Abstract
Background
The pathogenesis of non-alcoholic fatty liver disease (NAFLD) with a global prevalence of 30% is multifactorial and the involvement of gut bacteria has been recently proposed. However, finding robust bacterial signatures of NAFLD has been a great challenge, mainly due to its co-occurrence with other metabolic diseases.
Results
Here, we collected public metagenomic data and integrated the taxonomy profiles with in silico generated community metabolic outputs, and detailed clinical data, of 1206 Chinese subjects w/wo metabolic diseases, including NAFLD (obese and lean), obesity, T2D, hypertension, and atherosclerosis. We identified highly specific microbiome signatures through building accurate machine learning models (accuracy = 0.845–0.917) for NAFLD with high portability (generalizable) and low prediction rate (specific) when applied to other metabolic diseases, as well as through a community approach involving differential co-abundance ecological networks. Moreover, using these signatures coupled with further mediation analysis and metabolic dependency modeling, we propose synergistic defined microbial consortia associated with NAFLD phenotype in overweight and lean individuals, respectively.
Conclusion
Our study reveals robust and highly specific NAFLD signatures and offers a more realistic microbiome-therapeutics approach over individual species for this complex disease.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-024-01990-y.
Keywords: NAFLD, Gut microbiota, Metabolic diseases, Machine learning, Network analysis, Metabolomics, Microbial consortia
Background
Up to 40% of the general population in the western world has non-alcoholic fatty liver disease (NAFLD), making it the most frequent cause of chronic liver disease [1]. Despite numerous studies into NAFLD, its pathophysiology is still poorly understood and entails complicated interactions between variations in genetic susceptibility, environmental variables, insulin resistance, and the gut-liver axis [2]. Gut microbiota plays an essential role in the disruption of the gut–liver axis and the pathogenesis of NAFLD [2]. Based on the composition, functionality, and metabolic output of intestinal microbiota, we and others have developed microbiome-augmented models for the diagnosis of advanced fibrosis [3] and cirrhosis [4] and for the risk assessment for the development of liver disease [5] and NAFLD [6]. In addition, targeting the gut microbiota as a possible therapeutic avenue for NAFLD has been widely investigated, including manipulation of the microbiota by antibiotic therapy, prebiotics, probiotics, and synbiotics [2, 7].
Although NAFLD-associated microbial changes have been detected across various studies, suggesting robust and specific microbiome signatures for NAFLD remains a challenge, due to clinical, biological, and methodological reasons. First, metabolic diseases such as type 2 diabetes (T2D), hypertension, and cardiovascular diseases, usually co-occur with NAFLD creating obstacles in differentiating NAFLD-specific microbiome signatures from the other diseases [8]. Second, obesity is a well-recognized risk factor for and is inexorably related to NAFLD. However, around 20% of the patients with NAFLD are lean or non-overweight (BMI < 25), whom nevertheless exhibit similar cardiovascular- and cancer-related mortality compared to overweight (BMI > 25) NAFLD individuals and increased all-cause mortality risk [9]. Growing data, mostly based on 16S rRNA, suggests that lean people with NAFLD have a unique gut microbiome composition compared to overweight individuals with NAFLD, which is associated with clinical indices of NAFLD progression such as ALT, AST, GGT, and more [9, 10]. Third, multiple confounding factors of the study cohorts (age, gender, geographical location), drug intake, sequencing technologies, diet, and heterogeneity of analytic pipelines affecting the microbiome composition and annotation must be considered [8].
In addition, compared with individual species, resolving the complexity of NAFLD would benefit more from the investigation of the combinatorial effects of multiple microbial species in the context of a community, as shown before [11, 12]. Even though there might be species that are de facto beneficial or detrimental for a disease, the impact of others can be conditional and dependent of the particular microbiome constellation. The therapeutic potential of one individual species, even being backed up with preclinical models [13, 14], could be difficult to be replicated in humans. Indeed, though supplementation of, e.g., Akkermansia muciniphila has improved several metabolic parameters in a randomized controlled trial [15], such studies influencing the host phenotype using individual microbes are still scarce compared to the whole fecal microbiota transplantation (FMT) [16, 17]. Nevertheless, considering the recent risks associated with FMT [18, 19], moving to well-defined microbial consortia represents an attractive alternative [20].
Changes in the gut microbiota composition can lead to an alteration of the microbes-derived molecules and metabolites that influx in the systemic circulation [2]. Moreover, through the production of metabolites such as bile acids (BA) and short-chain fatty acids (SCFAs), gut microbiota impacts fat absorption in the liver and thus the development of a steatotic condition [2]. While the impact of the functional potential and especially the metabolic output of the microbiome on the development and progression of the disease is increasingly appreciated, the large variation in different metabolomics platforms and the challenges with cross-study integration have also hindered the identification of consistent microbiome-associated metabolic signatures related to NAFLD.
To understand the unique changes of gut microbiota in NAFLD compared to other metabolic diseases, we performed a large-scale meta-analysis of gut microbiota for 1206 subjects (same ethnicity: Chinese) with or without NAFLD or other metabolic diseases often co-occurring (obesity, T2D, hypertension, atherosclerosis) who had well-characterized clinical profiles. We analyzed shotgun metagenomics data generated using similar sequencing methodologies to reduce technical bias, and characterized in silico the metabolic output of all the microbiome communities involved to allow for the first time comparisons of these metabolic diseases at the microbial metabolite level. With a unified analytical framework, we proposed robust and highly specific NAFLD microbial signatures and microbial consortia with a potential role in driving or preventing disease development.
Methods
Study cohorts
In this study, we collected publicly available shotgun metagenomic sequencing data from 7 microbiome studies, together with one additional study for validation, which were all processed through the same pipeline. The studies related to NAFLD included (i) a NAFLD cohort diagnosed with MRS (N = 100) [7], (ii) a biopsy-proven cohort containing NAFLD (N = 81) and non-NAFLD (N = 10) subjects (BioProject ID: PRJNA732131), (iii) a non-NAFLD cohort diagnosed with ultrasound (N = 204) [6], and (iv) a biopsy-proven cirrhosis cohort (used for ML validation) containing NAFLD-Cirrhosis patients (N = 27) and non-NAFLD subjects (N = 54) [4]. The samples from the cirrhosis cohort with BMI < 25 (N = 30), and samples with T2D (N = 24) were excluded from the study, in an attempt to keep in the ML validation cohort the samples that exclusively had NAFLD. A combination of NAFLD diagnosis methods (biopsy and MRI-PDFF) has previously been used to shed light on associations between microbial species or metabolites with fibrosis [21, 22]. The studies related to other metabolic diseases included (i) a cohort related to hypertension with hypertension, pre-hypertension, and non-hypertension subjects (N = 185) [23], (ii) a cohort related to type 2 diabetes with obese and lean subjects along with their respective controls (N = 178) [24], (iii) a cohort containing prediabetic subjects (N = 69) [25], and (iv) an atherosclerosis cohort with atherosclerotic and non-atherosclerotic subjects (N = 405) [26]. Lastly, for the validation of species interaction, we used 928 samples from a population-based Health Professionals Follow-up Study (HPFS) [27], with their taxonomic data available in the R package curatedMetagenomicData (v3.6.2) [28]. For the cohorts that are not associated with a published study, ethics approvals were obtained by the Shanghai Jiao Tong University Affiliated Sixth People’s Hospital (approval no: 2015–65-(1)) and the University of Hong Kong/Hospital Authority Hong Kong West Cluster (approval no: UW 20–700) following the principles of the Declaration of Helsinki. Written informed consent was obtained from all participants.
Subjects diagnosed with NAFLD from the first 2 cohorts were categorized as NAFLD overweight (NAFLD-O) and NAFLD lean (NAFLD-L), based on BMI (overweight ≥ 25, lean < 25). The clinical data of the non-NAFLD subjects, from the NAFLD-related cohorts, were further evaluated. Therein, participants were separated into overweight and lean, based on the BMI criteria detailed above, with the overweight samples categorized as Control-NAFLD Overweight (CTRL-NAFLD-O). The lean participants were assessed against exhibiting any metabolic disease based on their metadata, specifically for hypertension (SBP ≥ 140 or DBP ≥ 90), pre-hypertension (SBP = 125–139 or DBP = 80–89) [29], or type 2 diabetes (HbA1c ≥ 6.5 or FBG ≥ 7) [24, 30], using the same diagnostic criteria as in the original studies. The participants that proved negative for all the above were categorized as Control-NAFLD Lean (CTRL-NAFLD-L). Participants who were found to exhibit only hypertension or pre-hypertension without diabetes were transferred to the hypertension and pre-hypertension cohort. The remaining participants who were diabetic with or without any form of hypertension were transferred to the type 2 diabetes lean group from the diabetic cohort.
Upon re-evaluating the diabetic cohort, participants initially categorized into the lean (T2D or non-T2D lean) or overweight groups (T2D or non-T2D overweight) that did not meet the BMI criteria were transferred to the relevant group. Participants from the cohorts of atherosclerosis and prediabetes remained unchanged and were subsequently categorized as atherosclerosis, control-atherosclerosis, and prediabetes. The number of samples and characteristics of each group, as well as the sample distribution per study, were shown in Tables 1 and S1–S3.
Table 1.
Summary of sequencing, microbiome, clinical, and anthropometric characteristics of NAFLD and control groups
| Groups | NAFLD-lean | Control-NAFLD-lean | P-value (Wilcoxon test) | NAFLD-overweight | Control-NAFLD-overweight | P-value (Wilcoxon test) |
|---|---|---|---|---|---|---|
| Sequencing information | ||||||
| Average read counts | 17,230,333 | 19,256,681 | ~ | 16,752,361 | 19,012,027 | ~ |
| Library preparation | 150 read length | 150 read length | ~ | 150 read length | 150 read length | ~ |
| Sequencing platform | Illumina | Illumina | ~ | Illumina | Illumina | ~ |
| Company | Novogene | Novogene | ~ | BGI, Novogene | BGI, Novogene | ~ |
| DNA extraction kits | PSP Spin Stool DNA | PSP Spin Stool DNA | ~ | PSP Spin Stool DNA | PSP Spin Stool DNA | ~ |
| Taxonomic and functional annotation | ||||||
| Metaphlan 3 | 67.61 ± 12.38 | 98.1 ± 17.63 | ~ | 72.45 ± 19.78 | 91.27 ± 18.81 | ~ |
| Kegg pathways | 154.39 ± 10.92 | 162.92 ± 5.71 | ~ | 156.53 ± 12.11 | 159.39 ± 7.26 | ~ |
| Anthropometric and clinical characteristics | ||||||
| Number of subjects | 18 | 39 | ~ | 163 | 84 | ~ |
| Ethnicity | Chinese (Han) | Chinese (Han) | ~ | Chinese (Han) | Chinese (Han) | ~ |
| Age | 39.11 ± 8.41 | 62.45 ± 3.88 | *** | 34.8 ± 9.08 | 58.74 ± 11.44 | *** |
| BMI (kg/m) | 23.75 ± 1.22 | 23.29 ± 1.01 | NS | 34.48 ± 7.5 | 27.86 ± 3.04 | *** |
| Fasting blood glucose (mmol/L) | 5.22 ± 0.65 | 5.75 ± 0.39 | ** | 5.73 ± 1.88 | 6.14 ± 1.14 | *** |
| Systolic blood pressure (mm Hg) | 117.56 ± 12.03 | 116.72 ± 5.92 | NS | 125.48 ± 13.79 | 130.71 ± 13.84 | *** |
| Diastolic blood pressure (mm Hg) | 79.28 ± 8.39 | 78 ± 3.4 | NS | 81.72 ± 10.49 | 81.19 ± 6.66 | NS |
| ALT (U/L) | 31.72 ± 25.85 | 16.03 ± 5.9 | * | 53.82 ± 45.86 | 17.38 ± 6.68 | *** |
| AST(U/L) | 24.5 ± 10.14 | 22.36 ± 5.97 | NS | 33.01 ± 23.36 | 21.42 ± 4.5 | *** |
| DBIL (μmol/L) | 4.16 ± 1.4 | NA | ~ | 3.66 ± 1.51 | NA | ~ |
| TBIL (μmol/L) | 13.46 ± 5.19 | 11.93 ± 3.98 | NS | 13.37 ± 5.1 | 11.41 ± 4.33 | *** |
| ASTALT | 0.96 ± 0.28 | 1.44 ± 0.25 | *** | 9.17 ± 107.64 | 1.33 ± 0.34 | *** |
| GGT (U/L) | 35.11 ± 26.8 | 22.92 ± 24.97 | ** | 46.08 ± 36.5 | 24.52 ± 16.86 | *** |
| TBA (μmol/L) | 2.84 ± 1.76 | NA | ~ | 3.57 ± 2.97 | NA | ~ |
| Triglycerides (mmol/L) | 1.83 ± 0.66 | 1.2 ± 0.61 | ** | 2.27 ± 2.26 | 1.56 ± 1.28 | *** |
| FLI | 37.59 ± 17.61 | 14.73 ± 9.59 | *** | 80.16 ± 20.78 | 38.6 ± 21.68 | *** |
| HOMAIR | 3.06 ± 1.18 | 1.28 ± 0.51 | *** | 6.25 ± 5.84 | 2.14 ± 1.82 | *** |
| Liver fat (%) | 3.95 ± 2.85 | 1.71 ± 0.69 | *** | 11.23 ± 10.57 | 3.6 ± 6.41 | *** |
P value < 0.0005 = ***, P value < 0.005 = **, P value < 0.05 = *, P value > 0.05 = NS
For each disease, the same exclusion criteria were used as in the original study except for 4 samples from the control-atherosclerotic cohort, which were excluded due to having a BMI < 17 (severe thinness). Importantly, the analyzed cohorts are all free of antibiotics usage and almost medication-free except for 33 subjects, including 19 atherosclerotic patients who had used metoprolol, 10 T2D-L, and 4 Control-NAFLD-O who had taken antidiabetic medication. The majority of the studies shared additional exclusion criteria namely heart failure, renal insufficiency, acute infectious disease, probiotics usage, and cancer.
