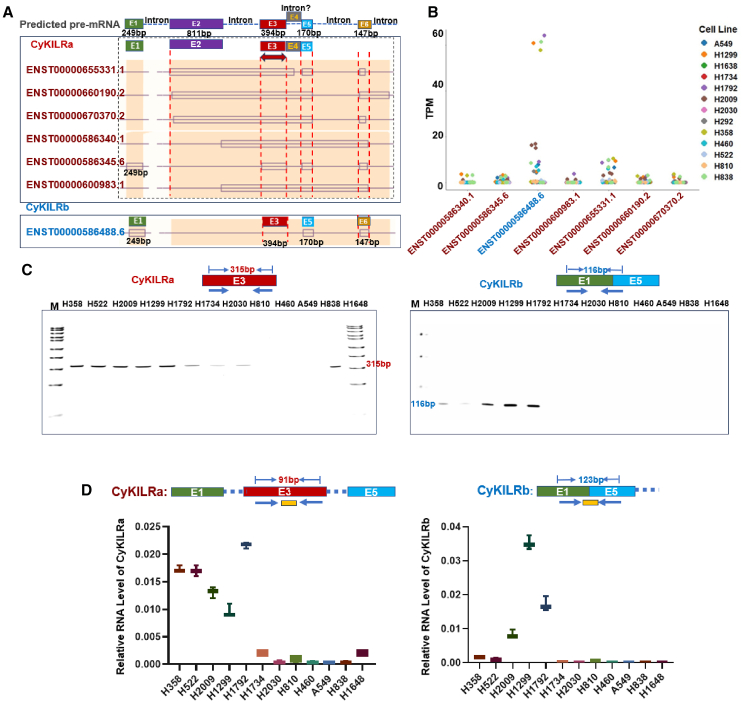
Figure 2.
CyKILR is characterized by two distinct isoform groups based on exon 3 inclusion/exclusion
Total RNA was collected from the indicated NSCLC cell lines. Short-read deep RNA-seq was conducted on these RNA samples. The R program was used for analysis of deep RNA-seq data. (A) The predicted pre-mRNA structure of CyKILR based on data from Ensembl. The CyKILR RNA splice variants were categorized into two distinct groups based on the inclusion or exclusion of exon 3 into the mature RNA transcript. Predicted exon and full-length CyKILR splice variants are annotated. Of note, the designated exon 6 is predicted to have multiple 5′ splice sites for several CyKILRa variants. (B) The expression level of CyKILR isoforms across the 12 cell lines from the deep RNA-seq conducted on RNA samples from these cell lines. The R program was used for analysis of deep RNA-seq data. (C) Relative RNA levels of CyKILRa (including exon 3) and CyKILRb (excluding exon 3) variants across a panel of 12 NSCLC cell lines using RT-PCR. (D) The RNA expression levels of CyKILRa and CyKILRb are depicted by using a customized quantitative primer and probe strategy for the detection of CyKILRa and CyKILRb variants by qRT-PCR and normalized to GAPDH mRNA levels. The data in (D) are presented as mean (SD) (n ≥ 3 replicates). −RT controls were included in all experiments and replicates with no signal observed.