Abstract
Background
According to the European Society for Clinical Oncology (ESMO) guidelines, the therapeutic algorithm for early-stage epithelial ovarian carcinoma (EOC) is primarily based on grading and histotype. Adjuvant chemotherapy is usually recommended for high-grade tumors and for the International Federation of Gynecology and Obstetrics (FIGO) stage IB-IC; however, overtreatment remains a concern. Conversely, patients truly at higher risk of recurrence currently lack access to additional therapeutic strategies.
Patients and methods
This study presents a descriptive analysis of early-stage EOC patients who were prospectively sequenced and stratified into high-, intermediate-, and low-risk groups based on clinicopathological features. Oncogenic alterations were identified using OncoKB and classified according to the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) Tier I-III. The prevalence of molecular findings was first reported for each risk subgroup, followed by an analysis on the cohort of patients who experienced relapse.
Results
A total of 180 patients with FIGO stage I-II EOC were enrolled between January 2022 and December 2023; 126 patients (70%) had at least one ESCAT Tier I-III alteration (including 51% high risk, 35% intermediate risk, and 14% low risk); among them, approximately one-quarter (26%, 95% confidence interval 19% to 35%) had an ESCAT Tier I alteration. BRCA1 and BRCA2 alterations were observed in about one-quarter of patients, with BRCA2 often co-altered with POLE mutations (55%, P = 2.1 × 10−4). Notably, almost all BRCA1 variants were found in high-risk patients. BRAF V600E mutation (ESCAT IC) was found in 2.4% of patients. PIK3CA variants were the most common Tier IIIA alterations found in 59% of patients. Among those who experienced recurrence, 60% had at least one ESCAT Tier I-III alteration, with PIK3CA mutations being the most frequent.
Conclusions
These findings highlight the potential for actionable alterations in most early-stage EOC patients and support the exploration of chemotherapy-free regimens for low- to intermediate-risk groups, as well as targeted maintenance therapy for high-risk individuals.
Key words: early-stage ovarian cancer, actionable alterations, ESCAT, target therapy
Highlights
-
•
Seventy percent of patients displayed at least one ESCAT Tier I-III alteration.
-
•
Twenty-eight percent of patients, primarily in the high-risk group, had an ESCAT Tier I alteration.
-
•
PIK3CA variants were the most common Tier IIIA alterations (59%).
-
•
Among patients who recurred, 60% had at least one ESCAT Tier I-III alteration, predominantly involving PIK3CA.
Introduction
The current standard of care for early-stage epithelial ovarian cancer (EOC) includes complete staging surgery with systematic pelvic and lumbo-aortic lymphadenectomy followed by platinum-based chemotherapy for patients at high risk of recurrence (HR).1,2 Adjuvant chemotherapy was reported to increase relapse-free survival (RFS) at 5 years by >10%, from 65% to 76%, suggesting that 100 patients need to be treated to prevent 12 recurrences.3 However, the role of chemotherapy in optimally staged patients remains controversial, as subgroup analyses fail to show a clear benefit in overall survival with adjuvant treatment.4 According to national and international guidelines, the therapeutic algorithm for adjuvant treatment in EOC is primarily guided by tumor grading and histotype. Adjuvant chemotherapy is mandatory for patients at HR which includes those with any stage high-grade serous and endometrioid carcinoma, stage IC2 and IC3 clear-cell carcinoma (CCC), stage II regardless of histotype. In contrast, there is a wide ‘gray area’ of patients at intermediate risk of recurrence (IR) including stage IB-IC low-grade serous and endometrioid carcinoma and stage IA-IC1 CCC where adjuvant chemotherapy is optional. For patients at low risk (LR) of recurrence, including those with FIGO stage IA low-grade serous and endometrioid carcinoma, clinical and radiological follow-up is recommended.1
The current definitions for HR patients are suboptimal as many of those patients do not experience recurrence, while some classified as IR and LR do.5 In addition, individuals truly at higher risk of recurrence lack access to additional therapeutic strategies. Conversely, adding adjuvant chemotherapy in IR patients radically cytoreduced potentially expose them to unnecessary toxicity compromising their quality of life without clear survival benefit. In this context, exploring actionable genomic alterations to inform additional or de-escalated therapeutic strategies could pave the way for future clinical trials. Comprehensive genomic profiling (CGP) holds the promise in addressing these challenges by offering a more precise disease characterization, which could ultimately redefine therapeutic approaches. The increasing adoption of CGP prompted the European Society for Clinical Oncology (ESMO) Translational Research and Precision Medicine Working Group to develop a structured approach for ranking molecular targets, prioritizing them based on the strength of evidence supporting their relevance as clinical targets [ESMO Scale for Clinical Actionability of molecular Targets (ESCAT)].6
In this article, we present a comprehensive genomic, pathological, and clinical characterization of an unselected series of prospectively clinically sequenced early-stage EOC patients from a large referral center with the objective of reporting the prevalence of potentially targetable alterations according to the ESCAT framework.
