cell Replicative History and Drive HSC Attrition. Cell Stem Cell26, 420–430.e426 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Gonzalez-Ibanez, A. M. et al. Erythroid Differentiation and Heme Biosynthesis Are Dependent on a Shift in the Balance of Mitochondrial Fusion and Fission Dynamics. Front Cell Dev. Biol.8, 592035 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Liang, R. et al. Restraining Lysosomal Activity Preserves Hematopoietic Stem Cell Quiescence and Potency. Cell Stem Cell26, 359–376.e357 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Vannini, N. et al. Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nat. Commun.7, 13125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Jin, G. et al. Atad3a suppresses Pink1-dependent mitophagy to maintain homeostasis of hematopoietic progenitor cells. Nat. Immunol.19, 29–40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Farruggia, P., Di Marco, F. & Dufour, C. Pearson syndrome. Expert Rev. Hematol.11, 239–246 (2018). [DOI] [PubMed] [Google Scholar]
- 366.Jacoby, E. et al. Mitochondrial augmentation of hematopoietic stem cells in children with single large-scale mitochondrial DNA deletion syndromes. Sci. Transl. Med14, eabo3724 (2022). [DOI] [PubMed] [Google Scholar]
- 367.Kjer, P., Jensen, O. A. & Klinken, L. Histopathology of eye, optic nerve and brain in a case of dominant optic atrophy. Acta Ophthalmol. (Copenh)61, 300–312 (1983). [DOI] [PubMed] [Google Scholar]
- 368.Pesch, U. E. et al. OPA1 mutations in patients with autosomal dominant optic atrophy and evidence for semi-dominant inheritance. Hum. Mol. Genet10, 1359–1368 (2001). [DOI] [PubMed] [Google Scholar]
- 369.Davies, V. J. et al. Opa1 deficiency in a mouse model of autosomal dominant optic atrophy impairs mitochondrial morphology, optic nerve structure and visual function. Hum. Mol. Genet16, 1307–1318 (2007). [DOI] [PubMed] [Google Scholar]
- 370.Yarosh, W. et al. The molecular mechanisms of OPA1-mediated optic atrophy in Drosophila model and prospects for antioxidant treatment. PLoS Genet4, e6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Bonneau, D. et al. Early-onset Behr syndrome due to compound heterozygous mutations in OPA1. Brain137, e301 (2014). [DOI] [PubMed] [Google Scholar]
- 372.Yu-Wai-Man, P. et al. Multi-system neurological disease is common in patients with OPA1 mutations. Brain133, 771–786 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Lei, Q., Xiang, K., Cheng, L. & Xiang, M. Human retinal organoids with an OPA1 mutation are defective in retinal ganglion cell differentiation and function. Stem Cell Rep.19, 68–83 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Quintana-Cabrera, R. & Scorrano, L. Determinants and outcomes of mitochondrial dynamics. Mol. Cell83, 857–876 (2023). [DOI] [PubMed] [Google Scholar]
- 375.Liao, C. et al. Dysregulated mitophagy and mitochondrial organization in optic atrophy due to OPA1 mutations. Neurology88, 131–142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Davis, C. H. et al. Transcellular degradation of axonal mitochondria. Proc. Natl. Acad. Sci. USA111, 9633–9638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Lin, Y. et al. Targeting DRP1 with Mdivi-1 to correct mitochondrial abnormalities in ADOA plus syndrome. JCI Insight9, e180582 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Zaninello, M. et al. Inhibition of autophagy curtails visual loss in a model of autosomal dominant optic atrophy. Nat. Commun.11, 4029 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Tezze, C. et al. Age-Associated Loss of OPA1 in Muscle Impacts Muscle Mass, Metabolic Homeostasis, Systemic Inflammation, and Epithelial Senescence. Cell Metab.25, 1374–1389.e1376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Frezza, C. et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell126, 177–189 (2006). [DOI] [PubMed] [Google Scholar]
- 381.Scorrano, L. et al. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev. Cell2, 55–67 (2002). [DOI] [PubMed] [Google Scholar]
- 382.Olichon, A. et al. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem.278, 7743–7746 (2003). [DOI] [PubMed] [Google Scholar]
- 383.Zhang, J. et al. A novel ADOA-associated OPA1 mutation alters the mitochondrial function, membrane potential, ROS production and apoptosis. Sci. Rep.7, 5704 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Kalogerou, M. et al. Omega-3 fatty acids promote neuroprotection, decreased apoptosis and reduced glial cell activation in the retina of a mouse model of OPA1-related autosomal dominant optic atrophy. Exp. Eye Res215, 108901 (2022). [DOI] [PubMed] [Google Scholar]
- 385.Varanita, T. et al. The OPA1-dependent mitochondrial cristae remodeling pathway controls atrophic, apoptotic, and ischemic tissue damage. Cell Metab.21, 834–844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Rodríguez-Nuevo, A. et al. Mitochondrial DNA and TLR9 drive muscle inflammation upon Opa1 deficiency. Embo j.37, e96553 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Naviaux, R. K. & Nguyen, K. V. POLG mutations associated with Alpers’ syndrome and mitochondrial DNA depletion. Ann. Neurol.55, 706–712 (2004). [DOI] [PubMed] [Google Scholar]
- 388.Huttenlocher, P. R., Solitare, G. B. & Adams, G. Infantile diffuse cerebral degeneration with hepatic cirrhosis. Arch. Neurol.33, 186–192 (1976). [DOI] [PubMed] [Google Scholar]
- 389.Bicknese, A. R., May, W., Hickey, W. F. & Dodson, W. E. Early childhood hepatocerebral degeneration misdiagnosed as valproate hepatotoxicity. Ann. Neurol.32, 767–775 (1992). [DOI] [PubMed] [Google Scholar]
- 390.Davidzon, G. et al. POLG mutations and Alpers syndrome. Ann. Neurol.57, 921–923 (2005). [DOI] [PubMed] [Google Scholar]
- 391.Hikmat, O. et al. The clinical spectrum and natural history of early-onset diseases due to DNA polymerase gamma mutations. Genet Med19, 1217–1225 (2017). [DOI] [PubMed] [Google Scholar]
- 392.Nguyen, K. V. et al. POLG mutations in Alpers syndrome. Neurology65, 1493–1495 (2005). [DOI] [PubMed] [Google Scholar]
- 393.Saneto, R. P., Cohen, B. H., Copeland, W. C. & Naviaux, R. K. Alpers-Huttenlocher syndrome. Pediatr. Neurol.48, 167–178 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Hakonen, A. H. et al. Recessive Twinkle mutations in early onset encephalopathy with mtDNA depletion. Brain130, 3032–3040 (2007). [DOI] [PubMed] [Google Scholar]
- 395.Scalais, E. et al. Polymerase gamma deficiency (POLG): clinical course in a child with a two stage evolution from infantile myocerebrohepatopathy spectrum to an Alpers syndrome and neuropathological findings of Leigh’s encephalopathy. Eur. J. Paediatr. Neurol.16, 542–548 (2012). [DOI] [PubMed] [Google Scholar]
- 396.Rahman, S. & Copeland, W. C. POLG-related disorders and their neurological manifestations. Nat. Rev. Neurol.15, 40–52 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Harding, B. N. et al. Progressive neuronal degeneration of childhood with liver disease (Alpers’ disease) presenting in young adults. J. Neurol. Neurosurg. Psychiatry58, 320–325 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Liang, K. X. et al. Disease-specific phenotypes in iPSC-derived neural stem cells with POLG mutations. EMBO Mol. Med12, e12146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Brandebura, A. N., Paumier, A., Onur, T. S. & Allen, N. J. Astrocyte contribution to dysfunction, risk and progression in neurodegenerative disorders. Nat. Rev. Neurosci.24, 23–39 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Smith, L. A. et al. Astrocytic pathology in Alpers’ syndrome. Acta Neuropathol. Commun.11, 86 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Liang, K. X. et al. Activation of Neurotoxic Astrocytes Due to Mitochondrial Dysfunction Triggered by POLG Mutation. Int. J. Biol. Sci.20, 2860–2880 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Chen, A. et al. Nicotinamide Riboside and Metformin Ameliorate Mitophagy Defect in Induced Pluripotent Stem Cell-Derived Astrocytes With POLG Mutations. Front Cell Dev. Biol.9, 737304 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Fiebig, C. et al. Mitochondrial Dysfunction in Astrocytes Impairs the Generation of Reactive Astrocytes and Enhances Neuronal Cell Death in the Cortex Upon Photothrombotic Lesion. Front Mol. Neurosci.12, 40 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Ignatenko, O. et al. Loss of mtDNA activates astrocytes and leads to spongiotic encephalopathy. Nat. Commun.9, 70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Li, S. et al. Valproic acid-induced hepatotoxicity in Alpers syndrome is associated with mitochondrial permeability transition pore opening-dependent apoptotic sensitivity in an induced pluripotent stem cell model. Hepatology61, 1730–1739 (2015). [DOI] [PubMed] [Google Scholar]
- 406.Joshi, C. N., Greenberg, C. R., Mhanni, A. A. & Salman, M. S. Ketogenic diet in Alpers-Huttenlocher syndrome. Pediatr. Neurol.40, 314–316 (2009). [DOI] [PubMed] [Google Scholar]
- 407.Fadic, R. et al. Sensory ataxic neuropathy as the presenting feature of a novel mitochondrial disease. Neurology49, 239–245 (1997). [DOI] [PubMed] [Google Scholar]
- 408.Van Goethem, G. et al. Recessive POLG mutations presenting with sensory and ataxic neuropathy in compound heterozygote patients with progressive external ophthalmoplegia. Neuromuscul. Disord.13, 133–142 (2003). [DOI] [PubMed] [Google Scholar]
- 409.Weiss, M. D. & Saneto, R. P. Sensory ataxic neuropathy with dysarthria and ophthalmoparesis (SANDO) in late life due to compound heterozygous POLG mutations. Muscle Nerve41, 882–885 (2010). [DOI] [PubMed] [Google Scholar]
- 410.Winterthun, S. et al. Autosomal recessive mitochondrial ataxic syndrome due to mitochondrial polymerase gamma mutations. Neurology64, 1204–1208 (2005). [DOI] [PubMed] [Google Scholar]
- 411.Hanisch, F. et al. SANDO syndrome in a cohort of 107 patients with CPEO and mitochondrial DNA deletions. J. Neurol. Neurosurg. Psychiatry86, 630–634 (2015). [DOI] [PubMed] [Google Scholar]
- 412.Hudson, G., Deschauer, M., Busse, K., Zierz, S. & Chinnery, P. F. Sensory ataxic neuropathy due to a novel C10Orf2 mutation with probable germline mosaicism. Neurology64, 371–373 (2005). [DOI] [PubMed] [Google Scholar]
- 413.Bugiardini, E. et al. Clinicopathologic and molecular spectrum of RNASEH1-related mitochondrial disease. Neurol. Genet3, e149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Cerritelli, S. M. & Crouch, R. J. Ribonuclease H: the enzymes in eukaryotes. Febs j.276, 1494–1505 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Cerritelli, S. M. et al. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell11, 807–815 (2003). [DOI] [PubMed] [Google Scholar]
- 416.Posse, V. et al. RNase H1 directs origin-specific initiation of DNA replication in human mitochondria. PLoS Genet15, e1007781 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Holmes, J. B. et al. Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication. Proc. Natl. Acad. Sci. USA112, 9334–9339 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Misic, J. et al. Mammalian RNase H1 directs RNA primer formation for mtDNA replication initiation and is also necessary for mtDNA replication completion. Nucleic Acids Res50, 8749–8766 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Tzoulis, C. et al. The spectrum of clinical disease caused by the A467T and W748S POLG mutations: a study of 26 cases. Brain129, 1685–1692 (2006). [DOI] [PubMed] [Google Scholar]
- 420.Hakonen, A. H. et al. Mitochondrial DNA polymerase W748S mutation: a common cause of autosomal recessive ataxia with ancient European origin. Am. J. Hum. Genet77, 430–441 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Hakonen, A. H. et al. Abundance of the POLG disease mutations in Europe, Australia, New Zealand, and the United States explained by single ancient European founders. Eur. J. Hum. Genet15, 779–783 (2007). [DOI] [PubMed] [Google Scholar]
- 422.Kang, Y. et al. Ancestral allele of DNA polymerase gamma modifies antiviral tolerance. Nature628, 844–853 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Stumpf, J. D., Saneto, R. P. & Copeland, W. C. Clinical and molecular features of POLG-related mitochondrial disease. Cold Spring Harb. Perspect. Biol.5, a011395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Cohen, B. H., Chinnery, P. F. & Copeland, W. C. POLG-Related Disorders. (University of Washington, Seattle Copyright © 1993-2024, University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved., 1993). [PubMed]
- 425.Koskinen, T., Sainio, K., Rapola, J., Pihko, H. & Paetau, A. Sensory neuropathy in infantile onset spinocerebellar ataxia (IOSCA). Muscle Nerve17, 509–515 (1994). [DOI] [PubMed] [Google Scholar]
- 426.Koskinen, T. et al. Infantile onset spinocerebellar ataxia with sensory neuropathy: a new inherited disease. J. Neurol. Sci.121, 50–56 (1994). [DOI] [PubMed] [Google Scholar]
- 427.Koskinen, T., Valanne, L., Ketonen, L. M. & Pihko, H. Infantile-onset spinocerebellar ataxia: MR and CT findings. AJNR Am. J. Neuroradiol.16, 1427–1433 (1995). [PMC free article] [PubMed] [Google Scholar]
- 428.Nikali, K. et al. Infantile onset spinocerebellar ataxia is caused by recessive mutations in mitochondrial proteins Twinkle and Twinky. Hum. Mol. Genet14, 2981–2990 (2005). [DOI] [PubMed] [Google Scholar]
- 429.Hakonen, A. H. et al. Infantile-onset spinocerebellar ataxia and mitochondrial recessive ataxia syndrome are associated with neuronal complex I defect and mtDNA depletion. Hum. Mol. Genet17, 3822–3835 (2008). [DOI] [PubMed] [Google Scholar]
- 430.Leung, T. C. S. et al. Mitochondrial damage and impaired mitophagy contribute to disease progression in SCA6. Acta Neuropathol.147, 26 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Barth, P. G. et al. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J. Neurol. Sci.62, 327–355 (1983). [DOI] [PubMed] [Google Scholar]
