Figure 1.
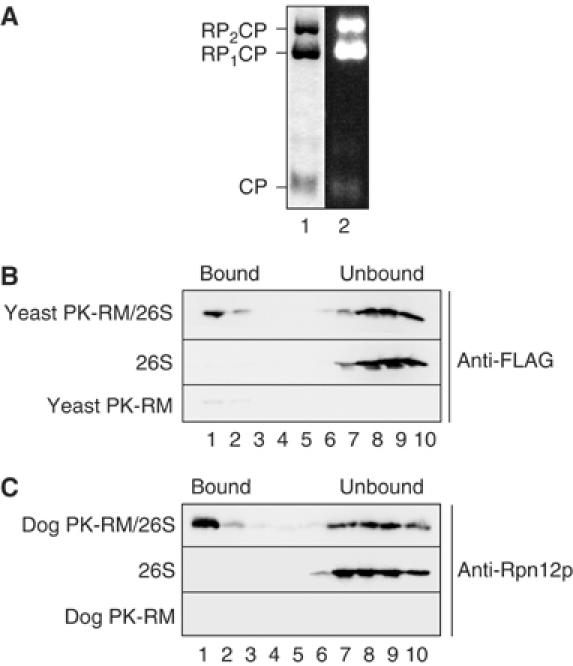
Proteasomes bind to a site conserved in yeast and mammalian ER membranes. (A) Purified yeast 26S proteasomes were separated by native gel electrophoresis and stained with Coomassie blue (lane 1), or the gel incubated with the fluorogenic proteasome substrate Suc-LLVY-AMC, and active proteasomes visualized on a UV transilluminator. CP, 20S core particle; RP1CP, 20S CP with one bound regulatory particle; RP2CP, 20S CP with two bound regulatory particles. (B) Yeast 26S proteasomes (2.5 pmol) were incubated with 10 eq of puromycin/high-salt-treated yeast microsomes (PK-RM) as described in Materials and methods. 1 eq equals 1 μl of microsomes of A280=50. The membranes were floated through 1.8 M sucrose cushions and the gradients fractionated from the top. Proteasomes in individual fractions were detected by SDS–PAGE and immunoblotting employing anti-FLAG antibody. Enhanced chemiluminescence and a CCD camera system (Raytest, Germany) were used as a detection system. The positions of proteasomes bound to membranes and unbound proteasomes are indicated. (C) Yeast 26S proteasomes (2.5 pmol) were bound to puromycin/high-salt-treated dog pancreas microsomes (10 eq). After binding, the samples were treated as in (B) and analyzed by SDS–PAGE and immunoblotting using anti-Rpn12p antibodies.