Sequencing, quality control, and taxonomic profiling
Details on sample collection and sequencing can be found in publications describing the original studies. For the quality control of the raw reads, the Sunbeam pipeline (v2.1) [31] was used. Human DNA contaminations were removed using BWA (v0.7.17) mem [32] against the human reference genome ucsc.hg19 and adaptors, low-quality reads, and bases were filtered using Trimmomatic (v 0.36) [33]. Samples above 20 million reads were subsampled to 20 million. The high-quality reads were taxonomically profiled at different taxonomic levels using MetaPhlAn 3.0 [34] with default settings, generating taxonomic relative abundances (total sum scaling normalization).
Functional profiling
Microbial gene family abundances were estimated using HUMAnN 3.0 [34] and were further mapped to the MetaCyc metabolic pathway database [35] and the KEGG database [36] to obtain the MetaCyc pathway abundances and KEGG Orthology (KO) abundances with species contribution. Tables of pathway and gene family abundances were normalized to copies per million (CPM), including unmapped and unintegrated read mass.
Microbiome alpha and beta diversity
Alpha diversity (Shannon index) for each sample was calculated at the species-level with R package vegan (v2.6–2) [37]. Wilcoxon rank-sum tests were used to test for significant differences. Weighted UniFrac and Bray–Curtis distances were used to calculate the beta diversity for species and functions, respectively. Canberra distance was used to calculate the beta diversity for metabolites. To analyze the overall change of each disease against its control, we normalized each disease group in the space of NMDS by calculating first the centroid of each respective control and then deducing it from the coordinates of relevant diseased samples. PERMANOVA was used for statistical comparison of beta diversity with adonis function in the R package vegan. Correction for multiple hypotheses testing was performed with the false-discovery rate (FDR) approach [38].
In silico metabolomic profiling of microbiota
The in silico approach MAMBO was used to profile the primary metabolic output of each individual sample, taking microbiota taxonomic profiles as input. In brief, MAMBO optimizes a high correlating metabolic profile to a given microbiota taxonomic relative abundance profile based on bacterial GEMs associated with the given metagenomic sample. We opted for using only GEMs associated with species from the metagenomics samples and downloaded 799 matching bacterial GEMs from the AGORA (https://vmh.life) [39] and CarveMe collection (https://github.com/cdanielmachado/embl_gems/tree/master/models) [40]. Optimizations were run in a Python environment (v3.7) using a high-performance cluster (192 cores, 1 TB RAM). The samples that produced very low prediction scores (less than 0.3) were excluded from further metabolites analysis. After removing metabolites that appeared in less than 80% of the samples, missing values of metabolite abundance were imputed with the R package miceRanger (v1.4.0) [41] using m = 1 and maxiter = 100 resulting in a final list of 510 metabolites.
Data integration
Our study consists of 8 disease groups originating from 7 distinct studies. While completely eliminating any potential cohort biases is challenging, we made efforts to minimize heterogeneity in the overall batch effects in our study, both within disease cohorts and throughout the analysis. During the study selection, we specifically chose to include only Chinese Han to ensure that any potential variations among different races were accounted for. When designing the disease-control groups we considered cohorts with comparable sequencing depth, read lengths, DNA extraction method, and sequencing platforms used, especially for NAFLD with it being the primary group of analysis. In addition, PERMONOVA was used in order to ascertain that there are no significant differences (P > 0.05) between the NAFLD participants in the MRS and biopsy cohorts (Fig. S4). To detect disease microbiome signatures reliably, we paired each disease with its corresponding control. In order to avoid biases in the results of different original studies due to various technologies, we processed all collected samples using a unified pipeline.
While our approach is not flawless, and in general meta-analysis methods have drawbacks, such as diminished statistical power, they have been widely employed to mitigate batch effects when combining genomic data from different studies, and have recently demonstrated their utility in microbiome studies [42, 43]. This is a proof-of-concept study that can offer the analytical framework for future study where all samples will be collected from subjects in the same region, then processed and sequenced using the same technology, together with detailed metadata that could be considered in a cross-disease analysis.
Random forest models for NAFLD prediction
We build Random Forest classifiers using R package caret (v6.0–93) [44] to discriminate NAFLD patients from control based on taxonomic, functional, and metabolic profiles. Firstly, we randomly split the data into 80% training set and 20% test set. Then, the training set was used to perform 100 random splits of 90% feature selection set and 10% validation set. For each split, feature selection was performed using the R package Boruta (v8.0.0) [45] on the feature selection set, then a Random Forest model was trained using the top 20 features and further tested on the corresponding 10% validation set. The receiver operating characteristic (ROC) curve and AUROC value for each of the 100 splits were calculated using the R package pROC (v1.18.0). From the 10 models with the highest AUROC values, the 20 features that were most frequently selected were then used to build a final Random Forest model on the initial 80% training set. Lastly, the final model was evaluated on the unseen 20% test set and the final AUROC was reported. Feature importance in the final model was calculated using the function varImp from R package caret and feature prediction was determined by computing Shapley values using R package fastshap (v0.0.7) [46]. The final model was then validated on an external biopsy-proven cohort that included NAFLD-Cirrhosis patients (N = 27) and non-NAFLD subjects (N = 54) [1]. To ensure the validation cohort focused solely on NAFLD, we excluded samples with BMI < 25 (N = 30) and those with type 2 diabetes (N = 24). Confusion matrices were built using function predict from R package caret (v6.0–93) and ROC curves and are under the curve (AUC) confidence intervals were calculated using the function roc from R package pROC (v1.18.0).
Portability and prediction rate analyses
Cross-study portability and prediction rate analyses were performed as described previously [47]. Briefly, we first computed the AUROC between the prediction vectors for cases in the test set and controls in the external data set and calculated cross-study portability as (|0.5—AUROC|)*2, obtaining a value ranging from 0 (indicating a complete loss of discriminatory power between cases and external controls) to 1 (meaning that the model can be used in another data set without losing discriminatory power). We then calculated the model’s prediction rate by assessing the percentage of samples from other metabolic diseases that were incorrectly classified as NAFLD-O when using a threshold set to maintain a 10% false positive rate (FPR) in the NAFLD-O vs. CTRL-NAFLD-O comparison. Specifically, after evaluating the model on NAFLD-O and CTRL-NAFLD-O samples, we identified the prediction score at which only 10% of CTRL-NAFLD-O samples were misclassified as NAFLD-O. This value served as our 10% FPR threshold, which we then applied when testing the model in other metabolic diseases. The prediction rate indicates the percentage of diseased samples with prediction scores above the threshold, showing how many were incorrectly predicted as NAFLD-O.
Differentially correlated species network
The R package MEGENA (v1.3.7) [48] was used to build correlation networks (module compactness P < 0.05) from differentially correlated species in NAFLD-O relative to CTRL-NAFLD-O and NAFLD-L relative to CTRL-NAFLD-L. Differential correlations were calculated using the R package DGCA (v1.0.2) [49] with the method of Spearman with 1000 permutations. Only species pairs with significant differential correlation (empirical P < 0.01) were included for analyses.
Associating species modules with NAFLD-related clinical data
Mantel test using the partial Spearman correlation coefficient, adjusting for age, gender, and BMI, with 9999 permutations from R package vegan was used to analyze the associations between species modules with clinical data related to NAFLD (AST, ALT, GGT, FLI, liver fat, TGs) and to determine whether the pairs of datasets were significantly correlated. FLI and liver fat values were computed as described previously [50, 51]. Bray–Curtis dissimilarity matrices based on species relative abundance and Euclidean distance matrices based on clinical data were computed to perform the test. Sparse canonical correlation analysis (sCCA) was then performed for each of the significant species modules against the clinical data using the R package PMA (v1.2.1) [52]. The CCA.permute function was used to select the L1 penalties for both datasets independently. The highest possible combination of penalties that lead to significant associations (P < 0.05) was selected, in order to gain insights into the associations for as many features as possible.
Mediation analysis
On a multivariate level, the R package MODIMA [53] was used to infer the mediation effects of the DA metabolites of NAFLD-O and NAFLD-L, for the interactions of NAFLD modules and NAFLD-related clinical parameters (P < 0.05). For individual microbial features, metabolites, and clinical parameters, we firstly checked whether species and metabolites were associated using a linear model using the lm function (P < 0.05) from the R package stats (v3.6.3). Next, we conducted mediation analysis using metabolites as mediators to assess their impact on the species-NAFLD clinical parameters relations using the mediate function from the R package mediation (v4.5.0) (P < 0.05) [54].
Bacterial synergistic communities
Genome-scale metabolic models (GSMMs) were downloaded from https://github.com/cdanielmachado/embl_gems/tree/master/models. All possible communities of bacterial species with two, three, four, and five members were considered. Next, the SMETANA (v1.2.0) approach [55] with Academic IBM CPLEX solver (v12.8.0.0) on the complete medium was applied to the communities using the downloaded GSMMs as input. Metabolic interaction potential (MIP)/metabolic resource overlap (MRO) scores were calculated and normalized by the size of each community, in order to account for the bias in the number of supporting metabolites detected due to community sizes.
Statistical analysis
Statistical analyses of clinical data, metagenomics data including taxonomy and functionality, and in silico metabolomics data were performed in R software version 3.6.3. Clinical data were statistically compared with the Wilcoxon rank-sum test. Spearman correlations were calculated using the R package stats.
Metagenomic data, including taxonomy and functional data were transformed into pseudo-count data by multiplying with 106 and analyzed using ANCOM-II (v2.1), without using th estructural zeroes option. Clinical data adjustments were used according to the dataset comparison: NAFLD-O and NAFLD-L (age-gender-BMI-HOMA-IR-SBP), ATH, T2D-O, PRE-T2D-O and T2D-L (age-gender), HYP and PRE-HYP (age-gender-BMI). Microbial features were considered statistically significant at the cutoff of 0.6.
Statistical comparison of metabolites followed three steps. Firstly, candidate metabolites were selected from the Wilcoxon rank-sum test using P < 0.1 as a cut-off. Secondly, the identified metabolites in the first step were investigated with an adaptive Lasso statistical design with a binomial distribution, adjusting for clinical data according to the dataset comparison, using R package glmnet [56] (v4.1) to identify important metabolites. Finally, we used a fixed Lasso design using R package selectiveInference (v1.2.5) [57] as post-selection inference method to identify the significance for each of the important metabolites (P ≤ 0.05).
Partial Spearman correlation was used to link significantly different metabolites in each comparison with liver-related clinical data, adjusting for age, gender, and BMI, using the function pcor.test from the R package ppcor (v1.1) [58]. For the metabolites found significantly different in non-NAFLD comparisons, due to lack of clinical data, the NAFLD (NAFLD-O + NAFLD-L) and Control (CTRL-NAFLD-O + CTRL-NAFLD-L) datasets were used to calculate the correlation.
All statistical analyses were performed with the R software and P value < 0.05 was deemed significant unless otherwise stated. Correction for multiple hypotheses testing was performed with the false-discovery rate (FDR) approach [38].
Data visualization
All figures were generated by R software (v3.6.3), using the R packages ggplot2 (v3.3.6) [59] and ComplexHeatmap (v2.2.0) [60], except the network plots that were made using Cytoscape software (v3.9) [61].