Patients and methods
Since January 2022, selected cancer patients at the Fondazione Policlinico Universitario Agostino Gemelli IRCCS (FPG) have been offered a tumor-only targeted next-generation sequencing panel as part of a CGP program (ClinicalTrials.gov Identifier: NCT06020625, Protocol ID: FPG500). The study received an institutional review board approval (Protocol U 00194/23, ID number: 3837) and all patients provided informed consent before participation. The program was conducted in accordance with the Declaration of Helsinki. Sequencing data from the EOC cohort were retrieved for the present study. Eligible patients for this analysis were required to be 18 years or older and have newly diagnosed histologically proven early-stage EOC [International Federation of Gynecology and Obstetrics (FIGO) stage I or II]7 with high-quality genomic and clinical data available. Patients with advanced-stage disease (stage III-IV), mucinous histotype, or incomplete clinical or genomic data were excluded.
Data collection
Demographic and clinical data of enrolled patients were collected through a customized electronic case report form (eCRF) by reviewing medical records. The eCRF was developed using RedCap (https://redcap-irccs.policlinicogemelli.it). This web application is fully compliant with EU guidelines for data protection and management (General Data Protection Regulation—GDPR—2016/679).
Histopathologic data
Histopathologic data were obtained from pathology reports generated by the gynecologic pathology unit, which operates with a standardized diagnostic approach. All EOC histologic types and FIGO 2014 stages were included and recorded. Estrogen and progesterone receptor status was classified as positive (>1% positive tumor cells) or negative (<1%). Before sequencing, histology re-review was carried out by one gynecologic pathologist (GZ) to assess tumor cell percentage.
Statistical analysis and sample size
Sample size was determined based on the consecutive recruitment of patients. No formal power calculations were carried out, as the study was designed as a descriptive analysis, including means and standard deviations for continuous variables and frequencies and percentages for categorical variables. RFS was defined as the time from diagnosis to the first radiological evidence of disease recurrence, according to RECIST 1.1 criteria, or death from any cause.8 Median follow-up was calculated using the inverted Kaplan–Meier technique.9 RFS was estimated using the Kaplan–Meier product limit method.10
Statistical analysis was carried out using IBM SPSS statistics for Windows, v.28.0 (IBM Corp, Armonk, NY). Survival plots were generated using R v.4.4.1 (https://www.R-Project.org). No imputation was carried out for missing data.
The evaluation of significant co-occurring or mutually exclusive genes was done by carrying out pairwise Fisher’s exact test. All tests were two-sided and a P value of 0.05 was considered for statistical significance.
Genomic data analysis
Details on the genomic methodologies used in this study have been previously published.11
Briefly, the TruSight Oncology 500 high-throughput panel (TSO500HT, Illumina Inc, San Diego, CA) was applied to analyze both DNA and RNA, covering up to 523 cancer-related genes. This assay detects single-nucleotide variants, insertions/deletions (indels), copy number variations (CNVs), as well as known and unknown fusions and splicing variants in 55 genes. In addition, it assesses genomic ‘signatures’ like microsatellite instability (MSI) and tumor mutational burden (TMB), which reflect the total number of somatic mutations in the sequenced genome.7 Raw sequencing data were processed using the Illumina software TSO500 v2.2 Local App and a custom analysis pipeline (https://github.com/lucianogiaco/lianne, accessed on 09 August 2022). Data were then sent to the Clinical Genomics Workspace software platform by Velsera for variant interpretation and reporting. Sequencing was carried out with a mean depth of >500×, with a minimum coverage of 100× for 90% of regions and 250× for hotspots.
Quality control was carried out using a custom tool (https://github.com/fernandoPalluzzi/VarHound, accessed on 09 August 2022). Variant call format (VCF) files were converted to mutation annotation format (MAF) and annotated using the Ensembl variant effect predictor (VEP) and vcf2maf (https://github.com/mskcc/vcf2maf.git), then further annotated with OncoKB (https://github.com/oncokb/oncokb-annotator.git). Data were filtered for non-synonymous, exonic variants with an allelic frequency (AF) below 0.04% in gnomAD v2 and a variant allele frequency (VAF) ≥5%. Subsequently, only mutations annotated as ‘Oncogenic’ or ‘Likely Oncogenic’ according to OncoKB were considered for the analysis. CNV files were first annotated with OncoKB, filtering out those not classified as ‘Oncogenic’ or ‘Likely Oncogenic’. Copy numbers were then discretized into categories (−2, −1, 0, 1, 2) with amplification defined as ≥5 copies and homozygous deletion.
To determine the clinical utility of the variants identified by the CGP, ESCAT scores were applied to all genetic variants detected. A comprehensive list of all considered variants is available in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2024.104090.
