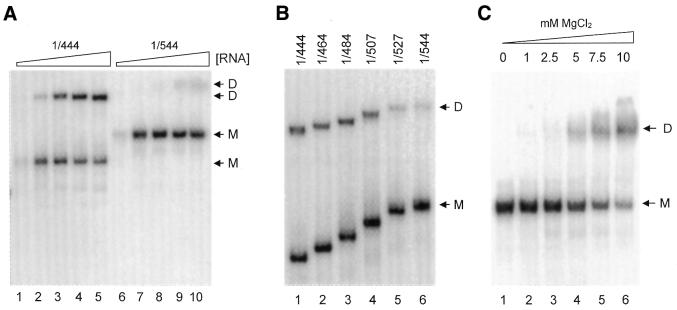
Figure 3.
Mapping of sequences 3′ of the HIV-2 DIS that inhibit dimerization. (A) Transcripts 1/444 and 1/544 were analyzed for their ability to dimerize at varying RNA concentrations. We incubated 10 ng (lanes 1 and 6), 50 ng (lanes 2 and 7), 250 ng (lanes 3 and 8), 500 ng (lanes 4 and 9) and 1000 ng (lanes 5 and 10) transcript at optimal dimerization conditions (65°C, 5 mM MgCl2), followed by gel electrophoresis on a non-denaturing 4% TBE gel and autoradiography. The monomer (M) and dimer (D) bands of both transcripts are marked. (B) Approximately 2 pmol of transcripts 1/444, 1/464, 1/484, 1/507, 1/527 and 1/544 was incubated at dimerization conditions as described in (A). Monomer and dimer bands are marked M and D. The dimerization efficiencies for the transcripts measured by phophorimager quantification are 43.6, 41.8, 44.8, 40.2, 25.0 and 17.9%, respectively. (C) Approximately 2 pmol of radiolabeled transcript 1/544 was incubated at optimal dimerization conditions (65°C, 5 mM MgCl2) in buffer containing 0, 1, 2.5, 5, 7.5 and 10 mM MgCl2.