Results
Characteristics of the study cohorts
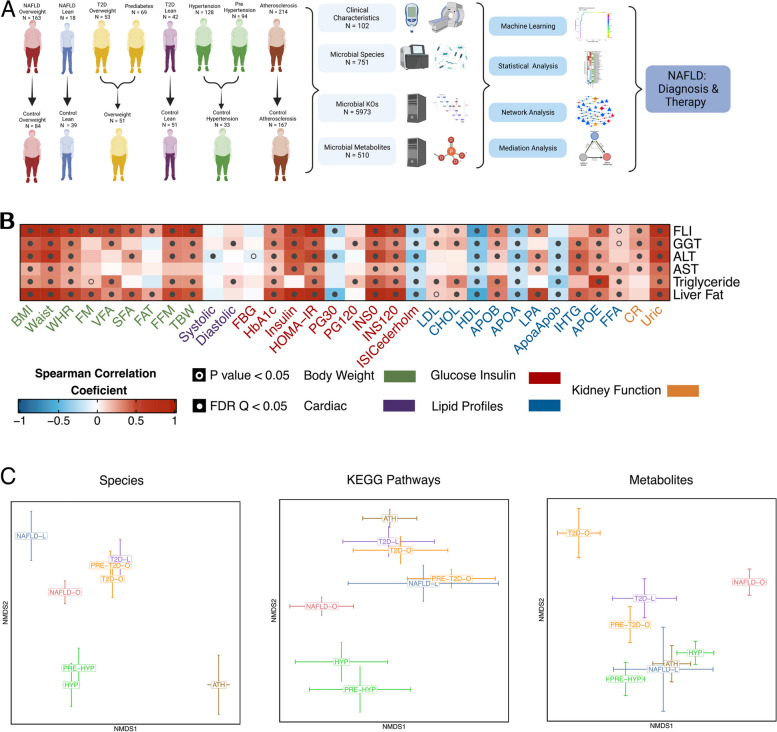
To investigate the NAFLD-specific gut microbiota changes in the context of other metabolic diseases, we collected publicly available shotgun metagenomics datasets covering NAFLD overweight (NAFLD-O), NAFLD lean (NAFLD-L), prediabetes overweight (PRE-T2D-O), T2D overweight (T2D-O), T2D lean (T2D-L), as well as cohorts with prehypertension (PRE-HYP), hypertension (HYP), and atherosclerosis (ATH), which were all at the overweight borderline (Fig. 1A). Their corresponding control subjects were also retrieved, and a wide range of clinical characteristics was further obtained to decipher the association between gut microbiota changes and disease-related clinical parameters (Tables 1, S1, S2). To avoid the confounding effect of ethnicity/country, we initially analyzed subjects from Chinese cohorts for defining the NAFLD signatures, which we further validated in a cohort of different ethnicities. After careful evaluation of the original clinical data (detailed in Methods), 1206 subjects were used to generate well-defined disease groups (Table S3), for the subsequent comparative analyses.
Fig. 1.
Study design overview and structure differences in microbiome among NAFLD and closely associated diseases. A A graphical representation summarizing the cohort information, collected data, and analysis performed in 1206 samples. Detailed criteria on the group formation can be found in the methods section. Created with Biorender.com. B High interconnection between NAFLD-related clinical data and other clinical measures representative of different cardiometabolic diseases. Spearman’s rank-based correlations were used. C Comparison of bacterial species profiles, KEGG pathway profiles, and metabolite profiles among all disease groups adjusted by their respective controls, using non-metric multidimensional scaling (NMDS) of weighted UniFrac, Bray Curtis, and Canberra distances, respectively. The error bars indicate the mean and standard errors of the mean. Significant differences were determined using PERMANOVA and were considered significant if P < 0.05
The diagnosis of NAFLD in our patient group was based on liver biopsy (44% of individuals), or magnetic resonance spectroscopy (MRS) (56%). We first examined the associations between NAFLD-related parameters and the clinical parameters characterizing the other metabolic diseases using all available subjects. Alanine aminotransferase (ALT), aspartate aminotransferase (AST), gamma-glutamyl transpeptidase (GGT), serum triglycerides (TGs), fatty liver index (FLI) [50], and liver fat percentage index [51] showed significant correlations with clinical data related to adiposity, blood pressure, glucose metabolism, lipid metabolism and kidney function, further demonstrating that NAFLD and other cardiometabolic diseases are highly interconnected (Fig. 1B).
To avoid technical biases [62, 63], the NAFLD groups (NAFLD-O [n = 163] and NAFLD-L [n = 18]) were matched with their respective controls (CTRL-NAFLD-O [n = 84] and CTRL-NAFLD-L [n = 39]) in both the DNA extraction method and the sequencing platform (Illumina paired-ended 150 bp), as well as in sequencing depth (Table 1). Compared with the non-NAFLD control subjects, the NAFLD patients, as expected, had significantly higher values of ALT, AST, GGT, TGs, FLI, and estimated liver fat percentages (Wilcoxon rank-sum test, P < 0.05). As for other clinical data, insulin resistance measured by the homeostatic model assessment of insulin resistance (HOMA-IR) was significantly higher in the two NAFLD groups compared to their respective controls; systolic blood pressure (SBP) was significantly different only in the overweight groups (slightly higher in CTRL-NAFLD-O) (Table 1). Since the two control groups had higher age than the corresponding NAFLD groups, and the NAFLD-O had a significantly higher BMI than the CTRL-NAFLD-O, these factors were considered and adjusted in downstream statistical analyses.
Comparative analysis reveals differences in the gut microbiome composition and metabolic output in NAFLD and other metabolic diseases
The collected shotgun metagenomics data were processed with MetaPhlAn3 and HUMANn3 [34], leading to the detection of in total of 751 microbial species and 5973 KEGG orthologs (KOs) across the entire cohort. At the community level, we found significantly lower alpha diversity (Shannon index) in both NAFLD-O and NAFLD-L groups than their respective controls (Wilcoxon rank-sum test, P < 0.05), but not between the two NAFLD groups (Fig. S1). Using weighted UniFrac distance for measuring microbiota species-level beta diversity, we revealed significantly different microbiota composition between each NAFLD group against their respective controls (PERMANOVA, adjusting for age, gender and BMI, P < 0.05, R2 = 0.02–0.05), but not between NAFLD-O and NAFLD-L, which might be related to the limited sample size of the NAFLD-L group (Fig. S2, Table S4). Interestingly, when comparing the changes of microbiome taxonomic profiles against the corresponding controls, the two NAFLD groups were clearly distinguished from other metabolic diseases (Fig. 1C). We then used KEGG pathways to evaluate and compare the microbiota functional diversity (Bray–Curtis dissimilarity). Unlike the species, the beta diversity of the KEGG pathways for the NAFLD groups against their respective controls was not found significantly different, nor was the comparison of NAFLD-O against NAFLD-L (PERMANOVA adjusting for age, gender, and BMI, P > 0.05) (Fig. S2, Table S4).
Given the current difficulty in integrating untargeted metabolomics data from multiple studies and in order to perform a large-scale comparative analysis of our cohorts at the microbial metabolites level, we used an in silico approach, namely MAMBO [64], to profile the primary metabolic output of each individual microbiota. Such metagenomics-predicted metabolomics has facilitated the identification of distinct microbial signatures for different types of colonic adenomas [65]. Taking taxonomic profiles as input and based on genome-scale metabolic modeling, MAMBO estimates the metabolic output of the whole microbial community without the confounding factors of human metabolism or food remnants as in experimental stool metabolomics. With this approach, we estimated the levels of 510 microbial metabolites for the 1098 subjects that generated reliable metabolite prediction scores. Similarly to the functional potential, the overall predicted metabolomic profiles of the two NAFLD groups showed no significant differences with their respective controls nor between them (PERMANOVA adjusting for age, gender, and BMI, P > 0.05) (Table S4). Despite this, the metabolites-based comparison of community changes from respective controls indicated different patterns than those of taxonomy- or functions-based, e.g., the two clusters formed for the T2D (T2D-O, T2D-L, PRE-T2D-O) and hypertension (HYP, PRE-HYP) groups in the species and pathway analysis, were distorted in the metabolite analysis (Fig. 1C).
In summary, by using a unified computational pipeline we revealed both similarities and differences in the overall microbiome structure, function, and metabolic output between NAFLD overweight and lean subjects and non-NAFLD controls or other metabolic diseases.
Machine learning identifies highly specific microbial signatures for NAFLD
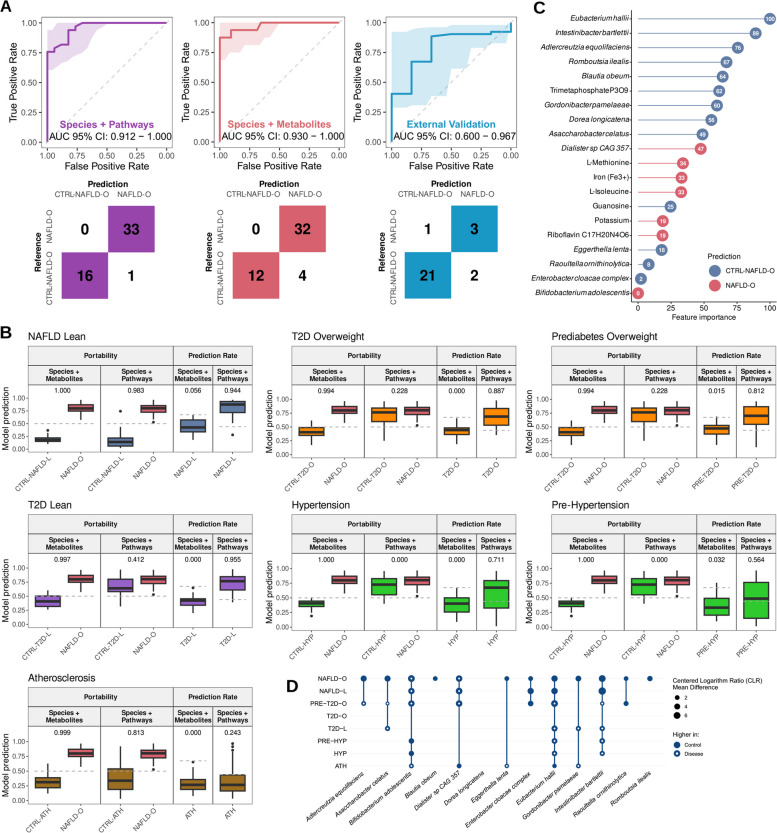
We next examined whether microbial features could be integrated into a machine learning (ML) model, which takes into account microbial interactions and the non-linear relationship with phenotypes, in order to identify key discriminative features associated to NAFLD. We focused on discriminating the NAFLD-O versus CTRL-NAFLD-O groups since the number of patients in the NAFLD-L group was low for the purpose of developing a reliable ML model. We first developed a random forest [66] classifier using either only microbial species or KEGG pathways, with accuracies being 0.824 and 0.588, respectively (Fig. S3). We proceeded in integrating species and KEGG pathways which greatly improved the accuracy to 0.998 (Fig. 2A, Table S5). Subsequently, the portability and prediction rate [47] of our model was evaluated against the other metabolic diseases. We first investigated whether the separation between NAFLD-O and CTRL-NAFLD-O would be maintained when using control samples from the different metabolic disease cohorts. We compared the true-positive rate (TPR) from our constructed model to the external false-positive rate (FPR) via a modified area under the receiver operating curve (AUROC) analysis, which was rescaled to range between 0, meaning a complete loss of separation, and 1, meaning that the model can be used in a different data set without losing discriminatory power. A low portability (portability = 0–0.412) was observed for the majority of data sets, indicating that the model would not be able to classify NAFLD-O subjects when using a different set of presumably non-NAFLD controls. The only exceptions were when the CTRL-NAFLD-L (portability = 0.983) and CTRL-ATH (portability = 0.813) subjects were used (Fig. 2B), which was in accordance with their similarities observed in the beta diversity analysis with the CTRL-NAFLD-O (Fig. 1C). Secondly, we estimated the prediction rate of the model by quantifying the percentage of samples from the other metabolic diseases that would be misclassified as NAFLD-O when a cut-off adjusted to maintain a FPR of 10% in our model was applied. A high prediction rate (prediction rate: 0.564–0.955) was found in most data sets suggesting a very low specificity for NAFLD-O of the constructed model (Fig. 2B). The only notable exception was the classification of the ATH subjects (prediction rate = 0.243), which could again be explained by the unique microbiome composition of ATH as shown in the beta diversity analysis (Fig. 1C). Taken together, our results suggest that there is a high similarity in terms of species and pathways that renders the discrimination between NAFLD-O and other metabolic diseases challenging.
Fig. 2.
