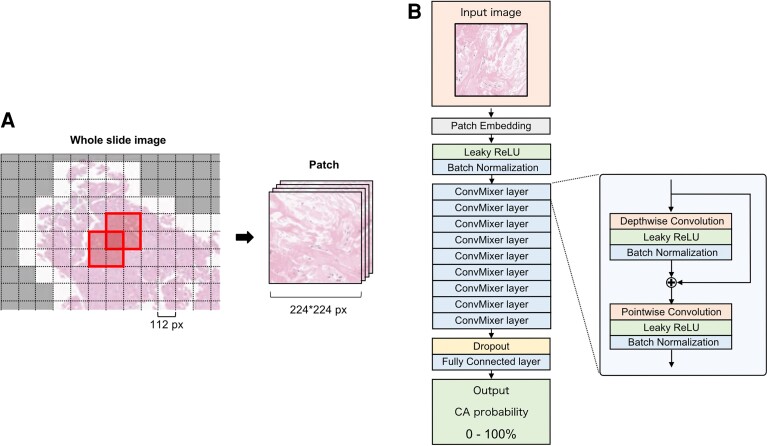
Figure 2.
(A) Each whole slide image of the specimen was cropped into multiple small patches of 224 × 224 px size and shifted by half (a patch is an area bordered by a bold line). Patches without myocardial tissue were removed (the white area was used for analysis, and the gray area was excluded). (B) The architecture of the deep learning model is based on ConvMixer. The input image is a patch (224 × 224 px) cropped from the whole slide image of myocardial specimens. And the output is the probability of CA. To improve the flexibility of the model and mitigate overfitting, the Leaky ReLU was used as the activation function, and a dropout layer was inserted at the end of the model. The hyperparameters of this model were determined using GS as follows: Kernel size = 5 (GS: 3, 5, 7, 9), patch size = 4 (GS: 2, 4, 8), number of filters = 128 (GS: 32, 64, 128, 256). GS, grid search.