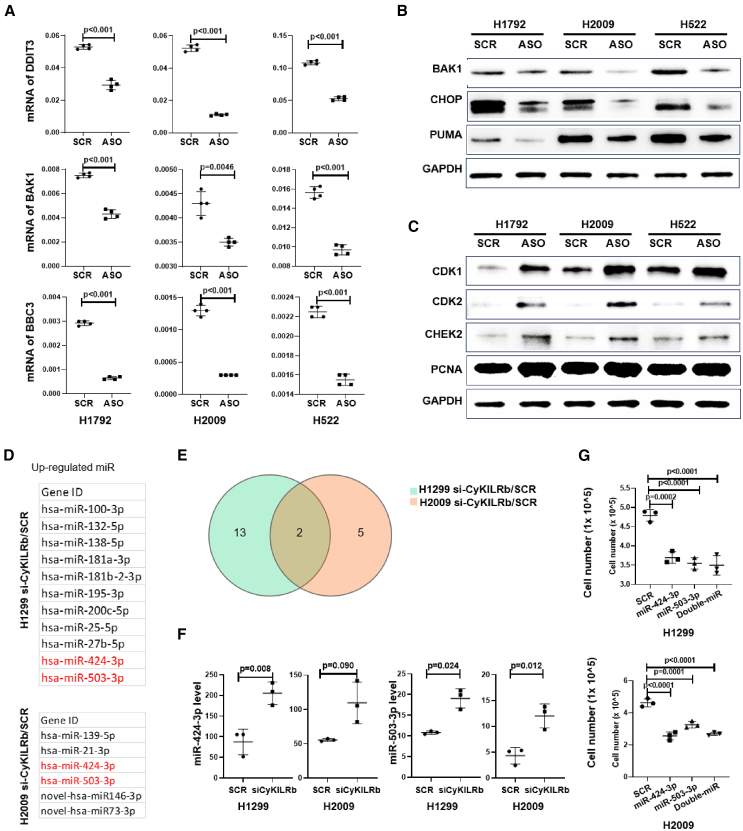
Figure 7.
CyKILRa and CyKILRb apposingly regulate cell growth signaling pathways
H2009 or H1792 cells were treated with either SCR ASO (SCR) or CyKILRa ASO (ASO), and H1299 and H2009 cells were treated with either siRNA-Con (SCR) or CyKILRb siRNA (siCyKILRb). Total RNA from cell subjected to these treatments underwent deep RNA-seq. Following data filtering and clean read alignment, gene expression levels and differential expression gene analysis were conducted using RSEM (v1.2.28) and DESeq2. Kyoto Encyclopedia of Genes and Genomes enrichment analysis of annotated differentially expressed genes was performed using R Phyper. The CyKILRa ASO-treated groups exhibited regulation of cell apoptosis and cell-cycle pathways compared with the control-treated groups (A–C). While the CyKILRb siRNA-treated groups exhibited the regulation of miRNAs (D–G). (A) RNA levels of key apoptosis markers were down-regulated by CyKILRa ASO in all cell lines as assessed by qRT-PCR. −RT controls were included in all experiments and replicates with no signal observed. (B) Protein levels of key apoptosis markers were down-regulated by CyKILRa ASO in all cell lines as assessed by western blotting. (C) Protein levels of key cell-cycle markers were up-regulated by CyKILRa ASO in all cell lines. (D) Up-regulated miRNAs by CyKILRb knockdown in both cell lines. (E) The Venn diagram displays the upregulated miRNAs resulting from CyKILRb knockdown in both H1299 and H2009 cell lines. (F) RNA levels of miR-424–3P and miR-503-3P in H1299 and H2009 cells treated with si-CyKILRb vs. scramble control (SCR). (G) Cell proliferation assay comparing miR-424-3P and miR-503-3P mimic-treated H1299 and H2009 cells vs. scramble-treated (SCR) cells 48 h after transfection. The data in (A), (F), and (G) are presented as boxplots (n ≥ 3 replicates), and statistical analysis was performed using the unpaired t test with Welch’s correction. Data in (B) and (C) are representative of n = 3.