Summary
Background
Liver involvement is a common complication of coronavirus disease 2019 (COVID-19), especially in hospitalized patients. However, the underlying mechanisms involved are not fully understood.
Methods
Immunohistochemistry (IHC) staining of SARS-CoV-2 spike (S) and nucleocapsid (N) proteins was conducted on liver tissues from six patients with COVID-19. The 10x Genomics Visium CytAssist Spatial Gene Assay was designed to analyze liver transcriptomics. TCR CDR3 sequences were analyzed in DNA from liver tissues. Liver function indicators were retrospectively studied in 650 hospitalized patients with COVID-19.
Findings
SARS-CoV-2 proteins were initially detected in the livers of naturally infected golden (Syrian) hamsters, prompting us to investigate the situation in clinical cases. Thus, we collected liver tissues from patients with abnormal liver biochemical values. Viral S and N proteins were detected in the livers of severe and deceased patients but not in those of moderate patients. We further demonstrated that hepatocytes and erythroid cells in hepatic sinusoids are major cells targeted by SARS-CoV-2. Immune cells, especially T cells, were enriched in surviving severe patients, characterized by enhanced CDR3α clonality and novel CDR3β recombination of the T-cell receptor. In contrast, hepatocyte apoptosis was triggered, and the transcription of albumin (ALB) was obviously impaired in the deceased patients. We then performed a retrospective study including patients with COVID-19. Serum aspartate aminotransferase (AST) and ALB levels at baseline significantly differed in the deceased cohort. However, AST regression did not decrease the risk of death. ALB recovery indicated clinical improvement, and declining or low serum ALB concentrations were associated with death.
Interpretation
This study provides clinical evidence for liver infection with SARS-CoV-2, insight into the impact of SARS-CoV-2 on the liver, and a potential way to evaluate the risk of death via assessing serum ALB concentration fluctuations in patients with COVID-19.
Funding
National Key R&D Program of China (2021YFC2300602), National Natural Science Foundation of China (92369110), National Natural Science Foundation of China (U23A20474), Shanghai Municipal Science and Technology Major Project (ZD2021CY001), Shanghai Jinshan District Medical and Health Technology Innovation Fund Project (2023-WS-31).
Keywords: SARS-COV-2, COVID-19, Liver, Spatial transcriptome, ALB
Research in context.
Evidence before this study
The pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused about seven million deaths worldwide. Infection of this virus can result mild, moderate or severe respiratory symptoms, known as coronavirus disease 2019 (COVID-19). Nevertheless, more evidences showed that SARS-COV-2 exposure results non-respiratory manifestations, including neurological disorders, kidney damage and gastrointestinal disease. The most described mechanism for this multi-organ dysfunction is the broad expression of angiotensin-converting enzyme 2 (ACE2), the main receptor for SARS-CoV-2 entry through interacting with receptor binding domain (RBD) on viral spike protein (S). Liver involvement is a nonnegligible clinical complication of COVID-19. Several studies described liver injury, manifested as increased alanine aminotransferase (ALT) or aspartate aminotransferase (AST). Since viral RNA is detectable in 69% of autopsy liver tissue, SARS-CoV-2 has been regarding to have hepatic tropism via ACE2. However, it is still debated whether SARS-CoV-2 can enter into hepatocytes, as hepatocytes expressing minimal ACE2 in liver, and only 2.5%–76.3% cases have enhanced ALT and AST as reports. Interestingly, liver dysfunction in some patients is believed to be caused by drug treatment. Detectable viral RNA might due to the blood circulation in livers. Recently, SARS-CoV-2 pseudovirus has been reported to infect primary hepatic parenchymal cells via ASGR1, rather than ACE2, but the situation in patients with COVID-19 remains unknown. These studies indicated that the liver tropism of SARS-CoV-2 is complicated. In addition, the impact of viruses on the liver still lacks in-depth study, especially in clinical patients.