Results
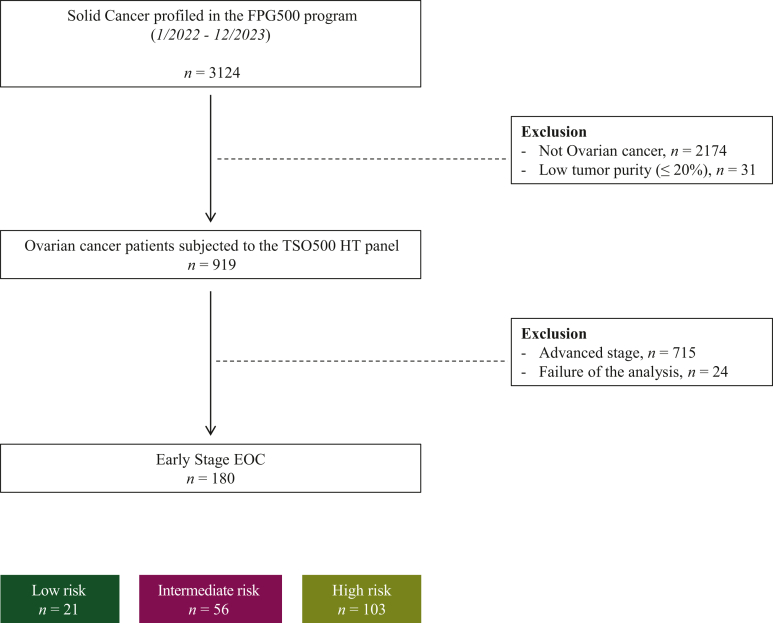
Out of 3125 patients enrolled in the institutional CGP program FPG500 from 1 January 2022 to 31 December 2023, 180 unique, high-quality EOC FIGO stage I-II cases were included in this study (Figure 1). In all cases, the primary tumor specimen was sequenced. Patients were categorized according to their risk of recurrence as previously mentioned (LR, IR, HR).
Figure 1.
STROBE diagram summarizing the early-stage EOC patients included in the study. EOC, epithelial ovarian cancer; FPG, Fondazione Policlinico Gemelli; HT, High-Throughput; TSO, TruSight Oncology 500.
The clinicopathological characteristics are detailed in Table 1. Among the 180 patients included in this study, 98.3% of patients were optimally staged according to international guidelines.10 The largest group of patients was identified as HR (57.2%), followed by IR (31.1%) and LR (11.7%). The overall median age of the population was 55 years (ranging from 47 to 63 years).
Table 1.
Clinical, pathological, and molecular characteristics of the study population (n = 180)
| Characteristics | All cases |
Low risk |
Intermediate risk |
High risk |
|---|---|---|---|---|
| N = 180 | n = 21 | n = 56 | n = 103 | |
| Age at diagnosis, years | 55 (47-63) | 55 (46-64) | 50 (43-56) | 58 (51-64) |
| BMI, kg/m2 | 25 (22-28) | 26 (22-29) | 24 (21-26) | 26 (22-28) |
| Histotype | ||||
| Serous high-grade carcinoma | 50 (27.8) | 0 (0.0) | 0 (0.0) | 50 (48.5) |
| Serous low-grade carcinoma | 5 (2.8) | 2 (9.5) | 3 (5.4) | 0 (0.0) |
| Endometroid low-grade carcinoma | 43 (23.9) | 19 (90.5) | 15 (26.8) | 9 (8.7) |
| Endometroid high-grade carcinoma | 18 (10.0) | 0 (0.0) | 6 (10.7) | 12 (11.7) |
| Clear cells carcinoma | 45 (25.0) | 0 (0.0) | 27 (48.2) | 18 (17.5) |
| Mixed | 12 (6.7) | 0 (0.0) | 3 (5.4) | 9 (8.7) |
| Other | 7 (3.9) | 0 (0.0) | 2 (3.6) | 5 (4.9) |
| Grading | ||||
| 1 | 48/179 (26.8) | 0/21 (0.0) | 25/56 (44.6) | 23/102 (22.5) |
| 2 | 14/179 (7.8) | 8/21 (38.1) | 3/56 (5.4) | 3/102 (2.9) |
| 3 | 41/179 (22.9) | 13/21 (61.9) | 19/56 (33.9) | 9/102 (8.8) |
| Not gradable | 76/179 (42.5) | 0/21 (0.0) | 9/56 (16.1) | 67/102 (65.7) |
| Stage | ||||
| IA | 51 (28.3) | 21 (100.0) | 16 (28.6) | 14 (13.6) |
| IB | 6 (3.3) | 0 (0.0) | 2 (3.6) | 4 (3.9) |
| IC1 | 33 (18.3) | 0 (0.0) | 27 (48.2) | 6 (5.8) |
| IC2 | 23 (12.8) | 0 (0.0) | 10 (17.9) | 13 (12.6) |
| IC3 | 7 (3.9) | 0 (0.0) | 1 (1.8) | 6 (5.8) |
| IIA | 25 (13.9) | 0 (0.0) | 0 (0.0) | 25 (24.3) |
| IIB | 35 (19.4) | 0 (0.0) | 0 (0.0) | 35 (34.0) |
| Residual tumor | ||||
| 0 | 177 (98.3) | 20 (95.2) | 56 (100.0) | 101 (98.1) |
| No staging | 3 (1.7) | 1 (4.8) | 0 (0.0) | 2 (1.9) |
| p53 status at IHC evaluation | ||||
| Wild type | 94 (52.2) | 20 (95.2) | 41 (73.2) | 33 (32.0) |
| Mutated | 60 (33.3) | 0 (0.0) | 4 (7.1) | 56 (54.4) |
| No Information | 26 (14.4) | 1 (4.8) | 11 (19.6) | 14 (13.6) |
| Estrogen receptor | ||||
| Negative | 33 (18.3) | 0 (0.0) | 15 (26.8) | 18 (17.5) |
| Positive | 126 (70) | 20 (95.2) | 31 (55.3) | 75 (72.8) |
| No Information | 21 (11.7) | 1 (4.8) | 10 (17.9) | 10 (9.7) |
| Progesterone receptor | ||||