- 432.Bione, S. et al. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat. Genet12, 385–389 (1996). [DOI] [PubMed] [Google Scholar]
- 433.Xu, Y., Malhotra, A., Ren, M. & Schlame, M. The enzymatic function of tafazzin. J. Biol. Chem.281, 39217–39224 (2006). [DOI] [PubMed] [Google Scholar]
- 434.Pang, J., Bao, Y., Mitchell-Silbaugh, K., Veevers, J. & Fang, X. Barth Syndrome Cardiomyopathy: An Update. Genes (Basel)13, 656 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Clarke, S. L. et al. Barth syndrome. Orphanet J. Rare Dis.8, 23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Schlame, M. et al. Deficiency of tetralinoleoyl-cardiolipin in Barth syndrome. Ann. Neurol.51, 634–637 (2002). [DOI] [PubMed] [Google Scholar]
- 437.Valianpour, F. et al. Monolysocardiolipins accumulate in Barth syndrome but do not lead to enhanced apoptosis. J. Lipid Res46, 1182–1195 (2005). [DOI] [PubMed] [Google Scholar]
- 438.Bowron, A. et al. Barth syndrome without tetralinoleoyl cardiolipin deficiency: a possible ameliorated phenotype. J. Inherit. Metab. Dis.38, 279–286 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Ghosh, S., Iadarola, D. M., Ball, W. B. & Gohil, V. M. Mitochondrial dysfunctions in barth syndrome. IUBMB Life71, 791–801 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.McKenzie, M., Lazarou, M., Thorburn, D. R. & Ryan, M. T. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J. Mol. Biol.361, 462–469 (2006). [DOI] [PubMed] [Google Scholar]
- 441.Lou, W. et al. Loss of tafazzin results in decreased myoblast differentiation in C2C12 cells: A myoblast model of Barth syndrome and cardiolipin deficiency. Biochim Biophys. Acta Mol. Cell Biol. Lipids1863, 857–865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Chu, C. T. et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol.15, 1197–1205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Hsu, P. et al. Cardiolipin remodeling by TAZ/tafazzin is selectively required for the initiation of mitophagy. Autophagy11, 643–652 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Zhang, J., Liu, X., Nie, J. & Shi, Y. Restoration of mitophagy ameliorates cardiomyopathy in Barth syndrome. Autophagy18, 2134–2149 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Zegallai, H. M. & Hatch, G. M. Barth syndrome: cardiolipin, cellular pathophysiology, management, and novel therapeutic targets. Mol. Cell Biochem476, 1605–1629 (2021). [DOI] [PubMed] [Google Scholar]
- 446.Dürr, A. et al. Clinical and genetic abnormalities in patients with Friedreich’s ataxia. N. Engl. J. Med335, 1169–1175 (1996). [DOI] [PubMed] [Google Scholar]
- 447.Harding, A. E. Friedreich’s ataxia: a clinical and genetic study of 90 families with an analysis of early diagnostic criteria and intrafamilial clustering of clinical features. Brain104, 589–620 (1981). [DOI] [PubMed] [Google Scholar]
- 448.Campuzano, V. et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science271, 1423–1427 (1996). [DOI] [PubMed] [Google Scholar]
- 449.Reetz, K. et al. Biological and clinical characteristics of the European Friedreich’s Ataxia Consortium for Translational Studies (EFACTS) cohort: a cross-sectional analysis of baseline data. Lancet Neurol.14, 174–182 (2015). [DOI] [PubMed] [Google Scholar]
- 450.Parkinson, M. H., Boesch, S., Nachbauer, W., Mariotti, C. & Giunti, P. Clinical features of Friedreich’s ataxia: classical and atypical phenotypes. J. Neurochem126(Suppl 1), 103–117 (2013). [DOI] [PubMed] [Google Scholar]
- 451.Alper, G. & Narayanan, V. Friedreich’s ataxia. Pediatr. Neurol.28, 335–341 (2003). [DOI] [PubMed] [Google Scholar]
- 452.Yandim, C., Natisvili, T. & Festenstein, R. Gene regulation and epigenetics in Friedreich’s ataxia. J. Neurochem126(Suppl 1), 21–42 (2013). [DOI] [PubMed] [Google Scholar]
- 453.Sakamoto, N. et al. Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich’s ataxia. Mol. Cell3, 465–475 (1999). [DOI] [PubMed] [Google Scholar]
- 454.Branda, S. S., Yang, Z. Y., Chew, A. & Isaya, G. Mitochondrial intermediate peptidase and the yeast frataxin homolog together maintain mitochondrial iron homeostasis in Saccharomyces cerevisiae. Hum. Mol. Genet8, 1099–1110 (1999). [DOI] [PubMed] [Google Scholar]
- 455.Pastore, A. & Puccio, H. Frataxin: a protein in search for a function. J. Neurochem126(Suppl 1), 43–52 (2013). [DOI] [PubMed] [Google Scholar]
- 456.Puccio, H. & Koenig, M. Recent advances in the molecular pathogenesis of Friedreich ataxia. Hum. Mol. Genet9, 887–892 (2000). [DOI] [PubMed] [Google Scholar]
- 457.Rötig, A. et al. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat. Genet17, 215–217 (1997). [DOI] [PubMed] [Google Scholar]
- 458.Read, A. D., Bentley, R. E., Archer, S. L. & Dunham-Snary, K. J. Mitochondrial iron-sulfur clusters: Structure, function, and an emerging role in vascular biology. Redox Biol.47, 102164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Pandolfo, M. & Hausmann, L. Deferiprone for the treatment of Friedreich’s ataxia. J. Neurochem126(Suppl 1), 142–146 (2013). [DOI] [PubMed] [Google Scholar]
- 460.Abeti, R. et al. Mitochondrial energy imbalance and lipid peroxidation cause cell death in Friedreich’s ataxia’. Cell Death Dis.7, e2237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Apolloni, S., Milani, M. & D’Ambrosi, N. Neuroinflammation in Friedreich’s Ataxia. Int. J. Mol. Sci.23, 6297 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Shidara, Y. & Hollenbeck, P. J. Defects in mitochondrial axonal transport and membrane potential without increased reactive oxygen species production in a Drosophila model of Friedreich ataxia. J. Neurosci.30, 11369–11378 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Chiang, S. et al. Mechanisms of impaired mitochondrial homeostasis and NAD(+) metabolism in a model of mitochondrial heart disease exhibiting redox active iron accumulation. Redox Biol.46, 102038 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.La Rosa, P. et al. The Nrf2 induction prevents ferroptosis in Friedreich’s Ataxia. Redox Biol.38, 101791 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Turchi, R. et al. Frataxin deficiency induces lipid accumulation and affects thermogenesis in brown adipose tissue. Cell Death Dis.11, 51 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Rouyer, A. et al. Long-term prognosis of fatty-acid oxidation disorders in adults: Optimism despite the limited effective therapies available. Eur. J. Neurol.31, e16138 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Wajner, M. & Amaral, A. U. Mitochondrial dysfunction in fatty acid oxidation disorders: insights from human and animal studies. Biosci. Rep.36, e00281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Gregersen, N. et al. Mitochondrial fatty acid oxidation defects–remaining challenges. J. Inherit. Metab. Dis.31, 643–657 (2008). [DOI] [PubMed] [Google Scholar]
- 469.Guerra, I. M. S. et al. Mitochondrial Fatty Acid β-Oxidation Disorders: From Disease to Lipidomic Studies-A Critical Review. Int J. Mol. Sci.23, 13933 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Lake, N. J., Compton, A. G., Rahman, S. & Thorburn, D. R. Leigh syndrome: One disorder, more than 75 monogenic causes. Ann. Neurol.79, 190–203 (2016). [DOI] [PubMed] [Google Scholar]
- 471.Rahman, J., Noronha, A., Thiele, I. & Rahman, S. Leigh map: A novel computational diagnostic resource for mitochondrial disease. Ann. Neurol.81, 9–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Ma, Y. Y. et al. Genetic and biochemical findings in Chinese children with Leigh syndrome. J. Clin. Neurosci.20, 1591–1594 (2013). [DOI] [PubMed] [Google Scholar]
- 473.Sonsalla, G. et al. Direct neuronal reprogramming of NDUFS4 patient cells identifies the unfolded protein response as a novel general reprogramming hurdle. Neuron112, 1117–1132.e1119 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Schubert Baldo, M. & Vilarinho, L. Molecular basis of Leigh syndrome: a current look. Orphanet. J. Rare Dis.15, 31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Patel, K. P., O’Brien, T. W., Subramony, S. H., Shuster, J. & Stacpoole, P. W. The spectrum of pyruvate dehydrogenase complex deficiency: clinical, biochemical and genetic features in 371 patients. Mol. Genet Metab.106, 385–394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Stenton, S. L. et al. Leigh Syndrome: A Study of 209 Patients at the Beijing Children’s Hospital. Ann. Neurol.91, 466–482 (2022). [DOI] [PubMed] [Google Scholar]
- 477.DeBrosse, S. D. et al. Spectrum of neurological and survival outcomes in pyruvate dehydrogenase complex (PDC) deficiency: lack of correlation with genotype. Mol. Genet Metab.107, 394–402 (2012). [DOI] [PubMed] [Google Scholar]
- 478.Elpeleg, O. et al. Deficiency of the ADP-forming succinyl-CoA synthase activity is associated with encephalomyopathy and mitochondrial DNA depletion. Am. J. Hum. Genet76, 1081–1086 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Van Hove, J. L. et al. Succinyl-CoA ligase deficiency: a mitochondrial hepatoencephalomyopathy. Pediatr. Res68, 159–164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Kistol, D. et al. Leigh Syndrome: Spectrum of Molecular Defects and Clinical Features in Russia. Int. J. Mol. Sci.24, 1597 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Zhao, Y. et al. Loss of succinyl-CoA synthase ADP-forming β subunit disrupts mtDNA stability and mitochondrial dynamics in neurons. Sci. Rep.7, 7169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Janer, A. et al. SLC25A46 is required for mitochondrial lipid homeostasis and cristae maintenance and is responsible for Leigh syndrome. EMBO Mol. Med8, 1019–1038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Cheramangalam, R. N. et al. Bendless is essential for PINK1-Park mediated Mitofusin degradation under mitochondrial stress caused by loss of LRPPRC. PLoS Genet19, e1010493 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Pulliam, D. A. et al. Complex IV-deficient Surf1(-/-) mice initiate mitochondrial stress responses. Biochem J.462, 359–371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Rolland, S. G. et al. Impaired complex IV activity in response to loss of LRPPRC function can be compensated by mitochondrial hyperfusion. Proc. Natl. Acad. Sci. USA110, E2967–E2976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Okamura, K., Santa, T., Nagae, K. & Omae, T. Congenital oculoskeletal myopathy with abnormal muscle and liver mitochondria. J. Neurol. Sci.27, 79–91 (1976). [DOI] [PubMed] [Google Scholar]
- 487.Hirano, M. et al. Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): clinical, biochemical, and genetic features of an autosomal recessive mitochondrial disorder. Neurology44, 721–727 (1994). [DOI] [PubMed] [Google Scholar]
- 488.Pacitti, D., Levene, M., Garone, C., Nirmalananthan, N. & Bax, B. E. Mitochondrial Neurogastrointestinal Encephalomyopathy: Into the Fourth Decade, What We Have Learned So Far. Front Genet9, 669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Halter, J. P. et al. Allogeneic haematopoietic stem cell transplantation for mitochondrial neurogastrointestinal encephalomyopathy. Brain138, 2847–2858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Papadimitriou, A. et al. Partial depletion and multiple deletions of muscle mtDNA in familial MNGIE syndrome. Neurology51, 1086–1092 (1998). [DOI] [PubMed] [Google Scholar]
- 491.Nishigaki, Y., Martí, R., Copeland, W. C. & Hirano, M. Site-specific somatic mitochondrial DNA point mutations in patients with thymidine phosphorylase deficiency. J. Clin. Invest111, 1913–1921 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Du, J. et al. Lysosomal dysfunction and overload of nucleosides in thymidine phosphorylase deficiency of MNGIE. J. Transl. Med22, 449 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Shaibani, A. et al. Mitochondrial neurogastrointestinal encephalopathy due to mutations in RRM2B. Arch. Neurol.66, 1028–1032 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Bonora, E. et al. Biallelic variants in LIG3 cause a novel mitochondrial neurogastrointestinal encephalomyopathy. Brain144, 1451–1466 (2021). [DOI] [PubMed] [Google Scholar]
- 495.Yadak, R., Sillevis Smitt, P., van Gisbergen, M. W., van Til, N. P. & de Coo, I. F. Mitochondrial Neurogastrointestinal Encephalomyopathy Caused by Thymidine Phosphorylase Enzyme Deficiency: From Pathogenesis to Emerging Therapeutic Options. Front Cell Neurosci.11, 31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Yadak, R. et al. Transplantation, gene therapy and intestinal pathology in MNGIE patients and mice. BMC Gastroenterol.18, 149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Vila-Julià, F. et al. Efficacy of adeno-associated virus gene therapy in a MNGIE murine model enhanced by chronic exposure to nucleosides. EBioMedicine62, 103133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Torres-Torronteras, J. et al. Long-Term Sustained Effect of Liver-Targeted Adeno-Associated Virus Gene Therapy for Mitochondrial Neurogastrointestinal Encephalomyopathy. Hum. Gene Ther.29, 708–718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Inbal, A. et al. Myopathy, lactic acidosis, and sideroblastic anemia: a new syndrome. Am. J. Med Genet55, 372–378 (1995). [DOI] [PubMed] [Google Scholar]
- 500.Bykhovskaya, Y., Casas, K., Mengesha, E., Inbal, A. & Fischel-Ghodsian, N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am. J. Hum. Genet74, 1303–1308 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Patton, J. R., Bykhovskaya, Y., Mengesha, E., Bertolotto, C. & Fischel-Ghodsian, N. Mitochondrial myopathy and sideroblastic anemia (MLASA): missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J. Biol. Chem.280, 19823–19828 (2005). [DOI] [PubMed] [Google Scholar]