Random forest model based on gut microbiota species and metabolites accurately and specifically predicts NAFLD-O. A Receiver operating characteristic (ROC) curves and confusion matrices evaluating the ability of random forest models to predict NAFLD. Each color represents the model performance using as features species and pathways (purple), or species and metabolites (red and blue). Blue indicates the model performance when being validated in an external US cohort. B Evaluation of model cross-study portability and prediction rate on NAFLD-L and other metabolic diseases. Models using species plus metabolites or species plus pathways as features were used. C Feature importance for the random forest model built with species and metabolites. The color indicates feature prediction as evaluated by Shapley values: blue for Control-NAFLD-O and red for NAFLD-O. D Abundance comparison between each disease and its control for the species selected in the model built with species and metabolites. ANCOM-II was used for statistical comparisons. Circle size corresponds to the mean difference, with a higher size/value indicating a stronger difference. Full circle: higher in control; empty circle: higher in disease. CLR: centered log ratio
In an attempt to explore whether we can increase the specificity of microbiome-based models and the involved microbial signatures for NAFLD-O, we built a RF classifier using species and in silico estimated metabolite abundances and we achieved an accuracy of 0.917 (Fig. 2A, C). Following the same approach as above, we observed a high portability (portability: 0.994–1) (the ML model is generalizable) and a low prediction rate (prediction rate: 0–0.056) (the ML model is specific for NAFLD-O and not generic for discriminating healthy from diseased subjects), suggesting that metabolites may provide a better discrimination between NAFLD-O and non-NAFLD-O samples (Fig. 2B). Interestingly, the prediction rate for NAFLD-L is very low (prediction rate = 0.056), indicating notable differences between overweight and lean NAFLD subjects in the microbiome metabolic output. Lastly, to rule out the possibility of overfitting during the ML development, we validated our hybrid model (species plus metabolites) in an external US cohort [4] consisting of healthy individuals and NAFLD patients and achieved an accuracy of 0.845, demonstrating that the model is highly accurate in a different ethnicity despite the most severe form of the disease (cirrhosis) in the external cohort (Fig. 2A).
Among the selected features in the ML hybrid model (13 species, 7 metabolites) we found that 12 species were significantly different in abundance (DA) in NAFLD-O compared to CTRL-NAFLD-O (ANCOM-II, cutoff = 0.6). However, only 5 of them were found DA in NAFLD-L compared to CTRL-NAFLD-L (ANCOM-II, cutoff = 0.6). Furthermore, out of the 12 species, 10 were also DA in other metabolic diseases (vs their respective controls), even though not always with the same directionality of change as in NAFLD-O (Fig. 2D). Eubacterium hallii, the top important feature in the ML model (Fig. 2C) and significantly higher in both CTRL-NAFLD groups compared to the NAFLD groups (Table S6, Fig. 2D), has been reported to alter the bile acid metabolism [67] and has been suggested as a potential probiotic candidate for treating dysbiosis-associated diseases [68]. Another species selected in the ML model as predictive of CTRL-NAFLD-O is Blautia obeum (Fig. 2C), which was only DA in NAFLD-O but not in any other metabolic diseases (Fig. 2D). This species has been previously found to have a lower abundance in subjects with steatosis and has been identified as a highly important feature in ML models classifying steatosis patients [10].
Selected features in the ML model also include several metabolites that have been previously associated with NAFLD (Fig. 2C), such as L-isoleucine, which has been found at a higher level in NAFLD patients compared to healthy individuals [69] and has been previously incorporated in models to predict NAFLD [70]. Moreover, riboflavin (vitamin B2), iron, and L-methionine all have been previously associated positively or negatively with NAFLD and fat accumulation in the liver [71–74].
In summary, by coupling microbial species and metabolites instead of the low-resolution functional potential of pathways, we were able to identify NAFLD microbiome signatures with high specificity and generalisability against other metabolic diseases. Its incapability for NAFLD-L diagnosis, together with the different distributions of model feature abundances in NAFLD-L and NAFLD-O, highlighted again the differences in the composition and metabolism of gut microbiome for the two NAFLD entities, which we explored below.
Gut ecological networks reveal structural differences of NAFLD-O and NAFLD-L patients
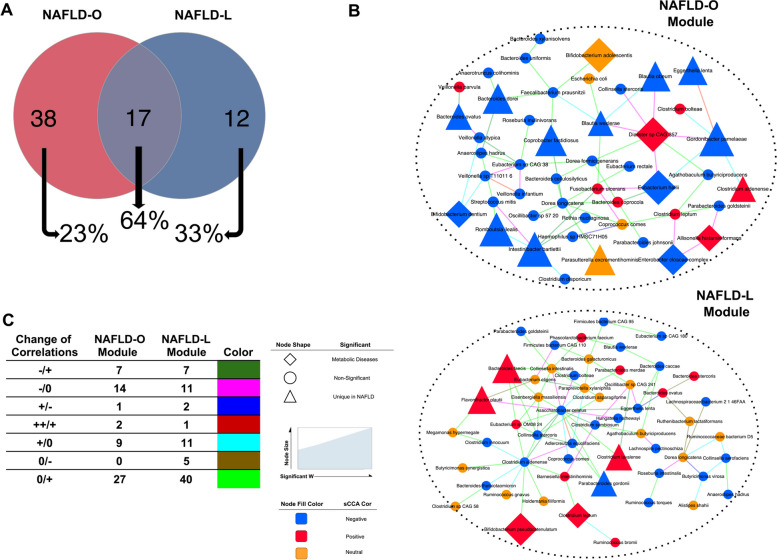
To reveal microbial consortia involved in the pathogenesis of NAFLD-O and NAFLD-L patients, we first performed DA species analysis. Using ANCOM-II (adjusting for age, gender, BMI, HOMA-IR and SBP, cutoff = 0.6), we found 55 DA species (32 increased, 23 decreased) in NAFLD-O compared to CTRL-NAFLD-O, and 29 (21 increased, 8 decreased) in NAFLD-L against CTRL-NAFLD-L, with 17 species in common (with same direction of change) (Fig. 3A, Table S6). From the 38 DA species found uniquely in NAFLD-O but not in NAFLD-L, 29 remained as NAFLD-O specific even when considering the complete pool of DA species from the comparisons of the other metabolic disease groups against their controls (Table S6). Similarly, 8 out of 12 NAFLD-L unique species (not DA species in NAFLD-O) were not found as DA in the other metabolic disease groups against their controls. Moreover, a set of 13 DA species shared between NAFLD-O and NAFLD-L were not found significant in the same direction (Table S6), in any of the other metabolic diseases and appear to be BMI-independent NAFLD-associated microbial changes. Among those, we found Intestinibacter bartlettii and Dialister sp. CAG 357, both selected in the ML model (Fig. 2C) and with previous evidence supporting their association with NAFLD [10].
Fig. 3.
Signature modules of species networks associated with NAFLD phenotype for NAFLD-O and NAFLD-L. A Venn diagram of significantly different in abundance species using ANCOM-II (cutoff = 0.6) adjusted by age gender BMI HOMA-IR and SBP for the comparisons of NAFLD-O vs CTRL-NAFLD-O and NAFLD-L vs CTRL-NAFLD-L. The amount of common significantly different species of each case, with other metabolic disease comparisons is displayed as a percentage under the Venns circles with arrows. Differentially abundant species analysis for metabolic diseases was done using ANCOM-II adjusting for a set of clinical data depending on the disease-control comparison (Methods). B Multiscale embedded correlation network analysis illustrates the differential correlation of species in NAFLD-O vs CTRL-NAFLD-O and NAFLD-L vs CTRL-NAFLD-L. Only species pairs with significant differential correlations (empirical P < 0.01) were included. The color of the link indicates the type of correlation change in control/NAFLD. Sparse canonical correlation analysis (sCCA) was used to evaluate the contribution of each species to the correlation between the module and NAFLD-related clinical parameters (P < 0.05), which is displayed by the color of the nodes (blue: negative, red: positive). ANCOM-II was used to find a significant difference in abundance species (cutoff = 0.6) adjusting for a set of clinical data depending on the disease-control comparison (Methods). Species found significantly different in abundance uniquely in NAFLD comparisons are marked with a triangle whereas a diamond is used if the species also appears significant in the comparison of any other metabolic diseases. The size of the node indicates the magnitude of the W statistic generated by ANCOM-II. C Table summarizing the numbers for each type of correlation change for the two modules in B. The types are depicted as from control to NAFLD (control/NAFLD) and colored differently as in B
We subsequently investigated using interaction networks whether defined microbial consortia with synergistic roles to NAFLD pathogenesis could be retrieved. An approach involving DGCA [49] and MEGENA [48] was employed to construct differential correlation networks between NAFLD subjects and controls and then to identify well-interconnected network modules. Applying this approach to both NAFLD-O and NAFLD-L comparisons (against their controls) generated 3 significant modules (module compactness P < 0.05) for each group. We subsequently examined the association of each module with NAFLD-related clinical parameters (ALT, AST, GGT, FLI, liver fat, and TGs) using a partial Mantel test adjusting for age, gender, and BMI. This revealed one NAFLD-O-related and one NAFLD-L-related species modules (partial Mantel test, P < 0.05) that are potentially associated with disease pathophysiology (Fig. 3B, Table S7). Sparse canonical correlation analysis (sCCA) was further used to identify subgroups of linear combinations of microbial species and NAFLD-related clinical parameters that are maximally correlated with each other, and to provide disease-association information for each individual species in the context of the microbial interaction network. This analysis showed that both NAFLD-O and NAFLD-L modules have significant species subgroups with associations (sCCA, P < 0.05) to most of the NAFLD clinical parameters (Fig. 3B and Table S8).
The module of NAFLD-O contains mostly species that were negatively associated to NAFLD-related parameters as inferred by sCCA. Most of the significant DA species involved in the module had higher abundance in the control and were uniquely found in the NAFLD-O vs CTRL-NAFLD-O comparison rather than being also DA in other metabolic diseases. We also observed that the beneficial species negatively associated to NAFLD in this cluster were mostly linked with light green (“0/ + ”) or light blue (“ + /0”) connections, indicating that either these species were concurrently reduced in NAFLD-O (thus positively correlated), or their interactions in the CTRL-NAFLD-O were disturbed and lost in NAFLD-O (Fig. 3B and C). Furthermore, the module contains a group of beneficial species negatively-associated to NAFLD (Gordonibacter pamelaeae, Eggertheila lenta, E. hallii, B. obeum, and Blautia wexlerae), among which four were predictive towards control subjects in the ML model (Fig. 2C). These species were connected by either ‘ + /0” suggesting disturbed interactions between these beneficial species from CTRL-NAFLD-O to NAFLD-O, or “ + + / + ” indicating weakened interactions between them in NAFLD-O. Moreover, their interactions with Dialister sp CAG 357 followed the same pattern from control to NAFLD-O (“ − /0″) (Fig. 3B and C).
Unlike NAFLD-O, the module of NAFLD-L represents a mixture of species that were both positively and negatively associated with NAFLD. The NAFLD-L module was larger (number of nodes) and contained a higher number of significantly changed microbial correlations (Fig. 3B). Two hub (highly connected) species, Asaccharobacter celatus, and Clostridium aldenense, were both negatively associated with NAFLD clinical indexes. A. celatus was connected with NAFLD-negatively-associated species in two ways: (1) weaker or disturbed connections, including Adlercreutzia equalifacies (“ + + / + ”), Coprococcus comes and Paraprevotella xylaniphila (“ + /0); (2) interactions suggestive of a concurrent reduction, involving Parabacteroides gordonii (“0/ + ”), Clostridium stercoris, Clostridium symbiosum, E. lenta and C. aldenense (“ − / + ”) (Fig. 3B and C). Notably, its negative correlation with Flavonifractor plautii, a species significantly higher and uniquely DA in NAFLD-L, was lost (“ − /0”) in the disease state.
In summary, employing differential correlation ecological networks, combined with DA species and ML, enabled us to disclose the coordinated structural changes in the gut microbiome of overweight and lean individuals with strong associations to NAFLD phenotype.
Mediation analysis reveals potential mechanistic links between species module signatures and NAFLD
We next investigated the metabolic output of the microbiota communities and performed DA analysis for NAFLD-O, NAFLD-L, and each of the other metabolic diseases against their corresponding control group. We identified 23 significant microbial metabolites (5 increased, 18 decreased) in NAFLD-O compared to CTRL-NAFLD-O and 31 (15 increased, 16 decreased) in NAFLD-L against CTRL-NAFLD-L (Wilcoxon rank-sum test and fixed Lasso after adjusting for age, gender, BMI, HOMA-IR and SBP, P < 0.05) (Fig. S3A, Table S6). Only 5 of those DA microbial metabolites were common between NAFLD-O and NAFLD-L groups (with abundance changes in the same direction; four higher in control and one higher in NAFLD), namely sulfate, potassium, protoheme, dihydroxyacetone, and L-histidine. When comparing with the DA microbial metabolites observed in other metabolic diseases, 21 out of the 23 were DA only in the NAFLD-O subjects and 23 out of the 31 were DA only in NAFLD-L subjects.