| Negative | 59 (32.8) | 1 (4.8) | 25 (44.6) | 33 (32.0) |
| Positive | 99 (55) | 19 (90.5) | 21 (37.5) | 59 (57.3) |
| No Information | 22 (12.2) | 1 (4.8) | 10 (17.9) | 11 (10.7) |
| Germline testing for BRCA | 12 (6.7) | 0 (0.0) | 3 (5.4) | 9 (8.7) |
| Wild type | 10/12 (83.3) | — | 3/3 (100.0) | 7/9 (77.8) |
| Mutated | 2/12 (16.7) | — | 0/3 (0.0) | 2/9 (22.2) |
| Chemotherapy | ||||
| No | 57 (31.7) | 21 (100.0) | 29 (51.8) | 7 (6.8) |
| Yes | 121 (67.2) | 0 (0.0) | 26 (46.4) | 95 (92.2) |
| No Information | 2 (1.1) | 0 (0.0) | 1 (1.8) | 1 (1.0) |
| Chemotherapy regimen | ||||
| Carboplatin | 11/121 (9.1) | — | 7/26 (26.9) | 4/95 (4.2) |
| Carboplatin taxol | 109/121 (90.1) | — | 19/26 (73.1) | 90/95 (94.7) |
| Other | 1/121 (0.8) | — | 0/26 (0.0) | 1/95 (1.1) |
Results are given as n/N (%) and as median (interquartile range) as appropriate.
Percentages for categorical variables are reported for patients without missing data.
BMI, body mass index; BRCA, breast cancer gene; IHC, immunohistochemistry.
Regarding histotypes, endometrioid carcinoma was the most common in the LR population (90.5%), while CCC was prevalent in the IR group (48.2%), and serous carcinoma in the HR one (49.5%).12 In the IR group, stage IC1 was the most common (48.2%), while stage IIB was most frequent in the HR group (34%). Estrogen and progesterone receptors (ER/PR) were positive in >90% of LR patients, ∼50% of IR patients, and around 70% of HR patients.
Overall, 67.2% of patients (21% IR and 79% HR) received adjuvant platinum-based chemotherapy. The median follow-up was 6.7 months.
Genomic landscape
Molecular findings from the overall cohort, including oncogenic/likely oncogenic variants, are shown in Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2024.104090. One hundred and seventy-six patients (98%) had at least one oncogenic/likely oncogenic variant. The most frequently altered genes were PIK3CA (n = 74, 41%), TP53 (n = 68, 38%), ARID1A (n = 59, 33%), and CTNNB1 (n = 39, 22%). When stratified by risk class, PIK3CA and ARID1A mutations were more prevalent in the LR (57% and 33%, respectively) and IR (63% and 46%, respectively) cohorts compared with the HR group (26% and 25%, respectively). Conversely, TP53 mutations were found in 57% of HR patients, compared with 11% in the IR group and 14% in the LR group. CTNNB1 mutations showed a decreasing trend from LR (62%) to IR (25%) and HR (12%).
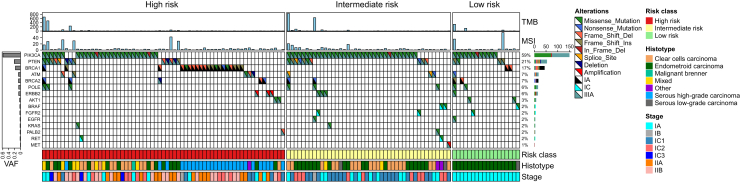
Out of 180 patients, 126 (70%) had at least one alteration classified as ESCAT Tier I-III (Figure 2). Considering risk groups, ESCAT Tier I-III variants were found in 51% of the HR subgroup, 35% of the IR group and 14% of the LR group. A single ESCAT alteration was identified in 93 patients (74%) while the remaining patients had two or more co-occurring alterations.
Figure 2.
Oncoplot of those patients with at least one alteration in a gene classified as level I-III according to ESCAT (n = 126). ESCAT, ESMO Scale for Clinical Actionability of molecular Targets; MSI, microsatellite instability; TMB, tumor mutational burden; VAF, variant allele frequency.