- 502.Wang, B. et al. Mitochondrial tRNA pseudouridylation governs erythropoiesis. Blood144, 657–671 (2024). [DOI] [PubMed] [Google Scholar]
- 503.Shi, D. et al. Pseudouridine synthase 1 regulates erythropoiesis via transfer RNAs pseudouridylation and cytoplasmic translation. iScience27, 109265 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Guzzi, N. et al. Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell173, 1204–1216.e1226 (2018). [DOI] [PubMed] [Google Scholar]
- 505.Riley, L. G. et al. Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia–MLASA syndrome. Am. J. Hum. Genet87, 52–59 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Jin, X. et al. An animal model for mitochondrial tyrosyl-tRNA synthetase deficiency reveals links between oxidative phosphorylation and retinal function. J. Biol. Chem.296, 100437 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Sengers, R. C., Trijbels, J. M., Willems, J. L., Daniels, O. & Stadhouders, A. M. Congenital cataract and mitochondrial myopathy of skeletal and heart muscle associated with lactic acidosis after exercise. J. Pediatr.86, 873–880 (1975). [DOI] [PubMed] [Google Scholar]
- 508.Mayr, J. A. et al. Lack of the mitochondrial protein acylglycerol kinase causes Sengers syndrome. Am. J. Hum. Genet90, 314–320 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Bektas, M. et al. A novel acylglycerol kinase that produces lysophosphatidic acid modulates cross talk with EGFR in prostate cancer cells. J. Cell Biol.169, 801–811 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Vukotic, M. et al. Acylglycerol Kinase Mutated in Sengers Syndrome Is a Subunit of the TIM22 Protein Translocase in Mitochondria. Mol. Cell67, 471–483.e477 (2017). [DOI] [PubMed] [Google Scholar]
- 511.Kang, Y. et al. Sengers Syndrome-Associated Mitochondrial Acylglycerol Kinase Is a Subunit of the Human TIM22 Protein Import Complex. Mol. Cell67, 457–470.e455 (2017). [DOI] [PubMed] [Google Scholar]
- 512.Ding, N. et al. AGK regulates the progression to NASH by affecting mitochondria complex I function. Theranostics12, 3237–3250 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Jenkinson, E. M. et al. Perrault syndrome: further evidence for genetic heterogeneity. J. Neurol.259, 974–976 (2012). [DOI] [PubMed] [Google Scholar]
- 514.Faridi, R. et al. New insights into Perrault syndrome, a clinically and genetically heterogeneous disorder. Hum. Genet141, 805–819 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Cole, A. et al. Inhibition of the Mitochondrial Protease ClpP as a Therapeutic Strategy for Human Acute Myeloid Leukemia. Cancer Cell27, 864–876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Jenkinson, E. M. et al. Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am. J. Hum. Genet92, 605–613 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Luo, B., Ma, Y., Zhou, Y., Zhang, N. & Luo, Y. Human ClpP protease, a promising therapy target for diseases of mitochondrial dysfunction. Drug Discov. Today26, 968–981 (2021). [DOI] [PubMed] [Google Scholar]
- 518.Szczepanowska, K. et al. A salvage pathway maintains highly functional respiratory complex I. Nat. Commun.11, 1643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Pryde, K. R., Taanman, J. W. & Schapira, A. H. A LON-ClpP Proteolytic Axis Degrades Complex I to Extinguish ROS Production in Depolarized Mitochondria. Cell Rep.17, 2522–2531 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Seiferling, D. et al. Loss of CLPP alleviates mitochondrial cardiomyopathy without affecting the mammalian UPRmt. EMBO Rep.17, 953–964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Rumyantseva, A., Popovic, M. & Trifunovic, A. CLPP deficiency ameliorates neurodegeneration caused by impaired mitochondrial protein synthesis. Brain145, 92–104 (2022). [DOI] [PubMed] [Google Scholar]
- 522.Mabanglo, M. F., Bhandari, V. & Houry, W. A. Substrates and interactors of the ClpP protease in the mitochondria. Curr. Opin. Chem. Biol.66, 102078 (2022). [DOI] [PubMed] [Google Scholar]
- 523.Pierce, S. B. et al. Mutations in mitochondrial histidyl tRNA synthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. Proc. Natl. Acad. Sci. USA108, 6543–6548 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Pierce, S. B. et al. Mutations in LARS2, encoding mitochondrial leucyl-tRNA synthetase, lead to premature ovarian failure and hearing loss in Perrault syndrome. Am. J. Hum. Genet92, 614–620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Dennerlein, S., Rozanska, A., Wydro, M., Chrzanowska-Lightowlers, Z. M. & Lightowlers, R. N. Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem J.430, 551–558 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Chatzispyrou, I. A. et al. A homozygous missense mutation in ERAL1, encoding a mitochondrial rRNA chaperone, causes Perrault syndrome. Hum. Mol. Genet26, 2541–2550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Guo, Y. et al. Mitochondrial dysfunction in aging. Ageing Res. Rev.88, 101955 (2023). [DOI] [PubMed] [Google Scholar]
- 528.Hou, Y. et al. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol.15, 565–581 (2019). [DOI] [PubMed] [Google Scholar]
- 529.Zong, Y. et al. Mitochondrial dysfunction: mechanisms and advances in therapy. Signal Transduct. Target Ther.9, 124 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.DeBalsi, K. L., Hoff, K. E. & Copeland, W. C. Role of the mitochondrial DNA replication machinery in mitochondrial DNA mutagenesis, aging and age-related diseases. Ageing Res Rev.33, 89–104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Kim, S. J. et al. A naturally occurring variant of SHLP2 is a protective factor in Parkinson’s disease. Mol. Psychiatry29, 505–517 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Gowda, P., Reddy, P. H. & Kumar, S. Deregulated mitochondrial microRNAs in Alzheimer’s disease: Focus on synapse and mitochondria. Ageing Res Rev.73, 101529 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Wang, W., Zhao, F., Ma, X., Perry, G. & Zhu, X. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Mol. Neurodegener.15, 30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Li, Y., Xia, X., Wang, Y. & Zheng, J. C. Mitochondrial dysfunction in microglia: a novel perspective for pathogenesis of Alzheimer’s disease. J. Neuroinflam.19, 248 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Henrich, M. T., Oertel, W. H., Surmeier, D. J. & Geibl, F. F. Mitochondrial dysfunction in Parkinson’s disease - a key disease hallmark with therapeutic potential. Mol. Neurodegener.18, 83 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Hu, Q. & Wang, G. Mitochondrial dysfunction in Parkinson’s disease. Transl. Neurodegener.5, 14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Lawrence, G., Holley, C. L. & Schroder, K. Parkinson’s disease: connecting mitochondria to inflammasomes. Trends Immunol.43, 877–885 (2022). [DOI] [PubMed] [Google Scholar]
- 538.Mandic, M. et al. No energy, no autophagy-Mechanisms and therapeutic implications of autophagic response energy requirements. J. Cell Physiol.10.1002/jcp.31366 (2024). [DOI] [PubMed]
- 539.Zhao, M. et al. Mitochondrial ROS promote mitochondrial dysfunction and inflammation in ischemic acute kidney injury by disrupting TFAM-mediated mtDNA maintenance. Theranostics11, 1845–1863 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Vela-Sebastián, A., Bayona-Bafaluy, P. & Pacheu-Grau, D. ISR pathway contribution to tissue specificity of mitochondrial diseases. Trends Endocrinol. Metab.10.1016/j.tem.2024.05.001 (2024). [DOI] [PubMed] [Google Scholar]
- 541.Burr, S. P. et al. Cell lineage-specific mitochondrial resilience during mammalian organogenesis. Cell186, 1212–1229.e1221 (2023). [DOI] [PubMed] [Google Scholar]
- 542.Roca-Portoles, A. & Tait, S. W. G. Mitochondrial quality control: from molecule to organelle. Cell Mol. Life Sci.78, 3853–3866 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Zhu, D., Li, X. & Tian, Y. Mitochondrial-to-nuclear communication in aging: an epigenetic perspective. Trends Biochem Sci.47, 645–659 (2022). [DOI] [PubMed] [Google Scholar]
- 544.Mancuso, M. et al. Diagnostic approach to mitochondrial disorders: the need for a reliable biomarker. Curr. Mol. Med9, 1095–1107 (2009). [DOI] [PubMed] [Google Scholar]
- 545.Parikh, S. et al. Diagnosis of ‘possible’ mitochondrial disease: an existential crisis. J. Med Genet56, 123–130 (2019). [DOI] [PubMed] [Google Scholar]
- 546.Hubens, W. H. G. et al. Blood biomarkers for assessment of mitochondrial dysfunction: An expert review. Mitochondrion62, 187–204 (2022). [DOI] [PubMed] [Google Scholar]
- 547.Parikh, S. et al. Diagnosis and management of mitochondrial disease: a consensus statement from the Mitochondrial Medicine Society. Genet Med.17, 689–701 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Suomalainen, A. et al. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol.10, 806–818 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Gill, E. L., Wang, J., Viaene, A. N., Master, S. R. & Ganetzky, R. D. Methodologies in Mitochondrial Testing: Diagnosing a Primary Mitochondrial Respiratory Chain Disorder. Clin. Chem.69, 564–582 (2023). [DOI] [PubMed] [Google Scholar]
- 550.Debray, F. G. et al. Diagnostic accuracy of blood lactate-to-pyruvate molar ratio in the differential diagnosis of congenital lactic acidosis. Clin. Chem.53, 916–921 (2007). [DOI] [PubMed] [Google Scholar]
- 551.Haas, R. H. et al. The in-depth evaluation of suspected mitochondrial disease. Mol. Genet Metab.94, 16–37 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Yatsuga, S. et al. Growth differentiation factor 15 as a useful biomarker for mitochondrial disorders. Ann. Neurol.78, 814–823 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Maresca, A. et al. Expanding and validating the biomarkers for mitochondrial diseases. J. Mol. Med (Berl.)98, 1467–1478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Miliotis, S., Nicolalde, B., Ortega, M., Yepez, J. & Caicedo, A. Forms of extracellular mitochondria and their impact in health. Mitochondrion48, 16–30 (2019). [DOI] [PubMed] [Google Scholar]
- 555.D’Acunzo, P. et al. Mitovesicles secreted into the extracellular space of brains with mitochondrial dysfunction impair synaptic plasticity. Mol. Neurodegener.19, 34 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Leggio, L. et al. Extracellular Vesicles as Novel Diagnostic and Prognostic Biomarkers for Parkinson’s Disease. Aging Dis.12, 1494–1515 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Ao, X. et al. Non-coding RNAs regulating mitochondrial function in cardiovascular diseases. J. Mol. Med (Berl.)101, 501–526 (2023). [DOI] [PubMed] [Google Scholar]
- 558.Indrieri, A. et al. miR-181a/b downregulation exerts a protective action on mitochondrial disease models. EMBO Mol. Med11, e8734 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Carrella, S. et al. miR-181a/b downregulation: a mutation-independent therapeutic approach for inherited retinal diseases. EMBO Mol. Med14, e15941 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Wang, W. et al. Identification of miRNA, lncRNA and mRNA-associated ceRNA networks and potential biomarker for MELAS with mitochondrial DNA A3243G mutation. Sci. Rep.7, 41639 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Finotti, A. et al. MicroRNAs and Long Non-coding RNAs in Genetic Diseases. Mol. Diagn. Ther.23, 155–171 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.McCully, J. D. et al. Injection of isolated mitochondria during early reperfusion for cardioprotection. Am. J. Physiol. Heart Circ. Physiol.296, H94–H105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Buzkova, J. et al. Metabolomes of mitochondrial diseases and inclusion body myositis patients: treatment targets and biomarkers. EMBO Mol. Med10, e9091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Esterhuizen, K., van der Westhuizen, F. H. & Louw, R. Metabolomics of mitochondrial disease. Mitochondrion35, 97–110 (2017). [DOI] [PubMed] [Google Scholar]
- 565.Ren, C. et al. Lipidomic profiling of plasma samples from patients with mitochondrial disease. Biochem. Biophys. Res Commun.500, 124–131 (2018). [DOI] [PubMed] [Google Scholar]
- 566.Ruiz, M. et al. Lipidomics unveils lipid dyshomeostasis and low circulating plasmalogens as biomarkers in a monogenic mitochondrial disorder. JCI Insight4, e123231 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Khan, S., Ince-Dunn, G., Suomalainen, A. & Elo, L. L. Integrative omics approaches provide biological and clinical insights: examples from mitochondrial diseases. J. Clin. Invest130, 20–28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Wortmann, S. B., Mayr, J. A., Nuoffer, J. M., Prokisch, H. & Sperl, W. A Guideline for the Diagnosis of Pediatric Mitochondrial Disease: The Value of Muscle and Skin Biopsies in the Genetics Era. Neuropediatrics48, 309–314 (2017). [DOI] [PubMed] [Google Scholar]
- 569.Mavraki, E. et al. Genetic testing for mitochondrial disease: the United Kingdom best practice guidelines. Eur. J. Hum. Genet31, 148–163 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Cui, H. et al. Comprehensive next-generation sequence analyses of the entire mitochondrial genome reveal new insights into the molecular diagnosis of mitochondrial DNA disorders. Genet Med15, 388–394 (2013). [DOI] [PubMed] [Google Scholar]
- 571.Macken, W. L., Vandrovcova, J., Hanna, M. G. & Pitceathly, R. D. S. Applying genomic and transcriptomic advances to mitochondrial medicine. Nat. Rev. Neurol.17, 215–230 (2021). [DOI] [PubMed] [Google Scholar]
- 572.Kremer, L. S. et al. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat. Commun.8, 15824 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Stark, R., Grzelak, M. & Hadfield, J. RNA sequencing: the teenage years. Nat. Rev. Genet20, 631–656 (2019). [DOI] [PubMed] [Google Scholar]
