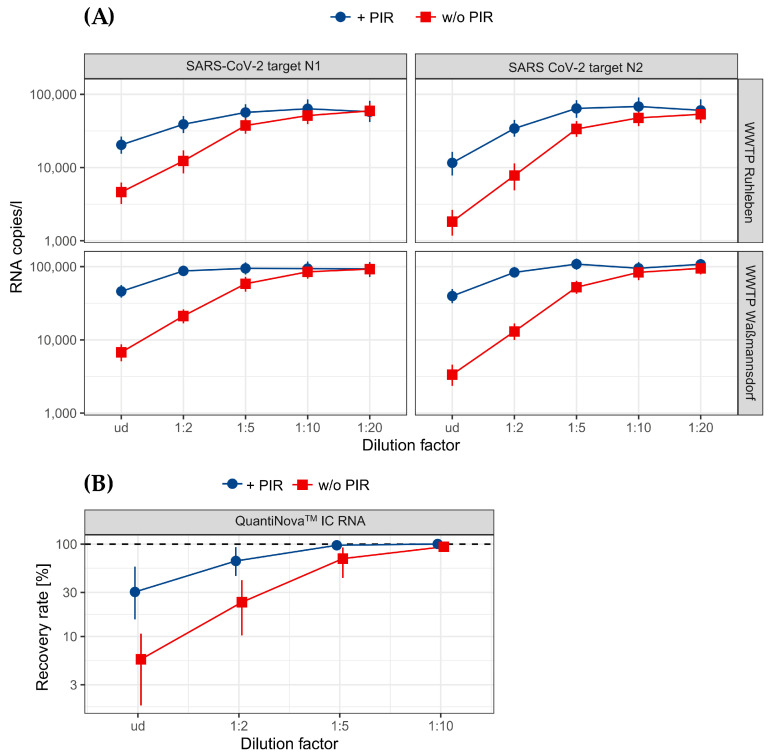
Figure 2.
PCR inhibitor removal (PIR) leads to improved dPCR runs and increased detection of target RNA copy numbers. (A) The data show comparative dPCR analyses using TNA extracts of the WWTPs Ruhleben (n = 21) and Waßmannsdorf (n = 19) from 4 February 2024 to 27 March 2024 with (blue, circle) or without (red, square) PIR treatment as templates. PIR was conducted using dilutions and the OneStepTM PCR inhibitor removal kit. (B) QuantiNovaTM IC RNA was spiked into dPCR mixtures in the presence of PIR-treated (blue, circle) or untreated (red, square) TNA extracts from WWTP Ruhleben samples (n = 4). The quantified QuantiNovaTM IC RNA copy number in the absence of WWTP TNA extracts was set to 100% and the data show the calculated relative amounts of amplified QuantiNovaTM IC RNA in the presence of WWTP TNA extracts.