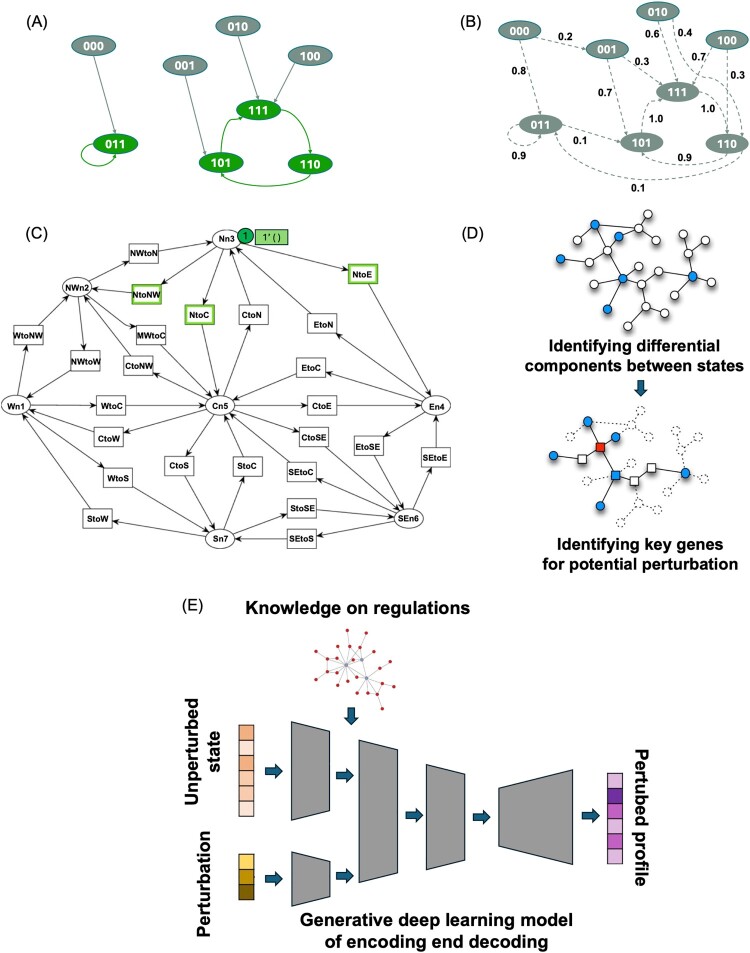
Figure 2.
Popular models for studying dynamics of cellular states. (A) An example of Boolean network of three genes. Each state is represented as a node with three 0 (inactive) or 1 (active) as values. Each state will go to another state in the next time step in a deterministic way. Green states are attractor (steady) states and gray states are unstable intermediate states. (B) An example of Probabilistic Boolean network of three genes. State transition occurs probabilistically based on the specified probability. (C) An example of a Petri-net model. Circles represent places and rectangles represent transitions. (D) Estimating key intervention points from static regulatory models. Static regulatory model is built from data of multiple cellular states. Regulatory components that are different between states are identified (blue-colored nodes). Based on identified regulatory components of difference, key biological components or genes (red-colored nodes) of high impact on the difference are identified as potential interventions that can drive differences between states. (E) Deep learning approach of estimating changed cellular state (omics profile) based on a current state and perturbation information. Prior knowledge on genetic regulations is often integrated in the process of learning and data embedding. Generative models can be used to construct a new profile of cellular state after perturbation.