- 574.Taanman, J. W. et al. Characterization of a novel TYMP splice site mutation associated with mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). Neuromuscul. Disord.19, 151–154 (2009). [DOI] [PubMed] [Google Scholar]
- 575.Pitceathly, R. D. et al. NDUFA4 mutations underlie dysfunction of a cytochrome c oxidase subunit linked to human neurological disease. Cell Rep.3, 1795–1805 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Oglesbee, D., Freedenberg, D., Kramer, K. A., Anderson, B. D. & Hahn, S. H. Normal muscle respiratory chain enzymes can complicate mitochondrial disease diagnosis. Pediatr. Neurol.35, 289–292 (2006). [DOI] [PubMed] [Google Scholar]
- 577.Petruzzella, V. et al. Extremely high levels of mutant mtDNAs co-localize with cytochrome c oxidase-negative ragged-red fibers in patients harboring a point mutation at nt 3243. Hum. Mol. Genet3, 449–454 (1994). [DOI] [PubMed] [Google Scholar]
- 578.Moraes, C. T., Ricci, E., Bonilla, E., DiMauro, S. & Schon, E. A. The mitochondrial tRNA(Leu(UUR)) mutation in mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes (MELAS): genetic, biochemical, and morphological correlations in skeletal muscle. Am. J. Hum. Genet50, 934–949 (1992). [PMC free article] [PubMed] [Google Scholar]
- 579.DiMauro, S., Schon, E. A., Carelli, V. & Hirano, M. The clinical maze of mitochondrial neurology. Nat. Rev. Neurol.9, 429–444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Sciacco, M., Bonilla, E., Schon, E. A., DiMauro, S. & Moraes, C. T. Distribution of wild-type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from patients with mitochondrial myopathy. Hum. Mol. Genet3, 13–19 (1994). [DOI] [PubMed] [Google Scholar]
- 581.Johnson, M. A., Turnbull, D. M., Dick, D. J. & Sherratt, H. S. A partial deficiency of cytochrome c oxidase in chronic progressive external ophthalmoplegia. J. Neurol. Sci.60, 31–53 (1983). [DOI] [PubMed] [Google Scholar]
- 582.Charles-Schoeman, C. & Verity, M. A. Nicotinamide adenine dinucleotide tetrazolium reductase identifies microvasculature activation in muscle from adult patients with dermatomyositis. J. Rheumatol.39, 94–99 (2012). [DOI] [PubMed] [Google Scholar]
- 583.van den Heuvel, L. P., Smeitink, J. A. & Rodenburg, R. J. Biochemical examination of fibroblasts in the diagnosis and research of oxidative phosphorylation (OXPHOS) defects. Mitochondrion4, 395–401 (2004). [DOI] [PubMed] [Google Scholar]
- 584.Spinazzi, M., Casarin, A., Pertegato, V., Salviati, L. & Angelini, C. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc.7, 1235–1246 (2012). [DOI] [PubMed] [Google Scholar]
- 585.Mantle, D., Millichap, L., Castro-Marrero, J. & Hargreaves, I. P. Primary Coenzyme Q10 Deficiency: An Update. Antioxid. (Basel)12, 1652 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Ma, Y. Y. et al. Analysis of the mitochondrial complex I-V enzyme activities of peripheral leukocytes in oxidative phosphorylation disorders. J. Child Neurol.26, 974–979 (2011). [DOI] [PubMed] [Google Scholar]
- 587.Takemura, G. et al. Electron Microscopic Findings Are an Important Aid for Diagnosing Mitochondrial Cardiomyopathy With Mitochondrial DNA Mutation 3243A>G. Circ. Heart Fail9, e003283 (2016). [DOI] [PubMed] [Google Scholar]
- 588.Imasawa, T. et al. Clinicopathologic Features of Mitochondrial Nephropathy. Kidney Int. Rep.7, 580–590 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Hazard, F. K., Ficicioglu, C. H., Ganesh, J. & Ruchelli, E. D. Liver pathology in infantile mitochondrial DNA depletion syndrome. Pediatr. Dev. Pathol.16, 415–424 (2013). [DOI] [PubMed] [Google Scholar]
- 590.Jaber, S. M., Yadava, N. & Polster, B. M. Mapping mitochondrial respiratory chain deficiencies by respirometry: Beyond the Mito Stress Test. Exp. Neurol.328, 113282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Divakaruni, A. S. & Jastroch, M. A practical guide for the analysis, standardization and interpretation of oxygen consumption measurements. Nat. Metab.4, 978–994 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Pesta, D. & Gnaiger, E. High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods Mol. Biol.810, 25–58 (2012). [DOI] [PubMed] [Google Scholar]
- 593.Avram, V. F. et al. Impairment of Mitochondrial Respiration in Metabolic Diseases: An Overview. Int. J. Mol. Sci.23, 8852 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Abu-Amero, K. K. & Bosley, T. M. Detection of mitochondrial respiratory dysfunction in circulating lymphocytes using resazurin. Arch. Pathol. Lab Med129, 1295–1298 (2005). [DOI] [PubMed] [Google Scholar]
- 595.Pecina, P. et al. Noninvasive diagnostics of mitochondrial disorders in isolated lymphocytes with high resolution respirometry. BBA Clin.2, 62–71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Westerlund, E. et al. Oxygen consumption in platelets as an adjunct diagnostic method for pediatric mitochondrial disease. Pediatr. Res83, 455–465 (2018). [DOI] [PubMed] [Google Scholar]
- 597.Koch, L. Clinical genetics: mitochondrial replacement techniques under the spotlight. Nat. Rev. Genet15, 516 (2014). [DOI] [PubMed] [Google Scholar]
- 598.Craven, L. et al. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature465, 82–85 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Sato, A. et al. Gene therapy for progeny of mito-mice carrying pathogenic mtDNA by nuclear transplantation. Proc. Natl. Acad. Sci. USA102, 16765–16770 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Dobler, R., Dowling, D. K., Morrow, E. H. & Reinhardt, K. A systematic review and meta-analysis reveals pervasive effects of germline mitochondrial replacement on components of health. Hum. Reprod. Update24, 519–534 (2018). [DOI] [PubMed] [Google Scholar]
- 601.Hyslop, L. A. et al. Towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease. Nature534, 383–386 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Yamada, M. et al. Genetic Drift Can Compromise Mitochondrial Replacement by Nuclear Transfer in Human Oocytes. Cell Stem Cell18, 749–754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Khrapko, K. Two ways to make an mtDNA bottleneck. Nat. Genet40, 134–135 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604.Wai, T., Teoli, D. & Shoubridge, E. A. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat. Genet40, 1484–1488 (2008). [DOI] [PubMed] [Google Scholar]
- 605.Fan, X. Y. et al. Reduction of mtDNA heteroplasmy in mitochondrial replacement therapy by inducing forced mitophagy. Nat. Biomed. Eng.6, 339–350 (2022). [DOI] [PubMed] [Google Scholar]
- 606.Herbert, M., Kalleas, D., Cooney, D., Lamb, M. & Lister, L. Meiosis and maternal aging: insights from aneuploid oocytes and trisomy births. Cold Spring Harb. Perspect. Biol.7, a017970 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Wu, K. et al. Mitochondrial replacement by pre-pronuclear transfer in human embryos. Cell Res27, 834–837 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Li, C. Y. et al. Generation of mitochondrial replacement monkeys by female pronucleus transfer. Zool. Res45, 292–298 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Wang, T. et al. Polar body genome transfer for preventing the transmission of inherited mitochondrial diseases. Cell157, 1591–1604 (2014). [DOI] [PubMed] [Google Scholar]
- 610.Reichmann, J. et al. Dual-spindle formation in zygotes keeps parental genomes apart in early mammalian embryos. Science361, 189–193 (2018). [DOI] [PubMed] [Google Scholar]
- 611.Woodson, J. D. & Chory, J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet9, 383–395 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Bayona-Bafaluy, M. P., Müller, S. & Moraes, C. T. Fast adaptive coevolution of nuclear and mitochondrial subunits of ATP synthetase in orangutan. Mol. Biol. Evol.22, 716–724 (2005). [DOI] [PubMed] [Google Scholar]
- 613.Ma, H. et al. Incompatibility between Nuclear and Mitochondrial Genomes Contributes to an Interspecies Reproductive Barrier. Cell Metab.24, 283–294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Neupane, J. et al. Assessment of nuclear transfer techniques to prevent the transmission of heritable mitochondrial disorders without compromising embryonic development competence in mice. Mitochondrion18, 27–33 (2014). [DOI] [PubMed] [Google Scholar]
- 615.Tachibana, M. et al. Towards germline gene therapy of inherited mitochondrial diseases. Nature493, 627–631 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Paull, D. et al. Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature493, 632–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Tesarik, J., Nagy, Z. P., Mendoza, C. & Greco, E. Chemically and mechanically induced membrane fusion: non-activating methods for nuclear transfer in mature human oocytes. Hum. Reprod.15, 1149–1154 (2000). [DOI] [PubMed] [Google Scholar]
- 618.Liao, X. et al. Significant decrease of maternal mitochondria carryover using optimized spindle-chromosomal complex transfer. PLoS Biol.21, e3002313 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Kang, E. et al. Mitochondrial replacement in human oocytes carrying pathogenic mitochondrial DNA mutations. Nature540, 270–275 (2016). [DOI] [PubMed] [Google Scholar]
- 620.Greenfield, A. et al. Assisted reproductive technologies to prevent human mitochondrial disease transmission. Nat. Biotechnol.35, 1059–1068 (2017). [DOI] [PubMed] [Google Scholar]
- 621.Li, Y. et al. Mitochondrial aggregation caused by cytochalasin B compromises the efficiency and safety of three-parent embryo. Mol. Hum. Reprod.28, gaac036 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622.Okamoto, R. et al. Aggregated chromosomes/chromatin transfer: a novel approach for mitochondrial replacement with minimal mitochondrial carryover: the implications of mouse experiments for human aggregated chromosome transfer. Mol. Hum. Reprod.29, gaad043 (2023). [DOI] [PubMed] [Google Scholar]
- 623.Bredenoord, A. L. & Appleby, J. B. Mitochondrial Replacement Techniques: Remaining Ethical Challenges. Cell Stem Cell21, 301–304 (2017). [DOI] [PubMed] [Google Scholar]
- 624.Wu, K. et al. Polar bodies are efficient donors for reconstruction of human embryos for potential mitochondrial replacement therapy. Cell Res27, 1069–1072 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Wang, Z. et al. Mitochondrial replacement in macaque monkey offspring by first polar body transfer. Cell Res31, 233–236 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626.Hou, X. et al. Effects of cytochalasin B on DNA methylation and histone modification in parthenogenetically activated porcine embryos. Reproduction152, 519–527 (2016). [DOI] [PubMed] [Google Scholar]
- 627.Hartl, F. U. & Neupert, W. Protein sorting to mitochondria: evolutionary conservations of folding and assembly. Science247, 930–938 (1990). [DOI] [PubMed] [Google Scholar]
- 628.Guy, J. et al. Rescue of a mitochondrial deficiency causing Leber Hereditary Optic Neuropathy. Ann. Neurol.52, 534–542 (2002). [DOI] [PubMed] [Google Scholar]
- 629.Cwerman-Thibault, H. et al. Nuclear expression of mitochondrial ND4 leads to the protein assembling in complex I and prevents optic atrophy and visual loss. Mol. Ther. Methods Clin. Dev.2, 15003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Manfredi, G. et al. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nat. Genet30, 394–399 (2002). [DOI] [PubMed] [Google Scholar]
- 631.Boominathan, A. et al. Stable nuclear expression of ATP8 and ATP6 genes rescues a mtDNA Complex V null mutant. Nucleic Acids Res44, 9342–9357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Lewis, C. J. et al. Codon optimization is an essential parameter for the efficient allotopic expression of mtDNA genes. Redox Biol.30, 101429 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Borna, N. N. et al. Identification of a novel MT-ND3 variant and restoring mitochondrial function by allotopic expression of MT-ND3 gene. Mitochondrion76, 101858 (2024). [DOI] [PubMed] [Google Scholar]
- 634.Wang, J. et al. Optimized allotopic expression of mitochondrial ND6 transgene restored complex I and apoptosis deficiencies caused by LHON-linked ND6 14484T > C mutation. J. Biomed. Sci.30, 63 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635.Entelis, N. S., Kolesnikova, O. A., Martin, R. P. & Tarassov, I. A. RNA delivery into mitochondria. Adv. Drug Deliv. Rev.49, 199–215 (2001). [DOI] [PubMed] [Google Scholar]
- 636.Kolesnikova, O. A. et al. Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum. Mol. Genet13, 2519–2534 (2004). [DOI] [PubMed] [Google Scholar]
- 637.Mahata, B., Mukherjee, S., Mishra, S., Bandyopadhyay, A. & Adhya, S. Functional delivery of a cytosolic tRNA into mutant mitochondria of human cells. Science314, 471–474 (2006). [DOI] [PubMed] [Google Scholar]
- 638.Qi, X., Sun, L., Lewin, A. S., Hauswirth, W. W. & Guy, J. The mutant human ND4 subunit of complex I induces optic neuropathy in the mouse. Invest Ophthalmol. Vis. Sci.48, 1–10 (2007). [DOI] [PubMed] [Google Scholar]
- 639.Koilkonda, R. et al. LHON gene therapy vector prevents visual loss and optic neuropathy induced by G11778A mutant mitochondrial DNA: biodistribution and toxicology profile. Invest Ophthalmol. Vis. Sci.55, 7739–7753 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Oca-Cossio, J., Kenyon, L., Hao, H. & Moraes, C. T. Limitations of allotopic expression of mitochondrial genes in mammalian cells. Genetics165, 707–720 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641.Perales-Clemente, E., Fernández-Silva, P., Acín-Pérez, R., Pérez-Martos, A. & Enríquez, J. A. Allotopic expression of mitochondrial-encoded genes in mammals: achieved goal, undemonstrated mechanism or impossible task? Nucleic Acids Res39, 225–234 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642.Chin, R. M., Panavas, T., Brown, J. M. & Johnson, K. K. Optimized Mitochondrial Targeting of Proteins Encoded by Modified mRNAs Rescues Cells Harboring Mutations in mtATP6. Cell Rep.22, 2818–2826 (2018). [DOI] [PubMed] [Google Scholar]