Following the DA analysis of the microbial metabolic output, we further attempted to consolidate them with microbial gene abundances (ECs). We initially used ANCOM-II (adjusting for age, gender, BMI, HOMA-IR, and SBP, cutoff = 0.6) to find DA ECs between the NAFLD groups and their corresponding controls and then linked them with the DA metabolites (Table S6) using the KEGG database [36]. Overall, the in silico estimated microbial metabolic output appears to be highly associated with the metagenomic functional data (ECs, KEGG Pathways). We successfully linked 18 DA microbial metabolites from the NAFLD-O and NAFLD-L groups, including 3,4-dihydroxyphenylacetate, dihydroxyacetone, and D-mannose-1-phopshate, with 101 unique DA ECs (Fig. S3B).
We then examined the possible biological implications of the DA microbial metabolites in NAFLD by analyzing their associations with NAFLD-related clinical parameters. We found 10 microbial metabolites significantly correlated (partial Spearman’s correlation adjusted by age, gender, and BMI; P < 0.05) with at least one of the clinical parameters in the NAFLD-O group (Fig. S3A). Notably, 3,4-dihydroxyphenylacetate and protoheme were found significantly higher in the CTRL-NAFLD-O group, and both negatively correlated with FLI, ALT, and TGs. Four DA microbial metabolites had significant correlations with the relevant clinical parameters in the NAFLD-L group. Specifically, adenine and 4-hydroxy-proline showed negative correlations with FLI or TGs, whereas D-tartrate and meso-2,6-diaminoheptanedioate (a precursor in the lysine biosynthesis pathway) were positively associated with liver enzymes (GGT and ALT, respectively).
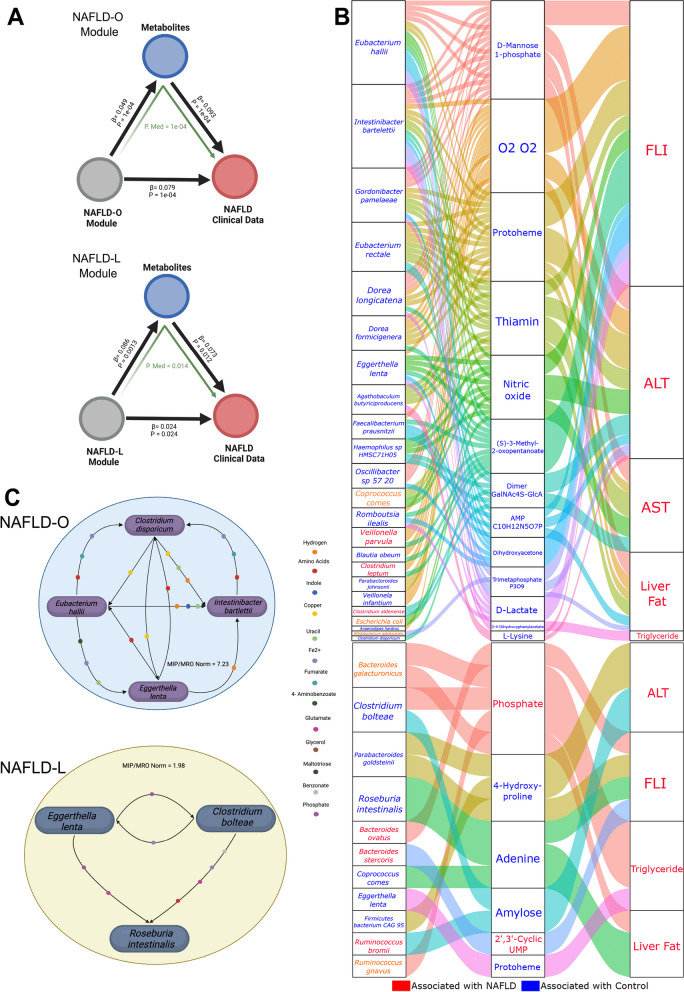
The analyses above resulted in microbial modules and metabolites that could be linked to either NAFLD-O or NAFLD-L using the liver-associated clinical parameters. We subsequently applied mediation analysis to investigate whether the groups of DA metabolites can mediate the impact of microbial modules (Fig. 3B) on the levels of NAFLD-related clinical parameters. Our analysis suggests that for both NAFLD-O and NAFLD-L, the DA metabolites significantly intercede the influence of the species modules on the NAFLD phenotypes (mediation P < 0.05) (Fig. 4A). Following the group level analysis, we examined, again with mediation analysis, which individual species from the microbial modules are associated with the levels of the clinical parameters through individual DA mediator metabolites. A total 130 linkages between 23 species from the NAFLD-O module and 13 DA metabolites were found, with most of them involving species negatively associated with clinical parameters in the module analysis (Figs. 4B and 3B). Notably, species such as I. bartelettii, E. hallii, G. pamelaeae, Dorea longicatena, E. lenta, Rombroutsia ilealis, and B. obeum were all selected as key features in the ML model and had the most links in the mediation analysis (Figs. 2C and 4B). Moreover, mediator metabolites, which were all higher in CTRL-NAFLD-O, such as S-3-methyl-2-oxopentanoate [75], protoheme [76], dihydroacetone [77], thiamin [78], and 3,4-dihydroxyphenylacetate [79] have all been previously associated negatively with NAFLD. On the contrary, only 15 linkages between species from the NAFLD-L modules and clinical parameters through the DA metabolites were significant. Species associated negatively with NAFLD were again the most prevalent (6) compared to those with positive (3) or no association (2). This result is in accordance with the profile of the NAFLD-L module (Fig. 3B), which consists of a mixture of species correlated positively or negatively with liver parameters. Phosphate and 2′,3′-cyclic UMP are two mediator metabolites found higher in NAFLD and have both been linked previously with NAFLD pathogenesis [80, 81].
Fig. 4.
Specific microbial metabolites mediate the associations between gut microbiota species modules and NAFLD. A Analysis of the significant (P < 0.05) effect of species modules on NAFLD-related clinical data mediated by the DA metabolites for NAFLD-O and NAFLD-L. B Parallel coordinates charts showing the 133 mediation effects of in silico estimated metabolites that were significant at P < 0.05. Upper chart: NAFLD-O; lower chart: NAFLD-L. Within each chart, it shows individual species from network modules (left), DA metabolites (middle), and NAFLD clinical data (right). The curved lines connecting the panels indicate the mediation effects, with line colors corresponding to different metabolites. The colors of feature names represent positive (red) or negative (blue) associations with NAFLD, as determined by sCCA for species and DA for metabolites and clinical data. C A graphical representation of bacterial synergistic communities generated from SMETANA for NAFLD-O and NAFLD-L. Metabolites exchanged with smetana score ≥ 1 are displayed. Created with Biorender.com
Identification of beneficial microbial consortia for NAFLD-O and NAFLD-L
Lastly, by integrating the different analytical steps above, we attempted to derive small synergistic bacterial consortia with potential value as therapeutics in NAFLD-O and NAFLD-L. We applied the SMETANA [55] method to identify small cooperative communities of species associated negatively with NAFLD. In brief, using genome-scale metabolic modeling (GSMM), SMETANA calculates how cooperative and competitive a target community is and the likelihood of exchanging metabolites between a pair of microbes in a community. The ability of SMETANA to successfully identify interspecies metabolite exchanges has been proved by the reproduction of experimentally mapped interactions within bacterial species communities in the lab [55]. Using species derived from the mediation analysis (Fig. 4C), we generated in total of 4928 possible species communities with up to 5 members for NAFLD-O and 352 for NAFLD-L, from which we uncovered 2 consortia of species that create a non-competitive and synergistic environment for each other. For NAFLD-O, a group of 4 species including E. hallii, I. bartlettii, E. lenta, and Clostridium disporicum was selected as the best community. Specifically, this consortium’s normalized metabolic interaction potential (MIP)/metabolic resource overlap (MRO) score (7.232) was 1.5 fold greater than the mean score of randomly generated communities and was the highest out of all beneficial species communities in our dataset (Fig. 4C) (Table S9). This community has in total 42 different metabolites being exchanged between its members (Table S9). The first 3 bacteria were all found significantly higher in CTRL-NAFLD-O subjects and were top features in the ML model contributing to CTRL-NAFLD-O (Table S6, Fig. 2C). In the differential correlation network analysis these 4 species were involved in 10 changes during the structural reshaping of the gut microbiota from the CTRL-NAFLD-O to NAFLD-O status. Moreover, all 4 species had negative associations with NAFLD clinical parameters (Fig. 3B) that were mediated by microbial metabolites (Fig. 4B). Lastly, both E. halli and I. bartlettii have previously been reported as species that can protect from the development of NAFLD [82–84]. Regarding the NAFLD-L, a set of 3 species was selected from SMETANA analysis, which included Clostridium bolteae, Roseburia intestinalis, and, as in NAFLD-O, E. lenta (Fig. 4C). In this consortium, 28 different metabolites are being exchanged (Table S9) making it a synergistic and non-competitive community even though the normalized MIP/MRO score (1.98) was lower than the one in NAFLD-O. None of the species was found in DA; however, the latter two had higher abundance in CTRL-NAFLD-L even though they did not reach statistical significance (possibly due to the small cohort size). Moreover, all 3 were associated negatively with NAFLD clinical parameters through sCCA (Fig. 3B) and were linked to DA metabolites from the mediation analysis (Fig. 4B). Interestingly, R. intestinalis is a thoroughly studied bacterial species, which has shown its beneficial activities against metabolic diseases and NAFLD specifically [85].
In order to validate the predicted potential of these bacteria to cooperate as a collective unit and establish co-abundance patterns with one another, we conducted a Spearman correlation analysis among the suggested species using 928 healthy samples in the population-based Health Professionals Follow-up Study (HPFS), which has been utilized previously to study the stability of the human fecal microbiome [27]. Notably, in both microbial consortia, the selected bacteria showed mostly significant positive correlations and synergistic trends (lower and upper quartile: ρ [0.04, 0.13], P [0.00003–0.049]) (Table S10). Moreover, we aimed to consolidate the associations of bacterial consortium with NAFLD-related clinical data by adding up the relative abundance of the NAFLD-O community members and correlating it with the metadata, both in our cohort and in an external cohort [4]. Overall, in both cohorts the selected microbial consortium was significantly correlated negatively with NAFLD clinical data, further reinforcing its NAFLD-alleviating potential (Table S10). The analysis described above was performed only in NAFLD-O community as there was no publically available dataset with detailed metadata for NAFLD-L.
In summary, by combining species interaction networks, DA analysis, interpretable machine learning, sCCA, and GSMM, we could reveal cooperative microbial consortia that consist of species specifically and negatively associated with NAFLD pathophysiology. Such small bacterial consortia hold the potential and feasibility to be further investigated for usage as novel microbiome-based therapeutics for NAFLD-O or NAFLD-L.
Discussion
Due to its close relationship with metabolic dysfunction, NAFLD often co-occurs with other metabolic diseases with typical examples being T2D, obesity, and in general cardiovascular conditions [8]. Hence, it is of great challenge to find which microbial changes are highly specific for NAFLD and which are shared with other metabolic diseases. To reveal robust and highly specific NAFLD microbiota signatures, we collected shotgun metagenomics data from both NAFLD patients and other metabolic diseases (including prediabetes, T2D, prehypertension, hypertension, and atherosclerosis), together with matched control groups for each corresponding disease, which summed up to > 1200 samples. The availability of well-characterized clinical profiles not only facilitated the identification of microbes associated with NAFLD, but also allowed to minimize the effect of confounding factors. Moreover, statistical adjustment (age, gender, BMI, HOMA-IR, and SBP) was performed to find specific NAFLD signatures accounting for the effect of T2D and hypertension. It is worth to note that our cohort is free of antibiotics and almost free of major medication usage except 33 subjects; 19 patients with atherosclerosis who had taken metoprolol, 10 T2D-L, and 4 Control-NAFLD-O who had taken antidiabetic medication. The non-negligible effect of medication on gut microbiome has been demonstrated [86, 87], including metoprolol [26, 88]. The majority of the studies shared additional exclusion criteria namely heart failure, renal insufficiency, acute infectious disease, and cancer.