Approximately one-quarter (26%, 95% confidence interval 19% to 35%) of the patients had an ESCAT Tier I alteration. Specifically, 24% displayed BRCA1 and BRCA2 alterations (n = 22 and 9, respectively; Tier IA) and 2%, BRAF V600E mutation (n = 3; Tier IC). Notably, BRCA2 variants were frequently co-altered with POLE mutations (55% of cases, P = 2.1 × 10−4). Nearly all BRCA1 variants, except for two, were identified in HR patients and accounted for the exclusive ESCAT alteration in 86% of these cases.
Eight patients (6%) were identified with POLE mutations (3 HR, 3 IR, and 2 LR). Most of them had stage IA (75%) with an endometrioid histotype (63%); nearly half were high-grade (G3). The median TMB for those patients was 404.8 mut/MB, consistent with an ultra-mutant phenotype. Co-alterations with three or more other ESCAT alterations were observed in all these patients, including mainly PIK3CA (75%), PTEN (63%), and BRCA2 (63%).
PIK3CA variants were the most common alterations within the ESCAT framework, occurring in 59% of cases (79% of IR patients, 67% of LR, and 42% of HR). Most of these were missense variants; eight patients had two concomitant hotspot variants. The most frequent hotspot variants were H1047R (30%), E542K (12%), and E545K (9%). In 48 patients PIK3CA was the unique ESCAT alteration, while in 26 cases (21%) it co-occurred with at least two other actionable variants (Figure 2). The most common co-occurrence was with PTEN (n = 14, 19%), ARID1A (n = 45, 61%), and CTNNB1 (n = 27, 36%). Conversely, BRCA1/PIK3CA (P = 1.7 × 10−8) and BRCA1/PTEN (P = 0.04) as well as TP53 and PIK3CA alterations were mutually exclusive.
Focusing on MSI status, 11 samples (9%) exhibited >20% unstable sites, classifying them as MSI-high.
Patients experiencing recurrence
At follow-up data cut-off (1 July 2024) all patients were alive, and 15 patients (8.3%) experienced disease recurrence (HR 87%, IR 13%). Among this subgroup, seven (47%) had grade 3 serous or endometrioid tumors, six (40%) had a clear-cell histotype, and six (40%) were diagnosed with FIGO stage II.
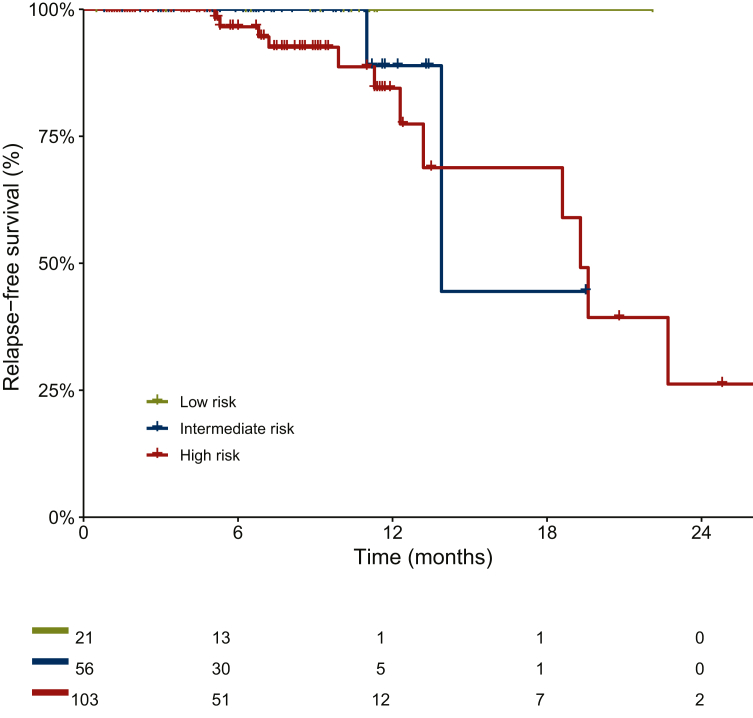
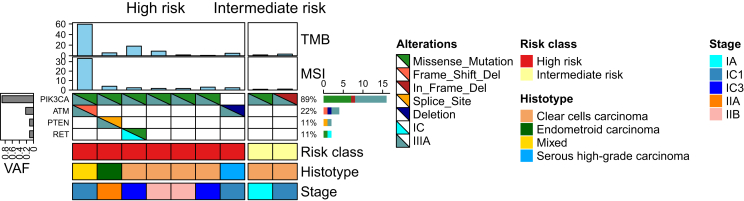
All clinical characteristics are reported in Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2024.104090. The RFS at 6 months was 100% and 96% in the IR and HR groups, respectively. At 12 months, the RFS was 89% in the IR group and 84% in the HR group (Figure 3). The majority of patients (86.7%; 93% HR, 7% IR) were referred for adjuvant chemotherapy. TP53 alterations accounted for 67%, PIK3CA for 53%, and ARID1A for 40% (Supplementary Figure S2, available at https://doi.org/10.1016/j.esmoop.2024.104090). In the two IR patients these alterations were the only ones found. ESCAT Tier I-III alterations were found in 9 out of 15 patients (60%) (Figure 4). PIK3CA was found in eight out of nine patients (89%), co-mutated with ATM, PTEN, and RET, respectively. Two cases exhibited high TMB values (59.5 and 18.2 mut/Mb, respectively) with one case also classified as MSI-high (33%), consistent with negative MSH2 and MSH6 staining observed at immunohistochemistry evaluation.