- 643.Colin, F. et al. Mammalian frataxin controls sulfur production and iron entry during de novo Fe4S4 cluster assembly. J. Am. Chem. Soc.135, 733–740 (2013). [DOI] [PubMed] [Google Scholar]
- 644.Perdomini, M. et al. Prevention and reversal of severe mitochondrial cardiomyopathy by gene therapy in a mouse model of Friedreich’s ataxia. Nat. Med20, 542–547 (2014). [DOI] [PubMed] [Google Scholar]
- 645.Chang, J. C. et al. AAV8 gene therapy reverses cardiac pathology and prevents early mortality in a mouse model of Friedreich’s ataxia. Mol. Ther. Methods Clin. Dev.32, 101193 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 646.Belbellaa, B., Reutenauer, L., Monassier, L. & Puccio, H. Correction of half the cardiomyocytes fully rescue Friedreich ataxia mitochondrial cardiomyopathy through cell-autonomous mechanisms. Hum. Mol. Genet28, 1274–1285 (2019). [DOI] [PubMed] [Google Scholar]
- 647.Huichalaf, C. et al. In vivo overexpression of frataxin causes toxicity mediated by iron-sulfur cluster deficiency. Mol. Ther. Methods Clin. Dev.24, 367–378 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648.Ling, Q., Rioux, M., Hu, Y., Lee, M. & Gray, S. J. Adeno-associated viral vector serotype 9-based gene replacement therapy for SURF1-related Leigh syndrome. Mol. Ther. Methods Clin. Dev.23, 158–168 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 649.Di Meo, I., Marchet, S., Lamperti, C., Zeviani, M. & Viscomi, C. AAV9-based gene therapy partially ameliorates the clinical phenotype of a mouse model of Leigh syndrome. Gene Ther.24, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Reynaud-Dulaurier, R. et al. Gene replacement therapy provides benefit in an adult mouse model of Leigh syndrome. Brain143, 1686–1696 (2020). [DOI] [PubMed] [Google Scholar]
- 651.Silva-Pinheiro, P., Cerutti, R., Luna-Sanchez, M., Zeviani, M. & Viscomi, C. A Single Intravenous Injection of AAV-PHP.B-hNDUFS4 Ameliorates the Phenotype of Ndufs4 (-/-) Mice. Mol. Ther. Methods Clin. Dev.17, 1071–1078 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 652.Liguore, W. A. et al. AAV-PHP.B Administration Results in a Differential Pattern of CNS Biodistribution in Non-human Primates Compared with Mice. Mol. Ther.27, 2018–2037 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 653.Hordeaux, J. et al. The Neurotropic Properties of AAV-PHP.B Are Limited to C57BL/6J Mice. Mol. Ther.26, 664–668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.Corrà, S., Cerutti, R., Balmaceda, V., Viscomi, C. & Zeviani, M. Double administration of self-complementary AAV9NDUFS4 prevents Leigh disease in Ndufs4-/- mice. Brain145, 3405–3414 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655.Parés, M. et al. Preclinical Assessment of a Gene-Editing Approach in a Mouse Model of Mitochondrial Neurogastrointestinal Encephalomyopathy. Hum. Gene Ther.32, 1210–1223 (2021). [DOI] [PubMed] [Google Scholar]
- 656.Cabrera-Pérez, R. et al. Alpha-1-Antitrypsin Promoter Improves the Efficacy of an Adeno-Associated Virus Vector for the Treatment of Mitochondrial Neurogastrointestinal Encephalomyopathy. Hum. Gene Ther.30, 985–998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Di Meo, I. et al. Effective AAV-mediated gene therapy in a mouse model of ethylmalonic encephalopathy. EMBO Mol. Med4, 1008–1014 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 658.Suzuki-Hatano, S. et al. Increased mtDNA Abundance and Improved Function in Human Barth Syndrome Patient Fibroblasts Following AAV-TAZ Gene Delivery. Int. J. Mol. Sci.20, 3416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 659.Suzuki-Hatano, S. et al. AAV-Mediated TAZ Gene Replacement Restores Mitochondrial and Cardioskeletal Function in Barth Syndrome. Hum. Gene Ther.30, 139–154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660.Wang, S. et al. AAV Gene Therapy Prevents and Reverses Heart Failure in a Murine Knockout Model of Barth Syndrome. Circ. Res126, 1024–1039 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661.Pacak, C. A. et al. One episode of low intensity aerobic exercise prior to systemic AAV9 administration augments transgene delivery to the heart and skeletal muscle. J. Transl. Med21, 748 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 662.Li, L. et al. Activation of Frataxin Protein Expression by Antisense Oligonucleotides Targeting the Mutant Expanded Repeat. Nucleic Acid Ther.28, 23–33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 663.Li, Y. et al. Premature transcription termination at the expanded GAA repeats and aberrant alternative polyadenylation contributes to the Frataxin transcriptional deficit in Friedreich’s ataxia. Hum. Mol. Genet31, 3539–3557 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Li, L., Matsui, M. & Corey, D. R. Activating frataxin expression by repeat-targeted nucleic acids. Nat. Commun.7, 10606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665.Shen, X. et al. Progress towards drug discovery for Friedreich’s Ataxia: Identifying synthetic oligonucleotides that more potently activate expression of human frataxin protein. Bioorg. Med Chem.28, 115472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 666.Li, Y. et al. Targeting 3’ and 5’ untranslated regions with antisense oligonucleotides to stabilize frataxin mRNA and increase protein expression. Nucleic Acids Res49, 11560–11574 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 667.Kilikevicius, A. et al. Difficulties translating antisense-mediated activation of Frataxin expression from cell culture to mice. RNA Biol.19, 364–372 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 668.Wang, F. et al. G-rich motifs within phosphorothioate-based antisense oligonucleotides (ASOs) drive activation of FXN expression through indirect effects. Nucleic Acids Res50, 12657–12673 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 669.Jackson, C. B., Turnbull, D. M., Minczuk, M. & Gammage, P. A. Therapeutic Manipulation of mtDNA Heteroplasmy: A Shifting Perspective. Trends Mol. Med26, 698–709 (2020). [DOI] [PubMed] [Google Scholar]
- 670.Muratovska, A. et al. Targeting peptide nucleic acid (PNA) oligomers to mitochondria within cells by conjugation to lipophilic cations: implications for mitochondrial DNA replication, expression and disease. Nucleic Acids Res29, 1852–1863 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 671.Falabella, M., Minczuk, M., Hanna, M. G., Viscomi, C. & Pitceathly, R. D. S. Gene therapy for primary mitochondrial diseases: experimental advances and clinical challenges. Nat. Rev. Neurol.18, 689–698 (2022). [DOI] [PubMed] [Google Scholar]
- 672.Gammage, P. A. et al. Genome editing in mitochondria corrects a pathogenic mtDNA mutation in vivo. Nat. Med24, 1691–1695 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 673.Bacman, S. R., Williams, S. L., Garcia, S. & Moraes, C. T. Organ-specific shifts in mtDNA heteroplasmy following systemic delivery of a mitochondria-targeted restriction endonuclease. Gene Ther.17, 713–720 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 674.Tanaka, M. et al. Gene therapy for mitochondrial disease by delivering restriction endonuclease SmaI into mitochondria. J. Biomed. Sci.9, 534–541 (2002). [DOI] [PubMed] [Google Scholar]
- 675.Srivastava, S. & Moraes, C. T. Manipulating mitochondrial DNA heteroplasmy by a mitochondrially targeted restriction endonuclease. Hum. Mol. Genet10, 3093–3099 (2001). [DOI] [PubMed] [Google Scholar]
- 676.Bayona-Bafaluy, M. P., Blits, B., Battersby, B. J., Shoubridge, E. A. & Moraes, C. T. Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease. Proc. Natl. Acad. Sci. USA102, 14392–14397 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 677.Bacman, S. R., Williams, S. L., Hernandez, D. & Moraes, C. T. Modulating mtDNA heteroplasmy by mitochondria-targeted restriction endonucleases in a ‘differential multiple cleavage-site’ model. Gene Ther.14, 1309–1318 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 678.Alexeyev, M. F. et al. Selective elimination of mutant mitochondrial genomes as therapeutic strategy for the treatment of NARP and MILS syndromes. Gene Ther.15, 516–523 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 679.Bacman, S. R., Williams, S. L., Duan, D. & Moraes, C. T. Manipulation of mtDNA heteroplasmy in all striated muscles of newborn mice by AAV9-mediated delivery of a mitochondria-targeted restriction endonuclease. Gene Ther.19, 1101–1106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 680.Reddy, P. et al. Selective elimination of mitochondrial mutations in the germline by genome editing. Cell161, 459–469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 681.Gammage, P. A. et al. Near-complete elimination of mutant mtDNA by iterative or dynamic dose-controlled treatment with mtZFNs. Nucleic Acids Res44, 7804–7816 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682.Minczuk, M., Papworth, M. A., Miller, J. C., Murphy, M. P. & Klug, A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res36, 3926–3938 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 683.Gammage, P. A., Rorbach, J., Vincent, A. I., Rebar, E. J. & Minczuk, M. Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO Mol. Med6, 458–466 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 684.Urnov, F. D. et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature435, 646–651 (2005). [DOI] [PubMed] [Google Scholar]
- 685.Yang, Y. et al. Targeted elimination of mutant mitochondrial DNA in MELAS-iPSCs by mitoTALENs. Protein Cell9, 283–297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 686.Bacman, S. R. et al. MitoTALEN reduces mutant mtDNA load and restores tRNA(Ala) levels in a mouse model of heteroplasmic mtDNA mutation. Nat. Med24, 1696–1700 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 687.Phillips, A. F. et al. Single-Molecule Analysis of mtDNA Replication Uncovers the Basis of the Common Deletion. Mol. Cell65, 527–538.e526 (2017). [DOI] [PubMed] [Google Scholar]
- 688.Persson, Ö. et al. Copy-choice recombination during mitochondrial L-strand synthesis causes DNA deletions. Nat. Commun.10, 759 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 689.Silva-Pinheiro, P. & Minczuk, M. The potential of mitochondrial genome engineering. Nat. Rev. Genet23, 199–214 (2022). [DOI] [PubMed] [Google Scholar]
- 690.Boch, J. & Bonas, U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev. Phytopathol.48, 419–436 (2010). [DOI] [PubMed] [Google Scholar]
- 691.Bacman, S. R. & Moraes, C. T. Mitochondrial DNA Base Editing: Good Editing Things Still Come in Small Packages. Mol. Cell79, 708–709 (2020). [DOI] [PubMed] [Google Scholar]
- 692.Carroll, D. Genome engineering with targetable nucleases. Annu Rev. Biochem83, 409–439 (2014). [DOI] [PubMed] [Google Scholar]
- 693.Valton, J. et al. Overcoming transcription activator-like effector (TALE) DNA binding domain sensitivity to cytosine methylation. J. Biol. Chem.287, 38427–38432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 694.Bultmann, S. et al. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res40, 5368–5377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 695.Mingozzi, F. & High, K. A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet12, 341–355 (2011). [DOI] [PubMed] [Google Scholar]
- 696.Moraes, C. T. A magic bullet to specifically eliminate mutated mitochondrial genomes from patients’ cells. EMBO Mol. Med6, 434–435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 697.Pereira, C. V. et al. mitoTev-TALE: a monomeric DNA editing enzyme to reduce mutant mitochondrial DNA levels. EMBO Mol. Med10, e8084 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 698.Hashimoto, M. et al. MitoTALEN: A General Approach to Reduce Mutant mtDNA Loads and Restore Oxidative Phosphorylation Function in Mitochondrial Diseases. Mol. Ther.23, 1592–1599 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 699.Yahata, N., Matsumoto, Y., Omi, M., Yamamoto, N. & Hata, R. TALEN-mediated shift of mitochondrial DNA heteroplasmy in MELAS-iPSCs with m.13513G>A mutation. Sci. Rep.7, 15557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 700.Jo, A. et al. Efficient Mitochondrial Genome Editing by CRISPR/Cas9. Biomed. Res Int.2015, 305716 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 701.Bi, R. et al. Direct evidence of CRISPR-Cas9-mediated mitochondrial genome editing. Innov. (Camb.)3, 100329 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 702.Wang, G. et al. PNPASE regulates RNA import into mitochondria. Cell142, 456–467 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 703.Chen, Y. et al. Synergistic engineering of CRISPR-Cas nucleases enables robust mammalian genome editing. Innov. (Camb.)3, 100264 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 704.Gammage, P. A., Moraes, C. T. & Minczuk, M. Mitochondrial Genome Engineering: The Revolution May Not Be CRISPR-Ized. Trends Genet34, 101–110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 705.Hathazi, D. & Horvath, R. Mitochondrial DNA editing with mitoARCUS. Nat. Metab.5, 2039–2040 (2023). [DOI] [PubMed] [Google Scholar]