Added value of this study
In this study, we stained liver tissues from COVID-19 cases with abnormal liver functions, and found viral proteins were detectable from deceased or severe cases, but not from moderate patients. We then combined the immunohistochemistry (IHC) staining with spatial transcriptomics analysis, and SARS-CoV-2 was found to mainly locate in clusters characterized as hepatocytes and erythroid cells. Additionally, ASGR1, but not ACE2, was high expressed in livers from both deceased and severe cases. Immune cells, especially T cells, were enriched in the survived severe cases, characterized by an enhanced CDR3α clonality and a novel CDR3β recombination of T cell receptor. In contrast, apoptosis of hepatocytes was triggered, and transcription of albumin (ALB) was obviously impaired in deceased cases. Using clinical cohorts, we confirmed that declined or persistent low level of serum ALB, rather than elevated aminotransferases, is closely related with outcomes of patients.
Implications of all the available evidence
This study showed clinical evidence for liver infection of SARS-CoV-2, suggested viral-induced apoptosis of hepatocytes leading decrease of ALB synthesis is associated with death, and gave a potential way to evaluate death risk via fluctuation of serum ALB in patients with COVID-19.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has caused approximately seven million deaths worldwide. Infection with this virus can result in mild, moderate, or severe respiratory symptoms, known as coronavirus disease 2019 (COVID-19). Increasing evidence has shown that SARS-CoV-2 exposure results in nonrespiratory manifestations, including neurological disorders, kidney damage, and gastrointestinal disease.1,2 The most commonly described mechanism for this multiorgan dysfunction is the broad expression of angiotensin-converting enzyme 2 (ACE2),3 the main receptor for SARS-CoV-2 entry.
Liver involvement, suggested by abnormal liver function, is common in hospitalized patients with COVID-19, and is a nonnegligible clinical complication of COVID-19. Several studies have described liver injury, manifesting as increased alanine aminotransferase (ALT) or aspartate aminotransferase (AST) levels.4 SARS-CoV-2 is known to have hepatic tropism via ACE2, with viral RNA detectable in 69% of autopsied liver tissue samples.5,6 More and more evidence have showed that SARS-CoV-2 can enter hepatocytes,5,7,8 although hepatocytes express minimal ACE2 in the liver,9 and only 2.5–76.3% of patients have increased ALT and AST levels.10,11 Interestingly, liver dysfunction in some patients is believed to be caused by drug treatment.12,13 The detection of viral RNA might be due to the blood circulation in the liver.14,15 The aforementioned studies indicate that the liver tropism of SARS-CoV-2 is complicated. In addition, the impact of viruses on the liver still lacks in-depth study, especially in clinical patients.
In this study, we stained liver tissues from patients infected by SARS-CoV-2 with abnormal liver function. We found that viral proteins were detectable in deceased patients or those with severe disease but not in those with moderate disease. We then combined immunohistochemistry (IHC) staining with spatial transcriptomics analysis and found that SARS-CoV-2 was located mainly in clusters characterized as hepatocytes and erythroid cells. Additionally, enhanced T-cell gene expression was found only in virus-positive clusters from the surviving patient with severe disease. Decreased albumin (ALB) transcription was observed in virus-positive clusters from the deceased patient. Using clinical cohorts, we confirmed that decreased or persistently low levels of serum ALB are closely related to outcomes of patients with COVID-19, and fluctuations in ALB could be used to evaluate the risk of death.
Methods
Animals
Golden (LVG) Syrian hamsters were purchased from Charles River Laboratories. Studies were carried out strictly according to the recommendations of the Guide for the Care and Use of Laboratory Animals. SARS-CoV-2 challenge experiments were conducted in the Animal Biosafety Level 3 (ABSL-3 in Shantou University) facility. Briefly, hamsters were inoculated with 1 × 104 PFU of SARS-CoV-2 virus in 100 μl of PBS through the intranasal route under anesthesia, and were sacrificed on day 5 post infection. Livers were fixed in 4% PFA perfusion, embedded with paraffin, and cut into 4 μm sections. The hamster studies were conducted under the approval of the institutional Animal Care and Use Committee of Xiaman University, and approved by the Medical Ethics Committee with number: SUCM2021-112.