Figure 3.
Relapse-free survival (RFS) curves of the entire cohort (n = 180).
Figure 4.
Oncoplot of those patients who recurred and had at least one alteration in a gene classified as level I-III according to ESCAT (n = 9). ESCAT, ESMO Scale for Clinical Actionability of molecular Targets; MSI, microsatellite instability; TMB, tumor mutational burden; VAF, variant allele frequency.
The only patient without PIK3CA alteration was classified as HR, diagnosed with HGSOC FIGO stage IC, and had an ATM deletion.
Discussion
This study provides a descriptive analysis of actionable mutations based on ESCAT criteria in a large single-center cohort of prospectively sequenced early-stage EOC patients.
The vast majority (70%) of patients in this early-stage EOC cohort had at least one ESCAT Tier I-III classified variant. Around one-quarter had an ESCAT Tier I alteration (BRCA1, BRCA 2, or BRAFV600E), most of which were found in the HR subgroup. Focusing on ESCAT II-III alterations, PIK3CA represented the most common findings regardless of risk group classification (42% in HR, 79% in IR, 67% in LR). Given that the majority of patients with disease progression were in the HR group and received platinum-based chemotherapy (81%), the potential actionability of PIK3CA mutations may offer additional therapeutic benefits and warrants further investigation. Similarly, PIK3CA alterations were identified in 79% of IR patients, a group where the benefit of chemotherapy is less certain. This highlights a potential starting point for exploring randomized trials focusing on chemo-free treatment strategies in this population. While data remain limited, it is important to note that co-alterations in PIK3CA and PTEN (mutations or deletions) or AKT (particularly E17K) could restore the function of the signaling axis, potentially reducing the effectiveness of PIK3CA-targeted therapies.13,14
The favorable prognosis of LR patients was confirmed in our analysis, since no recurrence events were observed. However, the impact of broad genomic profiling in this subgroup is limited by the short-term follow-up currently available. Definitive data on the prognostic significance of POLE mutations in EOC patients with endometrioid histotype, using molecular subtyping from endometrial cancer, are still missing.15 None the less, it is worth investigating whether patients with an ultramutated phenotype could be safely managed through follow-up alone or benefit from immunotherapy, regardless of histological features such as histotype and grade. From a biological perspective, other actionable variants co-occurring with pathogenic POLE mutations are likely passenger mutations and may not represent valuable therapeutic targets.
Within the HR group, recurrence occurred in 22% of clear-cell EOC, 17% of high-grade endometrioid, and 10% of high-grade serous cases. Both IR patients who experienced recurrence had a clear-cell histotype. All clear-cell cases that recurred had undergone adjuvant chemotherapy, consistent with existing evidence of the limited benefit of chemotherapy in this subgroup. Nevertheless, all of them displayed an actionable mutation according to ESCAT. The only two patients who recurred without adjuvant chemotherapy lacked ESCAT actionable alterations. Overall, these data are intended to generate hypotheses for agnostic drug administration, focusing on improving quality of life through chemotherapy de-escalation in LR/IR patients, while enhancing outcomes in HR patients through targeted maintenance therapies.
Pharmaceutical targeting of the potential, yet sometimes elusive, molecular Achilles’ heel of solid tumors through genomic profiling has already shown promise in advanced or recurrent settings.16 Large prospective multicenter randomized trials are awaited to establish whether tumor-agnostic targeting can produce a paradigm shift in oncology.17, 18, 19
At the same time, as broad genomic profiling becomes more widespread, the need for standardized methods in tumor-agnostic drug development has become more urgent. To address this, the ESMO Precision Working Group has recently introduced a practical framework for evaluating and validating the tumor-agnostic potential of molecularly targeted therapies.20 Moreover, the adoption of targeted agents in the context of early-stage disease is increasing. Osimertinib and alectinib are currently the only targeted therapies that have shown meaningful clinical benefits in early-stage non-small-cell lung cancer (NSCLC). They have been recently approved (2020/2021 and 2024, respectively) for adjuvant treatment in specific patient populations: osimertinib for stage IB-IIIA EGFR-mutated NSCLC (including EGFR exon 19 deletions or exon 21 L858R mutations) and alectinib for stage IB-IIIA ALK-positive NSCLC.21,22
Unlike NSCLC, EOC is known to have a low mutational load, high CNVs, and a high degree of genomic instability.23 While limited genomic data are available on early-stage disease, large datasets primarily focused on advanced-stage patients revealed that the most frequently mutated genes in high-grade serous histology included TP53 (96%) and BRCA1/2 (20%).24 In low-grade serous histology, common mutations included KRAS (33%), NRAS (11%), EIF1AX (10%), and BRAF (11%).25 For endometrioid histology, the prevalent mutations were CTNNB1 (43%), PIK3CA (43%), ARID1A (36%), PTEN (29%), KRAS (26%), TP53 (26%), and SOX8 (19%).26 Very few data are available on clear-cell EOC patients, showing pathogenic variants most frequently in ARID1A (54.3%), KRAS (19.8%), and TP53 (13.6%).27
Recent results from a multi-gene panel study of 168 EOC samples using the ESCAT framework were published. The study found that nearly all patients had ESCAT Tier I-II variants, including 17% with BRCA1/2 alterations, 9% with PIK3CA mutations, and 7% with KRAS mutations.28 Although direct comparisons are challenging due to differences in study design (such as the absence of FIGO stage data and the inclusion of TP53 alterations in ESCAT Tier II), the authors suggest that other ESCAT Tier II variants may offer potential new treatment options.