- 706.Antunes, M. S., Smith, J. J., Jantz, D. & Medford, J. I. Targeted DNA excision in Arabidopsis by a re-engineered homing endonuclease. BMC Biotechnol.12, 86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 707.Shoop, W. K. et al. Efficient elimination of MELAS-associated m.3243G mutant mitochondrial DNA by an engineered mitoARCUS nuclease. Nat. Metab.5, 2169–2183 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 708.Zekonyte, U. et al. Mitochondrial targeted meganuclease as a platform to eliminate mutant mtDNA in vivo. Nat. Commun.12, 3210 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 709.Gray, S. J. Timing of Gene Therapy Interventions: The Earlier, the Better. Mol. Ther.24, 1017–1018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 710.Mok, B. Y. et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature583, 631–637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 711.Wei, Y. et al. Human cleaving embryos enable efficient mitochondrial base-editing with DdCBE. Cell Discov.8, 7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 712.Chen, X. et al. DdCBE-mediated mitochondrial base editing in human 3PN embryos. Cell Discov.8, 8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 713.Silva-Pinheiro, P. et al. In vivo mitochondrial base editing via adeno-associated viral delivery to mouse post-mitotic tissue. Nat. Commun.13, 750 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 714.Tan, L. et al. A conditional knockout rat resource of mitochondrial protein-coding genes via a DdCBE-induced premature stop codon. Sci. Adv.9, eadf2695 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 715.Guo, J. et al. Precision modeling of mitochondrial diseases in zebrafish via DdCBE-mediated mtDNA base editing. Cell Discov.7, 78 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 716.Lee, H. et al. Mitochondrial DNA editing in mice with DddA-TALE fusion deaminases. Nat. Commun.12, 1190 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 717.Mok, Y. G. et al. Base editing in human cells with monomeric DddA-TALE fusion deaminases. Nat. Commun.13, 4038 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 718.Lim, K., Cho, S. I. & Kim, J. S. Nuclear and mitochondrial DNA editing in human cells with zinc finger deaminases. Nat. Commun.13, 366 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 719.Willis, J. C. W., Silva-Pinheiro, P., Widdup, L., Minczuk, M. & Liu, D. R. Compact zinc finger base editors that edit mitochondrial or nuclear DNA in vitro and in vivo. Nat. Commun.13, 7204 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 720.Lee, S. et al. Enhanced mitochondrial DNA editing in mice using nuclear-exported TALE-linked deaminases and nucleases. Genome Biol.23, 211 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 721.Roth, T. B., Woolston, B. M., Stephanopoulos, G. & Liu, D. R. Phage-Assisted Evolution of Bacillus methanolicus Methanol Dehydrogenase 2. ACS Synth. Biol.8, 796–806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 722.Mok, B. Y. et al. CRISPR-free base editors with enhanced activity and expanded targeting scope in mitochondrial and nuclear DNA. Nat. Biotechnol.40, 1378–1387 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 723.Guo, J. et al. A DddA ortholog-based and transactivator-assisted nuclear and mitochondrial cytosine base editors with expanded target compatibility. Mol. Cell83, 1710–1724.e1717 (2023). [DOI] [PubMed] [Google Scholar]
- 724.Wei, Y. et al. Enhanced C-To-T and A-To-G Base Editing in Mitochondrial DNA with Engineered DdCBE and TALED. Adv. Sci. (Weinh.)11, e2304113 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 725.Cho, S. I. et al. Targeted A-to-G base editing in human mitochondrial DNA with programmable deaminases. Cell185, 1764–1776.e1712 (2022). [DOI] [PubMed] [Google Scholar]
- 726.Qi, X. et al. Precision modeling of mitochondrial disease in rats via DdCBE-mediated mtDNA editing. Cell Discov.7, 95 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 727.Lee, S., Lee, H., Baek, G. & Kim, J. S. Precision mitochondrial DNA editing with high-fidelity DddA-derived base editors. Nat. Biotechnol.41, 378–386 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 728.Aushev, M. & Herbert, M. Mitochondrial genome editing gets precise. Nature583, 521–522 (2020). [DOI] [PubMed] [Google Scholar]
- 729.Tomoda, E. et al. Restoration of mitochondrial function through activation of hypomodified tRNAs with pathogenic mutations associated with mitochondrial diseases. Nucleic Acids Res51, 7563–7579 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 730.Ueda, S. et al. Mitochondrial haplotype mutation alleviates respiratory defect of MELAS by restoring taurine modification in tRNA with 3243A > G mutation. Nucleic Acids Res51, 7480–7495 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 731.Kirino, Y. et al. Acquisition of the wobble modification in mitochondrial tRNALeu(CUN) bearing the G12300A mutation suppresses the MELAS molecular defect. Hum. Mol. Genet15, 897–904 (2006). [DOI] [PubMed] [Google Scholar]
- 732.Asano, K. et al. Metabolic and chemical regulation of tRNA modification associated with taurine deficiency and human disease. Nucleic Acids Res46, 1565–1583 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 733.Fakruddin, M. et al. Defective Mitochondrial tRNA Taurine Modification Activates Global Proteostress and Leads to Mitochondrial Disease. Cell Rep.22, 482–496 (2018). [DOI] [PubMed] [Google Scholar]
- 734.Ohsawa, Y. et al. Taurine supplementation for prevention of stroke-like episodes in MELAS: a multicentre, open-label, 52-week phase III trial. J. Neurol. Neurosurg. Psychiatry90, 529–536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 735.Tischner, C. et al. MTO1 mediates tissue specificity of OXPHOS defects via tRNA modification and translation optimization, which can be bypassed by dietary intervention. Hum. Mol. Genet24, 2247–2266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 736.Bartsakoulia, M. et al. Cysteine Supplementation May be Beneficial in a Subgroup of Mitochondrial Translation Deficiencies. J. Neuromuscul. Dis.3, 363–379 (2016). [DOI] [PubMed] [Google Scholar]
- 737.Boczonadi, V. et al. Altered 2-thiouridylation impairs mitochondrial translation in reversible infantile respiratory chain deficiency. Hum. Mol. Genet22, 4602–4615 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 738.Meseguer, S. et al. microRNA-mediated differential expression of TRMU, GTPBP3 and MTO1 in cell models of mitochondrial-DNA diseases. Sci. Rep.7, 6209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 739.Chu, Y. et al. Mitochondrial tRNA Pseudouridylation Regulates Erythropoiesis Via the mTOR Signaling Pathway: Implications for Mlasa and Treatment Strategies. Blood142(Suppl 1), 139 (2023). [Google Scholar]
- 740.Ling, J. et al. Pathogenic mechanism of a human mitochondrial tRNAPhe mutation associated with myoclonic epilepsy with ragged red fibers syndrome. Proc. Natl. Acad. Sci. USA104, 15299–15304 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 741.Park, H., Davidson, E. & King, M. P. Overexpressed mitochondrial leucyl-tRNA synthetase suppresses the A3243G mutation in the mitochondrial tRNA(Leu(UUR)) gene. Rna14, 2407–2416 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 742.Sasarman, F., Antonicka, H. & Shoubridge, E. A. The A3243G tRNALeu(UUR) MELAS mutation causes amino acid misincorporation and a combined respiratory chain assembly defect partially suppressed by overexpression of EFTu and EFG2. Hum. Mol. Genet17, 3697–3707 (2008). [DOI] [PubMed] [Google Scholar]
- 743.Geromel, V. et al. Coenzyme Q(10) and idebenone in the therapy of respiratory chain diseases: rationale and comparative benefits. Mol. Genet Metab.77, 21–30 (2002). [DOI] [PubMed] [Google Scholar]
- 744.De la Mata, M. et al. Recovery of MERRF fibroblasts and cybrids pathophysiology by coenzyme Q10. Neurotherapeutics9, 446–463 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 745.Chen, Z. et al. Mild clinical manifestation and unusual recovery upon coenzyme Q₁₀ treatment in the first Chinese Leigh syndrome pedigree with mutation m.10197 G>A. Mol. Med Rep.11, 1956–1962 (2015). [DOI] [PubMed] [Google Scholar]
- 746.Ihara, Y., Namba, R., Kuroda, S., Sato, T. & Shirabe, T. Mitochondrial encephalomyopathy (MELAS): pathological study and successful therapy with coenzyme Q10 and idebenone. J. Neurol. Sci.90, 263–271 (1989). [DOI] [PubMed] [Google Scholar]
- 747.Aleo, S. J. et al. Genetic variants affecting NQO1 protein levels impact the efficacy of idebenone treatment in Leber hereditary optic neuropathy. Cell Rep. Med5, 101383 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 748.Haefeli, R. H. et al. NQO1-dependent redox cycling of idebenone: effects on cellular redox potential and energy levels. PLoS One6, e17963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 749.Yu-Wai-Man, P., Soiferman, D., Moore, D. G., Burté, F. & Saada, A. Evaluating the therapeutic potential of idebenone and related quinone analogues in Leber hereditary optic neuropathy. Mitochondrion36, 36–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 750.Heitz, F. D. et al. Idebenone protects against retinal damage and loss of vision in a mouse model of Leber’s hereditary optic neuropathy. PLoS One7, e45182 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 751.Klopstock, T. et al. A randomized placebo-controlled trial of idebenone in Leber’s hereditary optic neuropathy. Brain134, 2677–2686 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 752.van Everdingen, J. A. M. et al. Clinical outcomes of treatment with idebenone in Leber’s hereditary optic neuropathy in the Netherlands: A national cohort study. Acta Ophthalmol.100, 700–706 (2022). [DOI] [PubMed] [Google Scholar]
- 753.Carelli, V. et al. Idebenone treatment in Leber’s hereditary optic neuropathy. Brain134, e188 (2011). [DOI] [PubMed] [Google Scholar]
- 754.Yu-Wai-Man, P. et al. Therapeutic benefit of idebenone in patients with Leber hereditary optic neuropathy: The LEROS nonrandomized controlled trial. Cell Rep. Med5, 101437 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 755.Varela-Fernández, R., Lema-Gesto, M. I., González-Barcia, M. & Otero-Espinar, F. J. Design, development, and characterization of an idebenone-loaded poly-ε-caprolactone intravitreal implant as a new therapeutic approach for LHON treatment. Eur. J. Pharm. Biopharm.168, 195–207 (2021). [DOI] [PubMed] [Google Scholar]
- 756.Varela-Fernández, R. et al. Design, optimization, and in vitro characterization of idebenone-loaded PLGA microspheres for LHON treatment. Int. J. Pharm.616, 121504 (2022). [DOI] [PubMed] [Google Scholar]
- 757.Ikejiri, Y. et al. Idebenone improves cerebral mitochondrial oxidative metabolism in a patient with MELAS. Neurology47, 583–585 (1996). [DOI] [PubMed] [Google Scholar]
- 758.Haginoya, K. et al. Efficacy of idebenone for respiratory failure in a patient with Leigh syndrome: a long-term follow-up study. J. Neurol. Sci.278, 112–114 (2009). [DOI] [PubMed] [Google Scholar]
- 759.Di Prospero, N. A., Baker, A., Jeffries, N. & Fischbeck, K. H. Neurological effects of high-dose idebenone in patients with Friedreich’s ataxia: a randomised, placebo-controlled trial. Lancet Neurol.6, 878–886 (2007). [DOI] [PubMed] [Google Scholar]
- 760.Brière, J. J., Schlemmer, D., Chretien, D. & Rustin, P. Quinone analogues regulate mitochondrial substrate competitive oxidation. Biochem Biophys. Res Commun.316, 1138–1142 (2004). [DOI] [PubMed] [Google Scholar]
- 761.Varricchio, C. et al. The ying and yang of idebenone: Not too little, not too much - cell death in NQO1 deficient cells and the mouse retina. Free Radic. Biol. Med152, 551–560 (2020). [DOI] [PubMed] [Google Scholar]
- 762.Hargreaves, I. P. Coenzyme Q10 as a therapy for mitochondrial disease. Int. J. Biochem Cell Biol.49, 105–111 (2014). [DOI] [PubMed] [Google Scholar]
- 763.Auré, K. et al. Progression despite replacement of a myopathic form of coenzyme Q10 defect. Neurology63, 727–729 (2004). [DOI] [PubMed] [Google Scholar]
- 764.Enns, G. M. et al. Initial experience in the treatment of inherited mitochondrial disease with EPI-743. Mol. Genet Metab.105, 91–102 (2012). [DOI] [PubMed] [Google Scholar]
- 765.Pastore, A. et al. Glutathione: a redox signature in monitoring EPI-743 therapy in children with mitochondrial encephalomyopathies. Mol. Genet Metab.109, 208–214 (2013). [DOI] [PubMed] [Google Scholar]
- 766.Martinelli, D. et al. EPI-743 reverses the progression of the pediatric mitochondrial disease–genetically defined Leigh Syndrome. Mol. Genet Metab.107, 383–388 (2012). [DOI] [PubMed] [Google Scholar]
- 767.Chicani, C. F., Chu, E. R., Miller, G., Kelman, S. E. & Sadun, A. A. Comparing EPI-743 treatment in siblings with Leber’s hereditary optic neuropathy mt14484 mutation. Can. J. Ophthalmol.48, e130–e133 (2013). [DOI] [PubMed] [Google Scholar]
- 768.Peragallo, J. H. & Newman, N. J. Is there treatment for Leber hereditary optic neuropathy? Curr. Opin. Ophthalmol.26, 450–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 769.Haroon, S. et al. N-acetylcysteine and cysteamine bitartrate prevent azide-induced neuromuscular decompensation by restoring glutathione balance in two novel surf1-/- zebrafish deletion models of Leigh syndrome. Hum. Mol. Genet32, 1988–2004 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 770.Guha, S. et al. Combinatorial glucose, nicotinic acid and N-acetylcysteine therapy has synergistic effect in preclinical C. elegans and zebrafish models of mitochondrial complex I disease. Hum. Mol. Genet30, 536–551 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 771.Szeto, H. H. First-in-class cardiolipin-protective compound as a therapeutic agent to restore mitochondrial bioenergetics. Br. J. Pharm.171, 2029–2050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 772.Russo, S., De Rasmo, D., Rossi, R., Signorile, A. & Lobasso, S. SS-31 treatment ameliorates cardiac mitochondrial morphology and defective mitophagy in a murine model of Barth syndrome. Sci. Rep.14, 13655 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 773.Reid Thompson, W. et al. A phase 2/3 randomized clinical trial followed by an open-label extension to evaluate the effectiveness of elamipretide in Barth syndrome, a genetic disorder of mitochondrial cardiolipin metabolism. Genet Med23, 471–478 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 774.Thompson, W. R. et al. Long-term efficacy and safety of elamipretide in patients with Barth syndrome: 168-week open-label extension results of TAZPOWER. Genet Med26, 101138 (2024). [DOI] [PubMed] [Google Scholar]