Gut microbiota has been investigated for potential use as non-invasive diagnostic or prognostic tools for a wide variety of diseases, such as T1D [89] natural killer/T-cell lymphoma [90] and a multi-disease panel covering 8 diseases (colorectal cancer, Crohn’s disease, cardiovascular disease, etc.) [91]. Regarding NAFLD, analysis based on shotgun metagenomic sequencing revealed its potential for diagnosis of advanced fibrosis [3] and cirrhosis [4], while we utilized previously microbiome-derived features for early-stage risk assessment of NAFLD [6]. Despite their high predictive performance, the development and assessment of the machine learning models usually involve only the particular diseases being studied, where the model specificity has seldom been investigated. Recent studies have highlighted the existence of both disease-specific and shared microbe-disease associations [42] and host gene-microbiome associations [43]. Thus, from a clinical translational point of view, it is necessary to evaluate the model specificity for the diagnosis of target disease against other closely relevant diseases. Towards this direction, recently Wirbel et al. [47] and Kartal et al. [92] have identified microbial signatures that are specific for colorectal cancer and pancreatic cancer, respectively. Here we applied cross-disease portability and specificity evaluations, first described by Wirbel et al. [47], to our NAFLD ML models to identify NAFLD-specific microbial signatures. Using the metagenomics data, we found microbial taxonomic and functional signatures that are either specific to NAFLD-O or shared among different metabolic diseases and constructed an ML model with very high predictive performance but poor specificity. In contrast, when we coupled the metagenome with the in silico-predicted metabolome, the ML model not only reached an accuracy of 0.917 for the diagnosis of NAFLD-O, but also demonstrated much-enhanced specificity when being evaluated against the other metabolic diseases in the study. Notably, the metabolomic data used in this study were derived from in silico predictions as it is infeasible to integrate data from multiple studies and conduct a large-scale comparative analysis. In addition, the metabolic output provided by MAMBO is derived solely from microbial communities, independent of the human organism. Due to the limited number of patients in the NAFLD-L group, we focused only on building a ML model for NAFLD-O and its controls. Importantly, our model derived solely from the Chinese cohort in order to limit potential biases was validated in an independent US cohort with biopsy-confirmed NAFLD, reaching an accuracy of 0.845. Notably, the inability of the ML model to accurately predict NAFLD in lean individuals (Fig. 2B, prediction rate = 0.056) prompt our interest to conduct a more detailed investigation into microbiome variations in the two disease groups (NAFLD-O and NAFLD-L), at the level of both taxonomy and metabolic output.
By analyzing shotgun metagenomics data separately for NAFLD-O and NAFLD-L, our study reaches consistent findings with previous 16S rRNA-based studies [93, 94] that overweight and lean/non-overweight NAFLD differ in the gut microbiome composition, but further reveals species-level signatures and network modules specific to overweight or lean NAFLD. However, in the human gut, it is the interaction of different microbial species rather than individual microorganisms themselves that is responsible for maintaining the community structure and function and providing a stable habitat [95–97]. Therefore, the quantity of single bacteria, as most studies analyze, cannot characterize the ecosystem as a whole, let alone the shift from health to disease. Network analysis has been extensively utilized in numerous biological systems, and co-occurrence and correlation networks in particular, have contributed to our knowledge of the link between various species [98–100]. Thus, in our analysis, we attempted to focus on how the microbiome structural changes from healthy control to NAFLD are associated with NAFLD pathophysiology. Using DGCA, we found in the NAFLD-O group one network cluster of species associated with liver-related clinical parameters. This module was dominated by bacteria with a negative association to NAFLD, either by being significantly lower in abundance or being important features in the ML model. Their interactions were altered in NAFLD by either disturbing their positive correlations (+ /0) in healthy conditions or forming positive correlations due to the concurrent reduction of abundance in NAFLD (0/ +). Similarly, considerable changes in co-abundance patterns were also observed in the network module in the NAFLD-L group. Moreover, through a mediation analysis, we identified various in silico metabolites for NAFLD-O and NAFLD-L, that can mediate the impact of these network modules on the NAFLD phenotype.
Probiotics are living microorganisms that modify the intestinal microbiota and have positive health benefits for humans. In particular, they exert their effects through modulating the structural and functional composition of gut microbiota, generating antimicrobial compounds, enhancing epithelial barrier function, suppressing intestinal inflammation, and have demonstrated efficacy in preventing the development of NAFLD [101]. By summarizing all the information above, through metabolic modeling, we attempted to form two small synergistic microbial consortia that hold potential as microbiome therapeutics for NAFLD. Indeed, Eubacterium hallii, Eggerthela lenta, Clostridium bolteae, Intestinibacter bartlettii, and Roseburia intestinalis have been associated negatively with the onset of NAFLD [2–5], but also have been thoroughly established as producers of short-chain fatty acids (SCFAs) [2, 6–10]. SCFAs are a class of metabolites, produced by probiotics, which exhibit anti-inflammatory properties, promote a better gut flora, improve intestinal permeability, and regulate metabolism [11–14]. Their regulation of the inflammation effects has directly been associated with reduced levels of inflammation-related enzymes AST and ALT [11, 15].
Our analytical approach to investigating the differences between NAFLD and other metabolic diseases, even though it offers valuable insight, does not come without limitations. The metabolomics data used are based on in silico metabolic modeling with an emphasis on primary metabolism. Future studies may benefit from methodological advances in the integration of metabolomics data from different studies and get even stronger in the diagnosis or mechanistic understanding of NAFLD compared to other metabolic diseases. Moreover, information such as NAFLD status (in non-NAFLD studies), diet, and other metadata were not available, which prevented us from taking into account their confounding effects during group building and microbiome analysis. Furthermore, for the NAFLD Lean group, we had a relatively low sample size (18), compared to its lean controls (39) and the NAFLD overweight counterpart (163), even though no direct comparisons were made between the two NAFLD groups. Both the microbial signatures and metabolic signatures identified from our computational analyses warrant further experimental investigation in animal models of NAFLD.
Conclusion
In conclusion, this study integrated metagenomic data, detailed metadata, and an in silico metabolic output in order to identify specific microbiome signatures for NAFLD compared to other metabolic diseases. Moreover, we proposed synergistic microbial communities related to NAFLD phenotype in overweight and lean individuals, respectively. Ultimately, the goal is to provide further direction to the current research on creating live biotherapeutic products designed to counter the progression of NAFLD and provide beneficial conditions to salvage a dysbiotic state.
Supplementary Information
Additional file 1: Figure S1: Alpha diversity boxplots between NAFLD and controls for bacterial species. Significant differences were determined using Wilcoxon rank-sum test and were considered significant if P<0.05 (* = P <0.05, NS = Not Significant).
Additional file 2: Figure S2: Beta diversity between NAFLD and controls for bacterial species, KEGG pathways and metabolites, using NMDS of weighted UniFrac, Bray Curtis and Canberra distances, respectively. The error bars indicate the mean and standard errors of the mean. Significant differences were determined using PERMANOVA, adjusting for age, gender and BMI and were considered significant if P<0.05.
Additional file 3: Figure S3: ROC curves, confusion matrices and boxplots evaluating the ability of random forest models to predict NAFLD, using either species, kegg pathways or metabolites only. The lower and upper hinges of boxplots presented in the Figures correspond to the 25th and 75th percentiles, respectively. Bold horizontal lines denote median values and the whiskers are extending up to the most extreme points within 1.5-fold IQR. Data beyond the end of the whiskers are plotted individually.
Additional file 4: Figure S4: (A) Circos plot of significantly different metabolites for each metabolic disease comparison with their controls, together with correlation analysis with NAFLD clinical parameters (Partial spearman’s correlation adjusted by age, gender and BMI; P<0.05). (B) Representative examples of significantly different metabolites linked with significantly different ECs (ANCOM-II, cutoff = 0.6). Created with lucidchart.com.
Additional file 5: Figure S5: Beta diversity between the samples of the MRS-NAFLD study and those from the Biopsy-NAFLD study for bacterial species, using Principal Coordinate Analysis (PCoA) of weighted UniFrac. Significant differences were determined using PERMANOVA, and were considered significant if P<0.05.
Additional file 6: Table S1: Summary of the remaining clinical and anthropometric characteristics of NAFLD and control groups.
Additional file 7: Table S2: Summary of the sequencing, microbiome, clinical and anthropometric characteristics for other metabolic diseases and their controls.
Additional file 8: Table S3: Samples distribution across the study groups.
Additional file 9: Table S4: Summary of beta diversity results for species, KEGG pathways and metabolites for NAFLD against controls and metabolic diseases.
Additional file 10: Table S5: Features included in the species and pathways machine learning model.
Additional file 11: Table S6: Significantly different microbial species, ECs and metabolites lists generated for every disease against their control.
Additional file 12: Table S7: DGCA Networks for NAFLD-O and NAFLD-L significantly associated with NAFLD clinical parameters through Mantel test.
Additional file 13: Table S8: Sparse Canonical Correlation analysis coefficients between with NAFLD species modules and NAFLD-related clinical data.
Additional file 14: Table S9: SMETANA results for species communities and metabolites exchanged for the selected communities.
Additional file 15: Table S10: Spearman correlation analysis results for validating the prioritized species communities.
Aknowledgements
Not applicable
Authors’ contributions
E.N, G.P. and Y.N. conceptualized and designed the study. E.N., A.M.S., X.C. and M.M. collected, processed and analysed the data. E.N., Y.N., A.M.S. and G.P. wrote the original manuscript. A.X., H.Li, W.J. and R.L. provided data for analysis. G.P. and Y.N. lead and supervized the research work. G.P., Y.N., A.X., H.Li, W.J., H.B.N, M.N., R.L., E.N., A.M.S., X.C. and M.M reviewed and commented on the manuscript.
Funding
This work was supported by Marie Sklodowska-Curie Actions (MSCA), and Innovative Training Networks, H2020-MSCA-ITN-2018, 813781 “BestTreat” (G.P., E.N.), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy – EXC 2051 – Project ID 390713860 (G.P., Y.N.). M.N. is supported by a personal ZONMW-VICI grant 2020 (09150182010020). G.P. would like to thank the German Federal Ministry of Education and Research (BMBF) within the funding project PerMiCCion (project ID: 01KD2101A). R.L. receives funding support from NCATS (5UL1TR001442), NIDDK (U01DK061734, U01DK130190, R01DK106419, R01DK121378, R01DK124318, P30DK120515), NHLBI (P01HL147835), John C Martin Foundation (RP124).
Data availability
The accession numbers for shotgun sequencing datasets reported in this paper are publicly available at the NCBI Sequencing Read Archive, the European Genome-Phenome Archive and the European Bioinformatics Institute (EBI) database under IDs: PRJNA703757, PRJNA732131*, PRJNA728908, PRJNA686835, PRJEB13870, PRJNA454826, ERP023788, EGAS00001004600 and PRJNA354235. Codes and scripts developed in this study are all available at the GitHub repository (https://github.com/ManosNychas/NAFLD-Metabolic-Diseases).
Declarations
Ethics approval and consent to participate
For the cohorts that are not associated with a published study, ethics approvals were obtained by the Shanghai Jiao Tong University Affiliated Sixth People’s Hospital (approval no: 2015-65-(1)) and the University of Hong Kong / Hospital Authority Hong Kong West Cluster (approval no: UW 20-700) following the principles of the Declaration of Helsinki.
Consent for publication
Written informed consent was obtained from all participants from all the cohorts that are not associated with a published study.
Competing interests
M.N. is founder and scientific advisor of Caelus Health that is commercializing A. soehngenii for metabolic disease treatment. R.L. serves as a consultant to Aardvark Therapeutics, Altimmune, Arrowhead Pharmaceuticals, AstraZeneca, Cascade Pharmaceuticals, Eli Lilly, Gilead, Glympse bio, Inipharma, Intercept, Inventiva, Ionis, Janssen Inc., Lipidio, Madrigal, Neurobo, Novo Nordisk, Merck, Pfizer, Sagimet, 89 bio, Takeda, Terns Pharmaceuticals and Viking Therapeutics. In addition, his institution received research grants from Arrowhead Pharmaceuticals, Astrazeneca, Boehringer-Ingelheim, Bristol-Myers Squibb, Eli Lilly, Galectin Therapeutics, Gilead, Intercept, Hanmi, Intercept, Inventiva, Ionis, Janssen, Madrigal Pharmaceuticals, Merck, Novo Nordisk, Pfizer, Sonic Incytes and Terns Pharmaceuticals. Co-founder of LipoNexus Inc. All other authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Emmanouil Nychas and Andrea Marfil-Sánchez contributed equally to this work.
Contributor Information
Yueqiong Ni, Email: yueqiong.ni@connect.hku.hk.
Gianni Panagiotou, Email: gianni.panagiotou@leibniz-hki.de.