The main limitations of the present study are the relatively small sample size, single-center experience, and the short follow-up period. Although time is mature to explore a paradigm shift in treatment for clear-cell EOC patients, therapeutic changes for high-grade serous and endometrioid cases require better patient selection. Current risk subgroups do not yet allow for safe and effective treatment de-escalation/escalation. The overall favorable prognosis of these patients requires an accurate patient selection to avoid medical and financial toxicities. Improving the accuracy of prognostic biomarkers is crucial before fully leveraging the actionability of genomic data.
Conclusion
Our findings support the feasibility of targeting specific molecular vulnerabilities in a large cohort of early-stage EOC patients. Given the presence of multiple molecular alterations in some patients, the role of some variants in disease progression remains unclear, and resistance to targeted therapies may develop over time. However, EOC patients, especially those with rare histotypes, could benefit from this approach. This strategy has the potential not only to improve outcomes but also to spare selected patients from the adverse effects of unnecessary chemotherapy.
Acknowledgments
Funding
None declared.
Disclosure
CN declares travel support from MSD, Illumina, Menarini, and AZ; and honoraria from Veeva, GSK, MSD, AZ, Altems, Illumina, and Guardant Health. CM: consultant/advisory for Clovis, PharmaMar, GSK, AstraZeneca, and MSD; and received travel accommodations from PharmaMar and Roche. AF engages in relevant financial activities outside the submitted work that involves AstraZeneca/MSD, Johnson & Johnson, Fondazione Internazionale Menarini, and GlaxoSmithKline. GS reports research support from MSD and honoraria from Clovis Oncology, consultant for Tesaro and Johnson & Johnson. All other authors have declared no conflicts of interest.
Supplementary data
References
- 1.Colombo N., Sessa C., Bois A.D., et al. ESMO-ESGO consensus conference recommendations on ovarian cancer: pathology and molecular biology, early and advanced stages, borderline tumours and recurrent disease. Int J Gynecol Cancer. 2019 doi: 10.1136/ijgc-2019-000308. [DOI] [PubMed] [Google Scholar]
- 2.Ledermann J.A., Matias-Guiu X., Amant F., et al. ESGO-ESMO-ESP consensus conference recommendations on ovarian cancer: pathology and molecular biology and early, advanced and recurrent disease. Ann Oncol. 2024;35(3):248–266. doi: 10.1016/j.annonc.2023.11.015. [DOI] [PubMed] [Google Scholar]
- 3.Trimbos J.B., Parmar M., Vergote I., et al. International Collaborative Ovarian Neoplasm trial 1 and Adjuvant ChemoTherapy In Ovarian Neoplasm trial: two parallel randomized phase III trials of adjuvant chemotherapy in patients with early-stage ovarian carcinoma. J Natl Cancer Inst. 2003;95(2):105–112. [PubMed] [Google Scholar]
- 4.Kleppe M., van der Aa M.A., Van Gorp T., Slangen B.F., Kruitwagen R.F. The impact of lymph node dissection and adjuvant chemotherapy on survival: a nationwide cohort study of patients with clinical early-stage ovarian cancer. Eur J Cancer. 2016;66:83–90. doi: 10.1016/j.ejca.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Collinson F., Qian W., Fossati R., et al. Optimal treatment of early-stage ovarian cancer. Ann Oncol. 2014;25(6):1165–1171. doi: 10.1093/annonc/mdu116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mosele M.F., Westphalen C.B., Stenzinger A., et al. Recommendations for the use of next-generation sequencing (NGS) for patients with advanced cancer in 2024: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2024;35(7):588–606. doi: 10.1016/j.annonc.2024.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Mutch D.G., Prat J. 2014 FIGO staging for ovarian, fallopian tube and peritoneal cancer. Gynecol Oncol. 2014;133(3):401–404. doi: 10.1016/j.ygyno.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Eisenhauer E.A., Therasse P., Bogaerts J., et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 9.Schemper M., Smith T.L. A note on quantifying follow-up in studies of failure time. Control Clin Trials. 1996;17(4):343–346. doi: 10.1016/0197-2456(96)00075-x. [DOI] [PubMed] [Google Scholar]