- 775.Karanjia, R. & Sadun, A. A. Elamipretide Topical Ophthalmic Solution for the Treatment of Subjects with Leber Hereditary Optic Neuropathy: A Randomized Trial. Ophthalmology131, 422–433 (2024). [DOI] [PubMed] [Google Scholar]
- 776.Karaa, A. et al. Randomized dose-escalation trial of elamipretide in adults with primary mitochondrial myopathy. Neurology90, e1212–e1221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 777.Cotticelli, M. G., Crabbe, A. M., Wilson, R. B. & Shchepinov, M. S. Insights into the role of oxidative stress in the pathology of Friedreich ataxia using peroxidation resistant polyunsaturated fatty acids. Redox Biol.1, 398–404 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 778.Zesiewicz, T. et al. Randomized, clinical trial of RT001: Early signals of efficacy in Friedreich’s ataxia. Mov. Disord.33, 1000–1005 (2018). [DOI] [PubMed] [Google Scholar]
- 779.Lynch, D. R. et al. Double blind trial of a deuterated form of linoleic acid (RT001) in Friedreich ataxia. J. Neurol.270, 1615–1623 (2023). [DOI] [PubMed] [Google Scholar]
- 780.Beyrath, J. et al. KH176 Safeguards Mitochondrial Diseased Cells from Redox Stress-Induced Cell Death by Interacting with the Thioredoxin System/Peroxiredoxin Enzyme Machinery. Sci. Rep.8, 6577 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 781.de Haas, R. et al. Therapeutic effects of the mitochondrial ROS-redox modulator KH176 in a mammalian model of Leigh Disease. Sci. Rep.7, 11733 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 782.Klein Gunnewiek, T. M. et al. Sonlicromanol improves neuronal network dysfunction and transcriptome changes linked to m.3243A>G heteroplasmy in iPSC-derived neurons. Stem Cell Rep.16, 2197–2212 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 783.Janssen, M. C. H. et al. The KHENERGY Study: Safety and Efficacy of KH176 in Mitochondrial m.3243A>G Spectrum Disorders. Clin. Pharm. Ther.105, 101–111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 784.Jiang, Z. et al. Omaveloxolone inhibits IL-1β-induced chondrocyte apoptosis through the Nrf2/ARE and NF-κB signalling pathways in vitro and attenuates osteoarthritis in vivo. Front Pharm.13, 952950 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 785.Dinkova-Kostova, A. T. et al. Extremely potent triterpenoid inducers of the phase 2 response: correlations of protection against oxidant and inflammatory stress. Proc. Natl. Acad. Sci. USA102, 4584–4589 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 786.Abeti, R., Baccaro, A., Esteras, N. & Giunti, P. Novel Nrf2-Inducer Prevents Mitochondrial Defects and Oxidative Stress in Friedreich’s Ataxia Models. Front Cell Neurosci.12, 188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 787.Lynch, D. R. et al. Propensity matched comparison of omaveloxolone treatment to Friedreich ataxia natural history data. Ann. Clin. Transl. Neurol.11, 4–16 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 788.Madsen, K. L. et al. Safety and efficacy of omaveloxolone in patients with mitochondrial myopathy: MOTOR trial. Neurology94, e687–e698 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 789.Qureshi, M. Y. et al. Safety and efficacy of (+)-epicatechin in subjects with Friedreich’s ataxia: A phase II, open-label, prospective study. J. Inherit. Metab. Dis.44, 502–514 (2021). [DOI] [PubMed] [Google Scholar]
- 790.Lynch, D. R. et al. A0001 in Friedreich ataxia: biochemical characterization and effects in a clinical trial. Mov. Disord.27, 1026–1033 (2012). [DOI] [PubMed] [Google Scholar]
- 791.Clayton, R. et al. Safety, pharmacokinetics, and pharmacodynamics of nomlabofusp (CTI-1601) in Friedreich’s ataxia. Ann. Clin. Transl. Neurol.11, 540–553 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 792.Huang, Y. et al. The PPAR pan-agonist bezafibrate ameliorates cardiomyopathy in a mouse model of Barth syndrome. Orphanet J. Rare Dis.12, 49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 793.Schafer, C. et al. The Effects of PPAR Stimulation on Cardiac Metabolic Pathways in Barth Syndrome Mice. Front Pharm.9, 318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 794.da Rosa-Junior, N. T. et al. Antioxidant system disturbances and mitochondrial dysfunction induced by 3-methyglutaric acid in rat heart are prevented by bezafibrate. Eur. J. Pharm.924, 174950 (2022). [DOI] [PubMed] [Google Scholar]
- 795.Inak, G. et al. Defective metabolic programming impairs early neuronal morphogenesis in neural cultures and an organoid model of Leigh syndrome. Nat. Commun.12, 1929 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 796.Lyu, J. et al. Bezafibrate Rescues Mitochondrial Encephalopathy in Mice via Induction of Daily Torpor and Hypometabolic State. Neurotherapeutics19, 994–1006 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 797.Steele, H. et al. Metabolic effects of bezafibrate in mitochondrial disease. EMBO Mol. Med12, e11589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 798.Di Donfrancesco, A. et al. PPAR-gamma agonist pioglitazone recovers mitochondrial quality control in fibroblasts from PITRM1-deficient patients. Front Pharm.14, 1220620 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 799.Burgin, H. J., Lopez Sanchez, M. I. G., Smith, C. M., Trounce, I. A. & McKenzie, M. Pioglitazone and Deoxyribonucleoside Combination Treatment Increases Mitochondrial Respiratory Capacity in m.3243A>G MELAS Cybrid Cells. Int. J. Mol. Sci.21, 2139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 800.Rodríguez-Pascau, L. et al. PPAR gamma agonist leriglitazone improves frataxin-loss impairments in cellular and animal models of Friedreich Ataxia. Neurobiol. Dis.148, 105162 (2021). [DOI] [PubMed] [Google Scholar]
- 801.Ogasawara, E., Nakada, K. & Hayashi, J. Lactic acidemia in the pathogenesis of mice carrying mitochondrial DNA with a deletion. Hum. Mol. Genet19, 3179–3189 (2010). [DOI] [PubMed] [Google Scholar]
- 802.Barshop, B. A. et al. Chronic treatment of mitochondrial disease patients with dichloroacetate. Mol. Genet Metab.83, 138–149 (2004). [DOI] [PubMed] [Google Scholar]
- 803.Berendzen, K., Theriaque, D. W., Shuster, J. & Stacpoole, P. W. Therapeutic potential of dichloroacetate for pyruvate dehydrogenase complex deficiency. Mitochondrion6, 126–135 (2006). [DOI] [PubMed] [Google Scholar]
- 804.Böger, R. H. The pharmacodynamics of L-arginine. J. Nutr.137, 1650s–1655s (2007). [DOI] [PubMed] [Google Scholar]
- 805.Rodan, L. H. et al. L-Arginine Affects Aerobic Capacity and Muscle Metabolism in MELAS (Mitochondrial Encephalomyopathy, Lactic Acidosis and Stroke-Like Episodes) Syndrome. PLoS One10, e0127066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 806.Koga, Y. et al. Endothelial dysfunction in MELAS improved by l-arginine supplementation. Neurology66, 1766–1769 (2006). [DOI] [PubMed] [Google Scholar]
- 807.Siddiq, I., Widjaja, E. & Tein, I. Clinical and radiologic reversal of stroke-like episodes in MELAS with high-dose L-arginine. Neurology85, 197–198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 808.Koga, Y. et al. Therapeutic regimen of L-arginine for MELAS: 9-year, prospective, multicenter, clinical research. J. Neurol.265, 2861–2874 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 809.Stefanetti, R. J. et al. l-Arginine in Mitochondrial Encephalopathy, Lactic Acidosis, and Stroke-like Episodes: A Systematic Review. Neurology98, e2318–e2328 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 810.Wei, Y., Cui, L. & Pen, B. L-Arginine prevents stroke-like episodes but not brain atrophy: a 20-year follow-up of a MELAS patient. Neurol. Sci.40, 209–211 (2019). [DOI] [PubMed] [Google Scholar]
- 811.El-Hattab, A. W. et al. Impaired nitric oxide production in children with MELAS syndrome and the effect of arginine and citrulline supplementation. Mol. Genet Metab.117, 407–412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 812.Kakhlon, O. et al. Cell functions impaired by frataxin deficiency are restored by drug-mediated iron relocation. Blood112, 5219–5227 (2008). [DOI] [PubMed] [Google Scholar]
- 813.Pandolfo, M. et al. Deferiprone in Friedreich ataxia: a 6-month randomized controlled trial. Ann. Neurol.76, 509–521 (2014). [DOI] [PubMed] [Google Scholar]
- 814.Lim, C. K., Kalinowski, D. S. & Richardson, D. R. Protection against hydrogen peroxide-mediated cytotoxicity in Friedreich’s ataxia fibroblasts using novel iron chelators of the 2-pyridylcarboxaldehyde isonicotinoyl hydrazone class. Mol. Pharm.74, 225–235 (2008). [DOI] [PubMed] [Google Scholar]
- 815.Tomassini, B. et al. Interferon gamma upregulates frataxin and corrects the functional deficits in a Friedreich ataxia model. Hum. Mol. Genet21, 2855–2861 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 816.Luffarelli, R. et al. Interferon Gamma Enhances Cytoprotective Pathways via Nrf2 and MnSOD Induction in Friedreich’s Ataxia Cells. Int. J. Mol. Sci.24, 12687 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 817.Wyller, V. B. et al. Interferon gamma may improve cardiac function in Friedreich’s ataxia cardiomyopathy. Int. J. Cardiol.221, 376–378 (2016). [DOI] [PubMed] [Google Scholar]
- 818.Boesch, S. et al. Neurological effects of recombinant human erythropoietin in Friedreich’s ataxia: a clinical pilot trial. Mov. Disord.23, 1940–1944 (2008). [DOI] [PubMed] [Google Scholar]
- 819.Sturm, B. et al. Carbamylated erythropoietin increases frataxin independent from the erythropoietin receptor. Eur. J. Clin. Invest40, 561–565 (2010). [DOI] [PubMed] [Google Scholar]
- 820.Boesch, S. et al. Safety and tolerability of carbamylated erythropoietin in Friedreich’s ataxia. Mov. Disord.29, 935–939 (2014). [DOI] [PubMed] [Google Scholar]
- 821.Saada, A. et al. Mitochondrial deoxyribonucleoside triphosphate pools in thymidine kinase 2 deficiency. Biochem Biophys. Res Commun.310, 963–966 (2003). [DOI] [PubMed] [Google Scholar]
- 822.Garone, C. et al. Deoxypyrimidine monophosphate bypass therapy for thymidine kinase 2 deficiency. EMBO Mol. Med6, 1016–1027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 823.Lopez-Gomez, C. et al. Deoxycytidine and Deoxythymidine Treatment for Thymidine Kinase 2 Deficiency. Ann. Neurol.81, 641–652 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 824.Blázquez-Bermejo, C. et al. Age-related metabolic changes limit efficacy of deoxynucleoside-based therapy in thymidine kinase 2-deficient mice. EBioMedicine46, 342–355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 825.Domínguez-González, C. et al. Deoxynucleoside Therapy for Thymidine Kinase 2-Deficient Myopathy. Ann. Neurol.86, 293–303 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 826.Bermejo-Guerrero, L. et al. Remarkable clinical improvement with oral nucleoside treatment in a patient with adult-onset TK2 deficiency: A case report. Mitochondrion76, 101879 (2024). [DOI] [PubMed] [Google Scholar]
- 827.Lopez-Gomez, C. et al. Synergistic Deoxynucleoside and Gene Therapies for Thymidine Kinase 2 Deficiency. Ann. Neurol.90, 640–652 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 828.Murphy, J. L. et al. Resistance training in patients with single, large-scale deletions of mitochondrial DNA. Brain131, 2832–2840 (2008). [DOI] [PubMed] [Google Scholar]
- 829.Shoubridge, E. A., Johns, T. & Karpati, G. Complete restoration of a wild-type mtDNA genotype in regenerating muscle fibres in a patient with a tRNA point mutation and mitochondrial encephalomyopathy. Hum. Mol. Genet6, 2239–2242 (1997). [DOI] [PubMed] [Google Scholar]
- 830.Taivassalo, T. et al. Gene shifting: a novel therapy for mitochondrial myopathy. Hum. Mol. Genet8, 1047–1052 (1999). [DOI] [PubMed] [Google Scholar]
- 831.Zelissen, R. et al. Fusion of Wild-Type Mesoangioblasts with Myotubes of mtDNA Mutation Carriers Leads to a Proportional Reduction in mtDNA Mutation Load. Int. J. Mol. Sci.24, 2679 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 832.Sancricca, C. et al. Vessel-associated stem cells from skeletal muscle: From biology to future uses in cell therapy. World J. Stem Cells2, 39–49 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 833.Dellavalle, A. et al. Pericytes resident in postnatal skeletal muscle differentiate into muscle fibres and generate satellite cells. Nat. Commun.2, 499 (2011). [DOI] [PubMed] [Google Scholar]
- 834.Tedesco, F. S. & Cossu, G. Stem cell therapies for muscle disorders. Curr. Opin. Neurol.25, 597–603 (2012). [DOI] [PubMed] [Google Scholar]
- 835.van Tienen, F. et al. Healthy, mtDNA-mutation free mesoangioblasts from mtDNA patients qualify for autologous therapy. Stem Cell Res Ther.10, 405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 836.Nederveen, J. P. et al. The influence of capillarization on satellite cell pool expansion and activation following exercise-induced muscle damage in healthy young men. J. Physiol.596, 1063–1078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 837.Röeben, B. et al. Hemodialysis in MNGIE transiently reduces serum and urine levels of thymidine and deoxyuridine, but not CSF levels and neurological function. Orphanet. J. Rare Dis.12, 135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 838.Ariaudo, C. et al. Mitochondrial neurogastrointestinal encephalomyopathy treated with peritoneal dialysis and bone marrow transplantation. J. Nephrol.28, 125–127 (2015). [DOI] [PubMed] [Google Scholar]