References
- 1.Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat. Rev. Gastroenterol. Hepatol. 2013. p. 686–90. [DOI] [PubMed]
- 2.Kolodziejczyk AA, Zheng D, Shibolet O, Elinav E. The role of the microbiome in NAFLD and NASH. EMBO Mol Med. 2019;11:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017;25:1054-1062.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oh TG, Kim SM, Caussy C, Fu T, Guo J, Bassirian S, et al. A Universal Gut-Microbiome-Derived Signature Predicts Cirrhosis. Cell Metab. 2020;32:878-888.e6. Available from: 10.1016/j.cmet.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Méric G, Havulinna AS, Teo SM, Åberg F, Ruuskanen M, et al. Early prediction of incident liver disease using conventional risk factors and gut-microbiome-augmented gradient boosting. Cell Metab. 2022;34:719-730.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung H, Long X, Ni Y, Qian L, Nychas E, Siliceo SL, et al. Risk assessment with gut microbiome and metabolite markers in NAFLD development. Sci Transl Med. 2022;14(648):eabk0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni Y, Qian L, Siliceo SL, Long X, Nychas E, Liu Y, et al. Resistant starch decreases intrahepatic triglycerides in patients with NAFLD via gut microbiome alterations. Cell Metab. 2023;35:1530-1547.e8. [DOI] [PubMed] [Google Scholar]
- 8.Aron-Wisnewsky J, Vigliotti C, Witjes J, Le P, Holleboom AG, Verheij J, et al. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat Rev Gastroenterol Hepatol. Available from: 10.1038/s41575-020-0269-9. Cited 2022 Jan 28. [DOI] [PubMed]
- 9.Kuchay MS, Ignacio Martínez-Montoro J, Choudhary NS, Carlos Fernández-García J, Ramos-Molina B, Pericas JM. Non-Alcoholic Fatty Liver Disease in Lean and Non-Obese Individuals: Current and Future Challenges. biomedicines [Internet]. 2021; Available from: 10.3390/biomedicines9101346. [DOI] [PMC free article] [PubMed]
- 10.Zeybel M, Arif M, Li X, Altay O, Yang H, Shi M, et al. Multiomics Analysis Reveals the Impact of Microbiota on Host Metabolism in Hepatic Steatosis. Adv Sci. 2022;9:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yilmaz B, Juillerat P, Øyås O, Ramon C, Bravo FD, Franc Y, et al. Microbial network disturbances in relapsing refractory Crohn’s disease. Nat Med. 2019;25:323–36. [DOI] [PubMed] [Google Scholar]
- 12.Mac Aogáin M, Narayana JK, Tiew PY, Ali NABM, Yong VFL, Jaggi TK, et al. Integrative microbiomics in bronchiectasis exacerbations. Nat Med. 2021;27:688–99. [DOI] [PubMed] [Google Scholar]
- 13.Yuan J, Chen C, Cui J, Lu J, Yan C, Wei X, et al. Fatty Liver Disease Caused by High-Alcohol-Producing Klebsiella pneumoniae. Cell Metab. 2019;30:675–688.e7. Available from: 10.1016/j.cmet.2019.08.018. [DOI] [PubMed]
- 14.Seo B, Jeon K, Moon S, Lee K, Kim WK, Jeong H, et al. Roseburia spp. Abundance Associates with Alcohol Consumption in Humans and Its Administration Ameliorates Alcoholic Fatty Liver in Mice. Cell Host Microbe. 2020;27:25–40.e6. Available from: 10.1016/j.chom.2019.11.001. [DOI] [PubMed]
- 15.Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019;25:1096–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregory JC, Buffa JA, Org E, Wang Z, Levison BS, Zhu W, et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem. 2015;290:5647–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Lu Y, Yan Y, Tian S, Zheng D, Leng D, et al. Promising Treatment for Type 2 Diabetes: Fecal Microbiota Transplantation Reverses Insulin Resistance and Impaired Islets. Front Cell Infect Microbiol. 2020;9:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merrick B, Allen L, Masirah M Zain N, Forbes B, Shawcross DL, Goldenberg SD. Regulation, risk and safety of Faecal Microbiota Transplant. Infect Prev Pract. 2020;2:100069. Available from: 10.1016/j.infpip.2020.100069. [DOI] [PMC free article] [PubMed]
- 19.Safety alert regarding use of fecal microbiota for transplantation and risk of serious adverse events likely due to transmission of pathogenic organisms. Food and Drug Administration 2020. March 12,https://www.fda.gov/safety/medical-product-safety-informa.
- 20.van der Lelie D, Oka A, Taghavi S, Umeno J, Fan TJ, Merrell KE, et al. Rationally designed bacterial consortia to treat chronic immune-mediated colitis and restore intestinal homeostasis. Nat Commun. 2021;12:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017;25:1054–1062.e5. Available from: 10.1016/j.cmet.2017.04.001. [DOI] [PMC free article] [PubMed]
- 22.Caussy C, Hsu C, Lo M, Liu A, Bettencourt R, Veeral H, et al. Novel link between gut-microbiome derived metabolite and shared gene-effects with hepatic steatosis and fibrosis in NAFLD. 2019;68:918–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5:1–19. Available from: 10.1186/s40168-016-0222-x. [DOI] [PMC free article] [PubMed]
- 24.Zhang J, Ni Y, Qian L, Fang Q, Zheng T, Zhang M, et al. Decreased Abundance of Akkermansia muciniphila Leads to the Impairment of Insulin Secretion and Glucose Homeostasis in Lean Type 2 Diabetes. Adv Sci. 2021;8(16):e2100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Wang Y, Ni Y, Cheung CKY, Lam KSL, Wang Y, et al. Gut Microbiome Fermentation Determines the Efficacy of Exercise for Diabetes Prevention. Cell Metab. 2020;31:77–91.e5. Available from: 10.1016/j.cmet.2019.11.001. [DOI] [PubMed]
- 26.Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8:1–11. Available from: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed]
- 27.Mehta RS, Abu-ali GS, Drew DA, Lloyd-price J, Lochhead P, Joshi AD, et al. Stability of the human faecal microbiome in a cohort of adult men. 2018;3:347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasolli E, Schiffer L, Manghi P, Renson A, Obenchain V, Truong DT, et al. Accessible, curated metagenomic data through ExperimentHub. Nat Methods. 2017;14:1023–4. Available from: 10.1038/nmeth.4468. [DOI] [PMC free article] [PubMed]
- 29.Weber MA, Schiffrin EL, White WB, Mann S, Lindholm LH, Kenerson JG, et al. Clinical practice guidelines for the management of hypertension inthe community a statement by the American society of hypertension and the International Society of Hypertension. J Hypertens. 2014;32(1):3–15. [DOI] [PubMed] [Google Scholar]
- 30.WHO. Use of Glycated Haemoglobin (HbA1c) in the diagnosis of diabetes mellitus: abbreviated report of a WHO consultation. approved by the guidelines review committee. World Heal Organ. 2011;299–309. [PubMed]
- 31.Clarke EL, Taylor LJ, Zhao C, Connell A, Lee J, Fett B, et al. Sunbeam : an extensible pipeline for analyzing metagenomic sequencing experiments. Microbiome. 2019;7(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows – Wheeler transform. Bioinformatics. 2009;25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolger AM, Lohse M, Usadel B. Genome analysis Trimmomatic : a flexible trimmer for Illumina sequence data. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beghini F, McIver LJ, Blanco-Míguez A, Dubois L, Asnicar F, Maharjan S, et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife. 2021;10. Available from: PMC8096432. Cited 2022 Jan 28. [DOI] [PMC free article] [PubMed]
- 35.Caspi R, Billington R, Keseler IM, Kothari A, Krummenacker M, Midford PE, et al. The MetaCyc database of metabolic pathways and enzymes-a 2019 update. Nucleic Acids Res. 2020;48:D455–D453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27. Available from: PMC102409. Cited 2021 Nov 2. [DOI] [PMC free article] [PubMed]
- 37.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–30. Available from: http://doi.wiley.com/10.1111/j.1654-1103.2002.tb02049.x.
- 38.Benjamini Y, Hochberg Y, Benjamini, Yoav HY. Controlling The False Discovery Rate - A Practical And Powerful Approach To Multiple Testing. J. R. Stat. Soc. Ser. B. 1995. p. 289–300. Available from: http://www.stat.purdue.edu/~doerge/BIOINFORM.D/FALL06/Benjamini and Y FDR.pdf%5Cn http://engr.case.edu/ray_soumya/mlrg/controlling_fdr_benjamini95.pdf.
- 39.Magnúsdóttir S, Heinken A, Kutt L, Ravcheev DA, Bauer E, Noronha A, et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol. 2017;35:81–9. [DOI] [PubMed] [Google Scholar]
- 40.Machado D, Andrejev S, Tramontano M, Patil KR. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 2018;46:7542–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stekhoven DJ, Bühlmann P. Missforest-Non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28:112–8. [DOI] [PubMed] [Google Scholar]
- 42.Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun. 2017;8(1):1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Priya S, Burns MB, Ward T, Mars RAT, Adamowicz B, Lock EF, et al. Identification of shared and disease-specific host gene–microbiome associations across human diseases using multi-omic integration. Nat Microbiol. 2022;7:780–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Max A, Wing J, Weston S, Williams A, Keefer C, Engelhardt A, et al. Package ‘ caret ’ R topics documented. R Journal. 2022;223(7):48. [Google Scholar]
- 45.Kursa MB, Rudnicki WR. Feature selection with the boruta package. J Stat Softw. 2010;36:1–13. [Google Scholar]
- 46.Greenwell B. fastshap: Fast Approximate Shapley Values. R package. 2024. https://CRAN.R-project.org/package=fastshap.
- 47.Wirbel J, Zych K, Essex M, Karcher N, Kartal E, Salazar G, et al. Microbiome meta-analysis and cross-disease comparison enabled by the SIAMCAT machine learning toolbox. Genome Biol. 2021;22:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song W-M, Zhang B. Multiscale Embedded Gene Co-expression Network Analysis. PLOS Comput Biol. 2015;11:e1004574. Available from: https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004574. Cited 2021 Nov 3. [DOI] [PMC free article] [PubMed]
- 49.McKenzie AT, Katsyv I, Song W-M, Wang M, Zhang B. DGCA: A comprehensive R package for Differential Gene Correlation Analysis. BMC Syst Biol 2016 101. 2016 ;10:1–25. Available from: https://bmcsystbiol.biomedcentral.com/articles/10.1186/s12918-016-0349-1. Cited 2021 Nov 3. [DOI] [PMC free article] [PubMed]
- 50.Bedogni G, Bellentani S, Miglioli L, Masutti F, Passalacqua M, Castiglione A, et al. The fatty liver index: A simple and accurate predictor of hepatic steatosis in the general population. BMC Gastroenterol. 2006;6:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kotronen A, Peltonen M, Hakkarainen A, Sevastianova K, Bergholm R, Johansson LM, et al. Prediction of Non-Alcoholic Fatty Liver Disease and Liver Fat Using Metabolic and Genetic Factors. Gastroenterology. 2009;137:865–72. Available from: 10.1053/j.gastro.2009.06.005. [DOI] [PubMed]
- 52.Witten DM, Tibshirani R, Hastie T. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics. 2009;10:515–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamidi B, Wallace K, Alekseyenko AV. MODIMA, a method for multivariate omnibus distance mediation analysis, allows for integration of multivariate exposure–mediator–response relationships. Genes (Basel). 2019;10(7):524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yi Z. mediation: R Package for Causal Mediation Analysis. J Stat Softw. 2008;59:23–38. Available from: http://www.jstatsoft.org/.
- 55.Zelezniak A, Andrejev S, Ponomarova O, Mende DR, Bork P, Patil KR. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci U S A. 2015;112:6449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedman J, Tibshirani R, Hastie T. Regularization paths for generalized linear models via coordinate descent. J Stat Soft. 2010;33(1):1–22. 10.18637/jss.v033.i01. [PMC free article] [PubMed]
- 57.Reid S, Tibshirani R, Friedman J. A study of error variance estimation in lasso regression. Stat Sin. 2016;26:35–67. [Google Scholar]
- 58.Kim S. ppcor: An R package for a fast calculation to semi-partial correlation coefficients. Commun Stat Appl Methods. 2015;22(6):665–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wickham H. ggplot2: Elegant graphics for data analysis. Springer-Verlag New York; 2016. Available from: https://ggplot2.tidyverse.org.