- 10.Kaplan E.L., Meier P. Nonparametric estimation from incomplete observation. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 11.Giacò L., Palluzzi F., Guido D., et al. A computational framework for comprehensive genomic profiling in solid cancers: the analytical performance of a high-throughput assay for small and copy number variants. Cancers (Basel) 2022;14(24):6152. doi: 10.3390/cancers14246152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheffield B.S., Kos Z., Asleh-Aburaya K., et al. Molecular subtype profiling of invasive breast cancers weakly positive for estrogen receptor. Breast Cancer Res Treat. 2016;155(3):483–490. doi: 10.1007/s10549-016-3689-z. [DOI] [PubMed] [Google Scholar]
- 13.Clark A.S., Makhlin I., DeMichele A. Setting the pick: can PI3K inhibitors circumvent CDK4/6 inhibitor resistance? Clin Cancer Res. 2021;27(2):371–373. doi: 10.1158/1078-0432.CCR-20-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Razavi P., Dickler M.N., Shah P.D., et al. Alterations in PTEN and ESR1 promote clinical resistance to alpelisib plus aromatase inhibitors. Nat Cancer. 2020;1(4):382–393. doi: 10.1038/s43018-020-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cybulska P., Paula A.D.C., Tseng J., et al. Molecular profiling and molecular classification of endometrioid ovarian carcinomas. Gynecol Oncol. 2019;154(3):516–523. doi: 10.1016/j.ygyno.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krassilnikova S., Craig E.T., Craig T.J. Summary of the international multicenter prospective angioedema C1-inhibitor trials 1 and 2 (IMPACT1 and 2) Expert Rev Clin Immunol. 2010;6(3):327–334. doi: 10.1586/eci.10.19. [DOI] [PubMed] [Google Scholar]
- 17.Tsimberidou A.M., Hong D.S., Fu S., et al. Precision medicine: preliminary results from the Initiative for Molecular Profiling and Advanced Cancer Therapy 2 (IMPACT2) study. NPJ Precis Oncol. 2021;5(1):21. doi: 10.1038/s41698-021-00159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ClinicalTrials.gov A Study of Targeted Therapies or Immunotherapy in Participants With Advanced Solid Tumors (TAPISTRY). ClinicalTrials.gov identifier NCT04589845. Updated September 4, 2023. https://clinicaltrials.gov/study/NCT04589845?cond=tapistry&rank=1 Available at.
- 19.ClinicalTrials.gov The Rome Trial From Histology to Target: the Road to Personalize Target Therapy and Immunotherapy (ROME). ClinicalTrials.gov identifier NCT04591431. Updated September 4, 2023. https://www.clinicaltrials.gov/study/NCT04591431 Available at.
- 20.Kuzbari Z., Bandlamudi C., Loveday C., et al. Germline-focused analysis of tumour-detected variants in 49,264 cancer patients: ESMO Precision Medicine Working Group recommendations. Ann Oncol. 2023;34(3):215–227. doi: 10.1016/j.annonc.2022.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y.L., Tsuboi M., He J., et al. Osimertinib in resected EGFR-mutated non-small-cell lung cancer. N Engl J Med. 2020;383(18):1711–1723. doi: 10.1056/NEJMoa2027071. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y.L., Dziadziuszko R., Ahn J.S., et al. Alectinib in resected ALK-positive non-small-cell lung cancer. N Engl J Med. 2024;390(14):1265–1276. doi: 10.1056/NEJMoa2310532. [DOI] [PubMed] [Google Scholar]
- 23.Ciriello G., Miller M.L., Aksoy B.A., Senbabaoglu Y., Schultz N., Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45(10):1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning-Geist B., Gordhandas S., Liu Y.L., et al. MAPK pathway genetic alterations are associated with prolonged overall survival in low-grade serous ovarian carcinoma. Clin Cancer Res. 2022;28(20):4456–4465. doi: 10.1158/1078-0432.CCR-21-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollis R.L., Thomson J.P., Stanley B., et al. Molecular stratification of endometrioid ovarian carcinoma predicts clinical outcome. Nat Commun. 2020;11(1):4995. doi: 10.1038/s41467-020-18819-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanc-Durand F., Ngoi N., Rouleau E., et al. Unveiling the unique identity of clear-cell endometrial cancer (CCEC): a comprehensive comparative analysis with ovarian counterpart and other endometrial subtypes. J Clin Oncol. 2024;42(suppl 16) 5521-5521. [Google Scholar]
- 28.Fieuws C., Van Der Meulen J., Proesmans K., et al. Identification of potentially actionable genetic variants in epithelial ovarian cancer: a retrospective cohort study. NPJ Precis Oncol. 2024;8(1):71. doi: 10.1038/s41698-024-00565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.