- 839.Zaidman, I. et al. Hematopoietic stem cell transplantation for mitochondrial neurogastrointestinal encephalopathy: A single-center experience underscoring the multiple factors involved in the prognosis. Pediatr. Blood Cancer68, e28926 (2021). [DOI] [PubMed] [Google Scholar]
- 840.Torres-Torronteras, J. et al. Long-Term Restoration of Thymidine Phosphorylase Function and Nucleoside Homeostasis Using Hematopoietic Gene Therapy in a Murine Model of Mitochondrial Neurogastrointestinal Encephalomyopathy. Hum. Gene Ther.27, 656–667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 841.Torres-Torronteras, J. et al. Hematopoietic gene therapy restores thymidine phosphorylase activity in a cell culture and a murine model of MNGIE. Gene Ther.18, 795–806 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 842.Sicurelli, F. et al. Clinical and biochemical improvement following HSCT in a patient with MNGIE: 1-year follow-up. J. Neurol.259, 1985–1987 (2012). [DOI] [PubMed] [Google Scholar]
- 843.Halter, J. et al. Allogeneic hematopoietic SCT as treatment option for patients with mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): a consensus conference proposal for a standardized approach. Bone Marrow Transpl.46, 330–337 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 844.Ozek, G. et al. Hematopoietic stem cell transplantation with reduced toxicity conditioning regimen in mitochondrial neurogastrointestinal encephalopathy syndrome. Pediatr. Blood Cancer70, e30334 (2023). [DOI] [PubMed] [Google Scholar]
- 845.Filosto, M. et al. Course and management of allogeneic stem cell transplantation in patients with mitochondrial neurogastrointestinal encephalomyopathy. J. Neurol.259, 2699–2706 (2012). [DOI] [PubMed] [Google Scholar]
- 846.Meinders, M. et al. Expression and Retention of Thymidine Phosphorylase in Cultured Reticulocytes as a Novel Treatment for MNGIE. Mol. Ther. Methods Clin. Dev.17, 822–830 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 847.Levene, M. et al. Preclinical toxicity evaluation of erythrocyte-encapsulated thymidine phosphorylase in BALB/c mice and beagle dogs: an enzyme-replacement therapy for mitochondrial neurogastrointestinal encephalomyopathy. Toxicol. Sci.131, 311–324 (2013). [DOI] [PubMed] [Google Scholar]
- 848.Bax, B. E. et al. Clinical and biochemical improvements in a patient with MNGIE following enzyme replacement. Neurology81, 1269–1271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 849.De Vocht, C. et al. Assessment of stability, toxicity and immunogenicity of new polymeric nanoreactors for use in enzyme replacement therapy of MNGIE. J. Control Release137, 246–254 (2009). [DOI] [PubMed] [Google Scholar]
- 850.Boschetti, E. et al. Liver as a source for thymidine phosphorylase replacement in mitochondrial neurogastrointestinal encephalomyopathy. PLoS One9, e96692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 851.Kripps, K. et al. Successful liver transplantation in mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). Mol. Genet Metab.130, 58–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 852.De Giorgio, R. et al. Liver transplantation for mitochondrial neurogastrointestinal encephalomyopathy. Ann. Neurol.80, 448–455 (2016). [DOI] [PubMed] [Google Scholar]
- 853.Dionisi-Vici, C. et al. Liver transplant in ethylmalonic encephalopathy: a new treatment for an otherwise fatal disease. Brain139, 1045–1051 (2016). [DOI] [PubMed] [Google Scholar]
- 854.Zhou, G. P. et al. Compromised therapeutic value of pediatric liver transplantation in ethylmalonic encephalopathy: A case report. World J. Gastroenterol.26, 6295–6303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 855.Tzakis, A. G. et al. Liver, pancreas and kidney transplantation for the treatment of Wolcott-Rallison syndrome. Am. J. Transpl.15, 565–567 (2015). [DOI] [PubMed] [Google Scholar]
- 856.Nordström, J. et al. First European Case of Simultaneous Liver and Pancreas Transplantation as Treatment of Wolcott-Rallison Syndrome in a Small Child. Transplantation104, 522–525 (2020). [DOI] [PubMed] [Google Scholar]
- 857.Shimura, M. et al. Clinical and molecular basis of hepatocerebral mitochondrial DNA depletion syndrome in Japan: evaluation of outcomes after liver transplantation. Orphanet J. Rare Dis.15, 169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 858.Taivassalo, T. et al. The spectrum of exercise tolerance in mitochondrial myopathies: a study of 40 patients. Brain126, 413–423 (2003). [DOI] [PubMed] [Google Scholar]
- 859.Jeppesen, T. D. et al. Aerobic training is safe and improves exercise capacity in patients with mitochondrial myopathy. Brain129, 3402–3412 (2006). [DOI] [PubMed] [Google Scholar]
- 860.Taivassalo, T. et al. Endurance training and detraining in mitochondrial myopathies due to single large-scale mtDNA deletions. Brain129, 3391–3401 (2006). [DOI] [PubMed] [Google Scholar]
- 861.Taivassalo, T. et al. Aerobic conditioning in patients with mitochondrial myopathies: physiological, biochemical, and genetic effects. Ann. Neurol.50, 133–141 (2001). [DOI] [PubMed] [Google Scholar]
- 862.Taivassalo, T. et al. Combined aerobic training and dichloroacetate improve exercise capacity and indices of aerobic metabolism in muscle cytochrome oxidase deficiency. Neurology47, 529–534 (1996). [DOI] [PubMed] [Google Scholar]
- 863.Vainshtein, A., Tryon, L. D., Pauly, M. & Hood, D. A. Role of PGC-1α during acute exercise-induced autophagy and mitophagy in skeletal muscle. Am. J. Physiol. Cell Physiol.308, C710–C719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 864.Geng, T. et al. PGC-1alpha plays a functional role in exercise-induced mitochondrial biogenesis and angiogenesis but not fiber-type transformation in mouse skeletal muscle. Am. J. Physiol. Cell Physiol.298, C572–C579 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 865.Fiuza-Luces, C. et al. Physical Exercise and Mitochondrial Disease: Insights From a Mouse Model. Front Neurol.10, 790 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 866.Bhattacharya, S., Yin, J., Huo, W. & Chaum, E. Modeling of mitochondrial bioenergetics and autophagy impairment in MELAS-mutant iPSC-derived retinal pigment epithelial cells. Stem Cell Res Ther.13, 260 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 867.Mazzara, P. G. et al. Frataxin gene editing rescues Friedreich’s ataxia pathology in dorsal root ganglia organoid-derived sensory neurons. Nat. Commun.11, 4178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 868.Chen, J. R. et al. Nuclear modifier YARS2 allele correction restored retinal ganglion cells-specific deficiencies in Leber’s hereditary optic neuropathy. Hum. Mol. Genet32, 1539–1551 (2023). [DOI] [PubMed] [Google Scholar]
- 869.Guo, J. et al. A Combined Model of Human iPSC-Derived Liver Organoids and Hepatocytes Reveals Ferroptosis in DGUOK Mutant mtDNA Depletion Syndrome. Adv. Sci. (Weinh.)8, 2004680 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 870.Hong, Y. et al. The NAD(+) Precursor Nicotinamide Riboside Rescues Mitochondrial Defects and Neuronal Loss in iPSC derived Cortical Organoid of Alpers’ Disease. Int. J. Biol. Sci.20, 1194–1217 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 871.Wei, W., Gaffney, D. J. & Chinnery, P. F. Cell reprogramming shapes the mitochondrial DNA landscape. Nat. Commun.12, 5241 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 872.Deuse, T. et al. De novo mutations in mitochondrial DNA of iPSCs produce immunogenic neoepitopes in mice and humans. Nat. Biotechnol.37, 1137–1144 (2019). [DOI] [PubMed] [Google Scholar]
- 873.Yoshino, T. et al. Generation of ovarian follicles from mouse pluripotent stem cells. Science373, eabe0237 (2021). [DOI] [PubMed] [Google Scholar]
- 874.Liu, L. et al. MELAS-Derived Neurons Functionally Improve by Mitochondrial Transfer from Highly Purified Mesenchymal Stem Cells (REC). Int. J. Mol. Sci.24, 17186 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 875.Jain, I. H. et al. Leigh Syndrome Mouse Model Can Be Rescued by Interventions that Normalize Brain Hyperoxia, but Not HIF Activation. Cell Metab.30, 824–832.e823 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 876.Porcelli, S., Marzorati, M., Morandi, L. & Grassi, B. Home-based aerobic exercise training improves skeletal muscle oxidative metabolism in patients with metabolic myopathies. J. Appl. Physiol. (1985)121, 699–708 (2016). [DOI] [PubMed] [Google Scholar]
- 877.D’Angelo, R. et al. Liver transplantation in mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): clinical long-term follow-up and pathogenic implications. J. Neurol.267, 3702–3710 (2020). [DOI] [PubMed] [Google Scholar]
- 878.Palacios-González, C. Mexico and mitochondrial replacement techniques: what a mess. Br. Med. Bull.128, 97–107 (2018). [DOI] [PubMed] [Google Scholar]
- 879.Stewart, J. B. Current progress with mammalian models of mitochondrial DNA disease. J. Inherit. Metab. Dis.44, 325–342 (2021). [DOI] [PubMed] [Google Scholar]
- 880.Figueroa-Martínez, F. et al. What limits the allotopic expression of nucleus-encoded mitochondrial genes? The case of the chimeric Cox3 and Atp6 genes. Mitochondrion11, 147–154 (2011). [DOI] [PubMed] [Google Scholar]
- 881.Martikainen, M. H. & Majamaa, K. Incidence and prevalence of mtDNA-related adult mitochondrial disease in Southwest Finland, 2009-2022: an observational, population-based study. BMJ Neurol. Open6, e000546 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 882.Gorman, G. S. et al. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann. Neurol.77, 753–759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 883.Missen, S. et al. Mitochondrial disease in New Zealand: a nationwide prevalence study. Intern. Med. J.54, 388–397 (2024). [DOI] [PubMed] [Google Scholar]
- 884.Wong, T. S. et al. Mitochondrial diseases in Hong Kong: prevalence, clinical characteristics and genetic landscape. Orphanet. J. Rare Dis.18, 43 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 885.Ibayashi, K. et al. Estimation of the Number of Patients With Mitochondrial Diseases: A Descriptive Study Using a Nationwide Database in Japan. J. Epidemiol.33, 68–75 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 886.Bellusci, M. et al. The Genetic Landscape of Mitochondrial Diseases in Spain: A Nationwide Call. Genes (Basel)12, 1590 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 887.Castro-Gago, M. et al. Epidemiology of pediatric mitochondrial respiratory chain disorders in northwest Spain. Pediatr. Neurol.34, 204–211 (2006). [DOI] [PubMed] [Google Scholar]
- 888.Takano, F. et al. Incidence of Leber hereditary optic neuropathy in 2019 in Japan: a second nationwide questionnaire survey. Orphanet. J. Rare Dis.17, 319 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 889.Yu-Wai-Man, P. et al. The epidemiology of Leber hereditary optic neuropathy in the North East of England. Am. J. Hum. Genet72, 333–339 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 890.Puomila, A. et al. Epidemiology and penetrance of Leber hereditary optic neuropathy in Finland. Eur. J. Hum. Genet.15, 1079–1089 (2007). [DOI] [PubMed] [Google Scholar]
- 891.Rosenberg, T. et al. Prevalence and Genetics of Leber Hereditary Optic Neuropathy in the Danish Population. Invest Ophthalmol. Vis. Sci.57, 1370–1375 (2016). [DOI] [PubMed] [Google Scholar]
- 892.Lopez Sanchez, M. I. G. et al. Establishing risk of vision loss in Leber hereditary optic neuropathy. Am. J. Hum. Genet108, 2159–2170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 893.Spruijt, L. et al. Influence of mutation type on clinical expression of Leber hereditary optic neuropathy. Am. J. Ophthalmol.141, 676–682 (2006). [DOI] [PubMed] [Google Scholar]
- 894.Jančić, J. et al. Leber hereditary optic neuropathy in the population of Serbia. Eur. J. Paediatr. Neurol.18, 354–359 (2014). [DOI] [PubMed] [Google Scholar]
- 895.Yatsuga, S. et al. MELAS: a nationwide prospective cohort study of 96 patients in Japan. Biochim. Biophys. Acta1820, 619–624 (2012). [DOI] [PubMed] [Google Scholar]
- 896.Darin, N., Oldfors, A., Moslemi, A. R., Holme, E. & Tulinius, M. The incidence of mitochondrial encephalomyopathies in childhood: clinical features and morphological, biochemical, and DNA abnormalities. Ann. Neurol.49, 377–383 (2001). [PubMed] [Google Scholar]
- 897.Wedding, I. M. et al. Friedreich ataxia in Norway - an epidemiological, molecular and clinical study. Orphanet. J. Rare Dis.10, 108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 898.Leone, M. et al. Friedreich’s ataxia: a descriptive epidemiological study in an Italian population. Clin. Genet.38, 161–169 (1990). [DOI] [PubMed] [Google Scholar]
- 899.Yu-Wai-Man, P. et al. The prevalence and natural history of dominant optic atrophy due to OPA1 mutations. Ophthalmology117, 1538–1546 (2010). 1546.e1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 900.Kjer, B., Eiberg, H., Kjer, P. & Rosenberg, T. Dominant optic atrophy mapped to chromosome 3q region. II. Clinical and epidemiological aspects. Acta Ophthalmol. Scand.74, 3–7 (1996). [DOI] [PubMed] [Google Scholar]
- 901.Guillausseau, P. J. et al. Maternally inherited diabetes and deafness: a multicenter study. Ann. Intern Med.134, 721–728 (2001). [DOI] [PubMed] [Google Scholar]
- 902.Miller, P. C., Ren, M., Schlame, M., Toth, M. J. & Phoon, C. K. L. A Bayesian Analysis to Determine the Prevalence of Barth Syndrome in the Pediatric Population. J. Pediatr.217, 139–144 (2020). [DOI] [PubMed] [Google Scholar]
- 903.Alexander, C. et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet26, 211–215 (2000). [DOI] [PubMed] [Google Scholar]
- 904.Delettre, C. et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet26, 207–210 (2000). [DOI] [PubMed] [Google Scholar]
- 905.Wong, L. J. et al. Molecular and clinical genetics of mitochondrial diseases due to POLG mutations. Hum. Mutat.29, E150–E172 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]