- 60.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9. Available from: https://academic.oup.com/bioinformatics/article/32/18/2847/1743594. Cited 2021 Nov 2. [DOI] [PubMed]
- 61.Otasek D, Morris JH, Bouças J, Pico AR, Demchak B. Cytoscape Automation: empowering workflow-based network analysis. Genome Biol. 2019;20. Available from: https://pubmed.ncbi.nlm.nih.gov/31477170/. Cited 2021 Nov 2. [DOI] [PMC free article] [PubMed]
- 62.ten Hoopen P, Finn RD, Bongo LA, Corre E, Fosso B, Meyer F, et al. The metagenomic data life-cycle: Standards and best practices. Gigascience. 2017;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas V, Clark J, Doré J. Fecal microbiota analysis: An overview of sample collection methods and sequencing strategies. Future Microbiol. 2015;10:1485–504. [DOI] [PubMed] [Google Scholar]
- 64.Garza DR, Van Verk MC, Huynen MA, Dutilh BE. Towards predicting the environmental metabolome from metagenomics with a mechanistic model. Nat Microbiol. 2018;3:456–60. Available from: 10.1038/s41564-018-0124-8. [DOI] [PubMed]
- 65.Lee JWJ, Plichta DR, Asher S, Delsignore M, Jeong T, McGoldrick J, et al. Association of distinct microbial signatures with premalignant colorectal adenomas. Cell Host Microbe. 2023;31:827–838.e3. Available from: 10.1016/j.chom.2023.04.007. [DOI] [PMC free article] [PubMed]
- 66.Ho TK. Random Decision Forests. Proc 3rd Int Conf Doc Anal Recognit. 1995;
- 67.Udayappan S, Manneras-Holm L, Chaplin-Scott A, Belzer C, Herrema H, Dallinga-Thie GM, et al. Oral treatment with Eubacterium hallii improves insulin sensitivity in db/db mice. npj Biofilms Microbiomes. 2016;2. Available from: 10.1038/npjbiofilms.2016.9. [DOI] [PMC free article] [PubMed]
- 68.Almeida D, Machado D, Andrade JC, Mendo S, Gomes AM, Freitas AC. Evolving trends in next-generation probiotics: a 5W1H perspective. Crit Rev Food Sci Nutr. 2020;60:1783–96. Available from: 10.1080/10408398.2019.1599812. [DOI] [PubMed]
- 69.de Mello VD, Sehgal R, Männistö V, Klåvus A, Nilsson E, Perfilyev A, et al. Serum aromatic and branched-chain amino acids associated with NASH demonstrate divergent associations with serum lipids. Liver Int. 2021;41:754–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goffredo M, Santoro N, Tricò D, Giannini C, D’Adamo E, Zhao H, et al. A branched-chain amino acid-related metabolic signature characterizes obese adolescents with non-alcoholic fatty liver disease. Nutrients. 2017;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang XX, Di WJ, Mu JK, Liu X, Li FJ, Li YQ, et al. Mitochondrial metabolomic profiling for elucidating the alleviating potential of Polygonatum kingianum against high-fat diet-induced nonalcoholic fatty liver disease. World J Gastroenterol. 2019;25:6404–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mayneris-Perxachs J, Cardellini M, Hoyles L, Latorre J, Davato F, Moreno-Navarrete JM, et al. Iron status influences non-alcoholic fatty liver disease in obesity through the gut microbiome. Microbiome. 2021;9:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang Y, Chen X, Chen Q, Xiao J, Mi J, Liu Q, et al. Association of serum methionine metabolites with non-alcoholic fatty liver disease: a cross-sectional study. Nutr Metab. 2022;19:1–12. Available from: 10.1186/s12986-022-00647-7. [DOI] [PMC free article] [PubMed]
- 74.Ma P, Huang R, Jiang J, Ding Y, Li T, Ou Y. Potential use of C-phycocyanin in non-alcoholic fatty liver disease. Biochem Biophys Res Commun. 2020;526:906–12. [DOI] [PubMed] [Google Scholar]
- 75.Ferrannini E, Iervasi G, Cobb J, Ndreu R, Nannipieri M. Insulin resistance and normal thyroid hormone levels: prospective study and metabolomic analysis. Am J Physiol Endocrinol Metab. 2017;312:429–36. Available from: http://www.ajpendo.org. [DOI] [PubMed]
- 76.Tang SY, Cheah IKM, Ng PE, Hoi A, Jenner AM. Heme Consumption Reduces Hepatic Triglyceride and Fatty Acid Accumulation in a Rat Model of NAFLD Fed Westernized Diet. ISRN Oxidative Med. 2014;2014:1–7. [Google Scholar]
- 77.Carbajo-Pescador S, Porras D, Garcia-Mediavilla MV, Martinez-Florez S, Juarez-Fernandez M, Cuevas MJ, et al. Beneficial effects of exercise on gut microbiota functionality and barrier integrity, and gut-liver crosstalk in an in vivo model of early obesity and non-alcoholic fatty liver disease. DMM Dis Model Mech. 2019;12(5):dmm039206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kalyesubula M, Mopuri R, Asiku J, Rosov A, Yosefi S, Edery N, et al. High-dose vitamin B1 therapy prevents the development of experimental fatty liver driven by overnutrition. DMM Dis Model Mech. 2021;14(3):dmm048355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu T, Zhou J, Zhu J, Zhang S, Zhang N, Zhao Y, et al. Carnosic acid protects non-alcoholic fatty liver-induced dopaminergic neuron injury in rats. Metab Brain Dis. 2017;32:483–91. [DOI] [PubMed] [Google Scholar]
- 80.Shin JY, Kim MJ, Kim ES, Mo EY, Moon SD, Han JH, et al. Association between serum calcium and phosphorus concentrations with non-alcoholic fatty liver disease in Korean population. J Gastroenterol Hepatol. 2015;30:733–41. [DOI] [PubMed] [Google Scholar]
- 81.Huang ZR, Chen M, Guo WL, Li TT, Liu B, Bai WD, et al. Monascus purpureus-fermented common buckwheat protects against dyslipidemia and non-alcoholic fatty liver disease through the regulation of liver metabolome and intestinal microbiome. Food Res Int. 2020;136:109511. [DOI] [PubMed] [Google Scholar]
- 82.Witjes JJ, Smits LP, Pekmez CT, Prodan A, Meijnikman AS, Troelstra MA, et al. Donor Fecal Microbiota Transplantation Alters Gut Microbiota and Metabolites in Obese Individuals With Steatohepatitis. Hepatol Commun. 2020;4:1578–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grabherr F, Grander C, Effenberger M, Adolph TE, Tilg H. Gut Dysfunction and Non-alcoholic Fatty Liver Disease. Front Endocrinol (Lausanne). 2019;10:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brahe LK, Le Chatelier E, Prifti E, Pons N, Kennedy S, Hansen T, et al. Specific gut microbiota features and metabolic markers in postmenopausal women with obesity. Nutr Diabetes. 2015;5:e159–7. Available from: 10.1038/nutd.2015.9. [DOI] [PMC free article] [PubMed]
- 85.Nie K, Ma K, Luo W, Shen Z, Yang Z, Xiao M, et al. Roseburia intestinalis: A Beneficial Gut Organism From the Discoveries in Genus and Species. Front Cell Infect Microbiol. 2021;11:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rogers MAM, Aronoff DM. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect. 2016;22:178.e1-178.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Le Bastard Q, Al-Ghalith GA, Grégoire M, Chapelet G, Javaudin F, Dailly E, et al. Systematic review: human gut dysbiosis induced by non-antibiotic prescription medications. Aliment Pharmacol Ther. 2018;47:332–45. [DOI] [PubMed] [Google Scholar]
- 88.Brocker CN, Velenosi T, Flaten HK, McWilliams G, McDaniel K, Shelton SK, et al. Metabolomic profiling of metoprolol hypertension treatment reveals altered gut microbiota-derived urinary metabolites. Hum Genomics. 2020;14:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuan X, Wang R, Han B, Sun CJ, Chen R, Wei H, et al. Functional and metabolic alterations of gut microbiota in children with new-onset type 1 diabetes. Nat Commun. 2022;13:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shi Z, Hu G, Li MW, Zhang L, Li X, Li L, et al. Gut microbiota as non-invasive diagnostic and prognostic biomarkers for natural killer/T-cell lymphoma. Gut. 2022;72(10):1999–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Su Q, Liu Q, Lau RI, Zhang J, Xu Z, Yeoh YK, et al. Faecal microbiome-based machine learning for multi-class disease diagnosis. Nat Commun. 2022;13:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kartal E, Schmidt TSB, Molina-Montes E, Rodríguez-Perales S, Wirbel J, Maistrenko OM, et al. A faecal microbiota signature with high specificity for pancreatic cancer. Gut. 2022;71:1359–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Iwaki M, Kessoku T, Ozaki A, Kasai Y, Kobayashi T, Nogami A, et al. Gut microbiota composition associated with hepatic fibrosis in non-obese patients with non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2021;36:2275–84. [DOI] [PubMed] [Google Scholar]
- 94.Duarte SMB, Stefano JT, Miele L, Ponziani FR, Souza-Basqueira M, Okada LSRR, et al. Gut microbiome composition in lean patients with NASH is associated with liver damage independent of caloric intake: A prospective pilot study. Nutr Metab Cardiovasc Dis. 2018;28:369–84. Available from: 10.1016/j.numecd.2017.10.014. [DOI] [PubMed]
- 95.Banerjee S, Walder F, Büchi L, Meyer M, Held AY, Gattinger A, et al. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 2019;13:1722–36. Available from: 10.1038/s41396-019-0383-2. [DOI] [PMC free article] [PubMed]
- 96.Rao C, Coyte KZ, Bainter W, Geha RS, Martin CR, Rakoff-Nahoum S. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature. 2021;591:633–8. Available from: 10.1038/s41586-021-03241-8. [DOI] [PMC free article] [PubMed]
- 97.Xiao L, Wang J, Zheng J, Li X, Zhao F. Deterministic transition of enterotypes shapes the infant gut microbiome at an early age. Genome Biol. 2021;22:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cheng R, Wang L, Le S, Yang Y, Zhao C, Zhang X, et al. A randomized controlled trial for response of microbiome network to exercise and diet intervention in patients with nonalcoholic fatty liver disease. Nat Commun. 2022;13(1):2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Samara J, Moossavi S, Alshaikh B, Ortega VA, Pettersen VK, Ferdous T, et al. Supplementation with a probiotic mixture accelerates gut microbiome maturation and reduces intestinal inflammation in extremely preterm infants. Cell Host Microbe. 2022;30:696-711.e5. [DOI] [PubMed] [Google Scholar]
- 100.Liu H, Liao C, Wu L, Tang J, Chen J, Lei C, et al. Ecological dynamics of the gut microbiome in response to dietary fiber. ISME J. 2022;16(8):2040–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Iacono A, Raso GM, Canani RB, Calignano A, Meli R. Probiotics as an emerging therapeutic strategy to treat NAFLD: Focus on molecular and biochemical mechanisms. J Nutr Biochem. 2011;22:699–711. Available from: 10.1016/j.jnutbio.2010.10.002. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1: Alpha diversity boxplots between NAFLD and controls for bacterial species. Significant differences were determined using Wilcoxon rank-sum test and were considered significant if P<0.05 (* = P <0.05, NS = Not Significant).
Additional file 2: Figure S2: Beta diversity between NAFLD and controls for bacterial species, KEGG pathways and metabolites, using NMDS of weighted UniFrac, Bray Curtis and Canberra distances, respectively. The error bars indicate the mean and standard errors of the mean. Significant differences were determined using PERMANOVA, adjusting for age, gender and BMI and were considered significant if P<0.05.
Additional file 3: Figure S3: ROC curves, confusion matrices and boxplots evaluating the ability of random forest models to predict NAFLD, using either species, kegg pathways or metabolites only. The lower and upper hinges of boxplots presented in the Figures correspond to the 25th and 75th percentiles, respectively. Bold horizontal lines denote median values and the whiskers are extending up to the most extreme points within 1.5-fold IQR. Data beyond the end of the whiskers are plotted individually.
Additional file 4: Figure S4: (A) Circos plot of significantly different metabolites for each metabolic disease comparison with their controls, together with correlation analysis with NAFLD clinical parameters (Partial spearman’s correlation adjusted by age, gender and BMI; P<0.05). (B) Representative examples of significantly different metabolites linked with significantly different ECs (ANCOM-II, cutoff = 0.6). Created with lucidchart.com.
Additional file 5: Figure S5: Beta diversity between the samples of the MRS-NAFLD study and those from the Biopsy-NAFLD study for bacterial species, using Principal Coordinate Analysis (PCoA) of weighted UniFrac. Significant differences were determined using PERMANOVA, and were considered significant if P<0.05.
Additional file 6: Table S1: Summary of the remaining clinical and anthropometric characteristics of NAFLD and control groups.
Additional file 7: Table S2: Summary of the sequencing, microbiome, clinical and anthropometric characteristics for other metabolic diseases and their controls.
Additional file 8: Table S3: Samples distribution across the study groups.
Additional file 9: Table S4: Summary of beta diversity results for species, KEGG pathways and metabolites for NAFLD against controls and metabolic diseases.
Additional file 10: Table S5: Features included in the species and pathways machine learning model.
Additional file 11: Table S6: Significantly different microbial species, ECs and metabolites lists generated for every disease against their control.
Additional file 12: Table S7: DGCA Networks for NAFLD-O and NAFLD-L significantly associated with NAFLD clinical parameters through Mantel test.
Additional file 13: Table S8: Sparse Canonical Correlation analysis coefficients between with NAFLD species modules and NAFLD-related clinical data.
Additional file 14: Table S9: SMETANA results for species communities and metabolites exchanged for the selected communities.
Additional file 15: Table S10: Spearman correlation analysis results for validating the prioritized species communities.
Data Availability Statement
The accession numbers for shotgun sequencing datasets reported in this paper are publicly available at the NCBI Sequencing Read Archive, the European Genome-Phenome Archive and the European Bioinformatics Institute (EBI) database under IDs: PRJNA703757, PRJNA732131*, PRJNA728908, PRJNA686835, PRJEB13870, PRJNA454826, ERP023788, EGAS00001004600 and PRJNA354235. Codes and scripts developed in this study are all available at the GitHub repository (https://github.com/ManosNychas/NAFLD-Metabolic-Diseases).