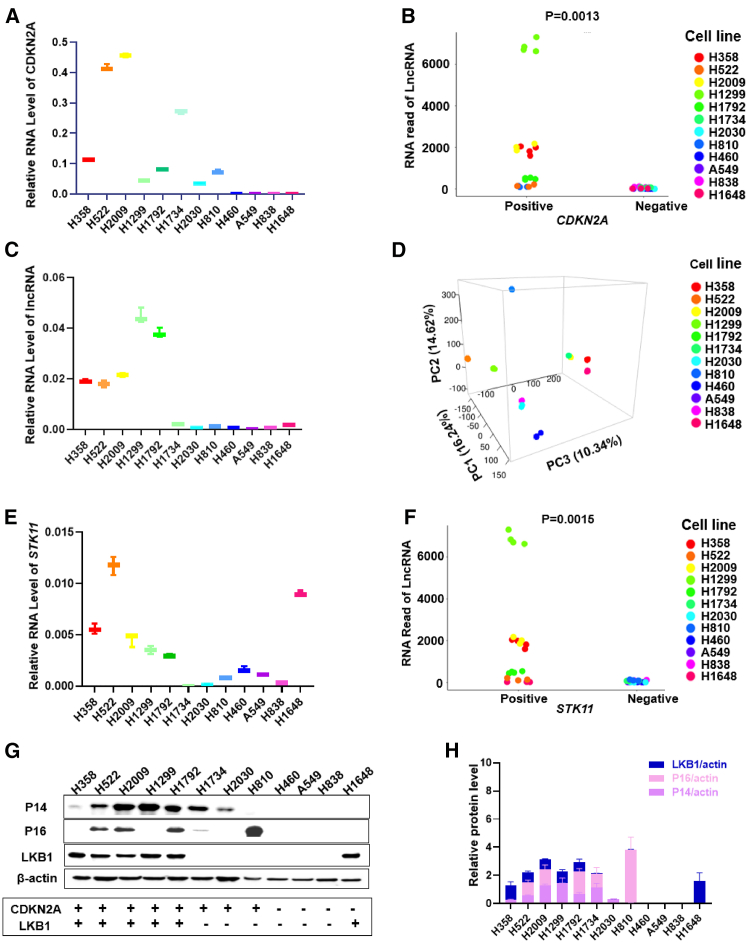
Figure 1.
The association of CyKILR expression with an active, WT STK11, and CDKN2A genes in NSCLCs
Total RNA was collected from the indicated NSCLC cell lines and reverse transcribed (RT). qRT-PCR and deep RNA-seq were conducted. The R program was used for analysis of deep RNA-seq data. Western blotting was conducted on 12 NSCLC lines to assess the expression of CDKN2A and STK11 gene products with CyKILR expression. (A) The mRNA level of CDKN2A across the 12 NSCLC lines normalized to GAPDH mRNA levels. (B) Correlation plot of deep RNA-seq data displaying the relationship between CDKN2A gene status and the expression of the lncRNA, CyKILR. (C) CyKILR RNA levels by qRT-PCR across the 12 NSCLC lines normalized to GAPDH mRNA levels. Data in (A) and (C) are presented as mean (SD) (n ≥ 3 replicates). (D) Three-dimensional PCA plot representing the variation of CDKN2A and CyKILR expression for the 12 NSCLC lines. (E) The mRNA level of the STK11 gene product, LKB1, across the 12 NSCLC lines normalized to GAPDH mRNA levels. Data in (E) are presented as mean ± SD (n ≥ 3 replicates). (F) Correlation plot of deep RNA-seq data to an active, WT STK11 gene displaying the relationship between STK11 mRNA expression and CyKILR expression. (G) Relative protein levels of the indicated factors across a panel of 12 cell lines with β-actin used for normalization. Data are representative of n = 3. (H) The protein levels of the CDKN2A gene products, p14 and p16, the STK11 gene product, LKB1, across the 12 NSCLC lines with β-actin used for normalization. Data in (H) are presented as mean ± SD (n ≥ 3 replicates). Data in (B) and (F) were analyzed by unpaired t test with Welch’s correction. Reverse transcription (RT) “minus” controls (−RT) were included in all RT-PCR experiments and replicates with no signal observed.