Human subjects
Liver tissues were separated from deceased individuals with COVID-19, fixed with formalin and embedded in paraffin within 48 h after death in Shanghai Public Health Clinical Center in the latter half year of 2022. Formalin-fixed, paraffin-embedded (FFPE) liver biopsy sections from respiratory SARS-CoV-2 RNA positive cases with abnormal liver functions were collected in Huashan Hospital in the year of 2023. Patients are infected by SARS-CoV-2 strain sequenced as Omicron BA.5 or BF.7. Their clinical indicators were shown in Supplementary Table S1. Cohorts used for retrospective study of liver function indicators were randomly gathered from the hospitalized COVID-19 cases in December 2022 and January 2023 from Huashan Hospital. Studied were viewed and proved by The Ethics Committee of Huashan Hospital (Fudan University) with number: KY-2022-721. Informed consent from all participants was obtained.
Immunohistochemistry and immunofluorescence staining
Mouse anti-SARS-CoV-2 N protein (Clone: 15A7) and S protein (Clone: 39C2) antibodies developed by in Xiamen University were used in immunohistochemistry staining.16 Briefly, primary antibodies were diluted to 2 μg/ml, and added on liver sections incubating overnight on 4 °C after blocking. Goat anti-Mouse IgG H&L (HRP) pre-adsorbed (Abcam, Cat#: ab97040) was used as the second antibody. 3,3′-diaminobenzidine was used as the substrate, and hematoxylin was used to stain cell nuclear. Sections were observed via Olympus BX63 microscope, images were generated via DP22 digital camera. In immunofluorescence (IF) staining, antibodies targeting CD3 (Abcam Cat#: ab16669, RRDI: AB_443425) and Cleaved caspase-3 (CST Cat#: 9664, RRID: AB_2070042), and Alexa Fluor Tyramide SuperBoost kits from Thermo Fisher Scientific were used according to the manufacturer's instructions. IF was performed with the technical support from Yuanxibio (Shanghai, China). For whole scanned IHC and IF image, prior H101F ProScan® Flat Top Stage combining with microscope and camera were used.
Spatial transcription analysis
10x Genomics Visium CytAssist Spatial Gene Assay was designed to analyze viral RNA and mRNA isolated from FFPE liver tissue sections from human subjects, and was conducted by Shanghai Xuran Biotechnology. To detect SARS-CoV-2 RNA, three pairs of probes targeting viral nucleocapsid domain were designed as shown in Supplementary Table S2. Spatial Barcodes are used to associate the reads back to the tissue section images for spatial mapping of gene expression. Figure images were generated via Loupe Browser 6.5.0 downloaded from the website of 10x Genomics Inc. The raw data is available in NCBI GEO database (GSE279405).
TCR CDR3 sequence
DNA samples were isolated from FFPE liver tissues, and analyzed by HTS of T cell receptor (TCR) chains using the ImmuHub® TCR profiling system at a deep level (ImmuQuad Biotech, Hangzhou, China). Sequencing was performed on an Illumina NovaSeq® system with PE150 mode. The raw sequencing data were then aligning with NCBI VDJ database with IgBLAST and PCR amplification and sequencing errors correction based on clone frequency. The resulting sequences of TCRα and TCRβ CDR3 were determined. We further defined amounts of each TCR clonotype by adding numbers of TCR clones sharing the same sequence of CDR3.
Statistical analysis
Data were analyzed using GraphPad Prism 10.0.2. Statistical significance was assessed by Mann–Whitney test or unpaired/paired two-tailed t tests. Error bars in figures presented the median with interquartile range. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Role of funders
The funders did not have any role in study design, data collection, data analysis, data interpretation, or writing of report.
Results
Detecting viral proteins in SARS-CoV-2-infected livers
Golden (Syrian) hamsters constitute a natural SARS-CoV-2 infection model17,18 that is broadly used in the study of pathogenic mechanisms and drug development for COVID-19.19,20 Previously, we used this model to evaluate the protective effects of antibodies and vaccines.16,21 After sacrificing the hamsters, we surprisingly detected the SARS-CoV-2 S and N proteins in their livers (Supplementary Figure S1a, b), prompting us to investigate the situation in clinical cases.
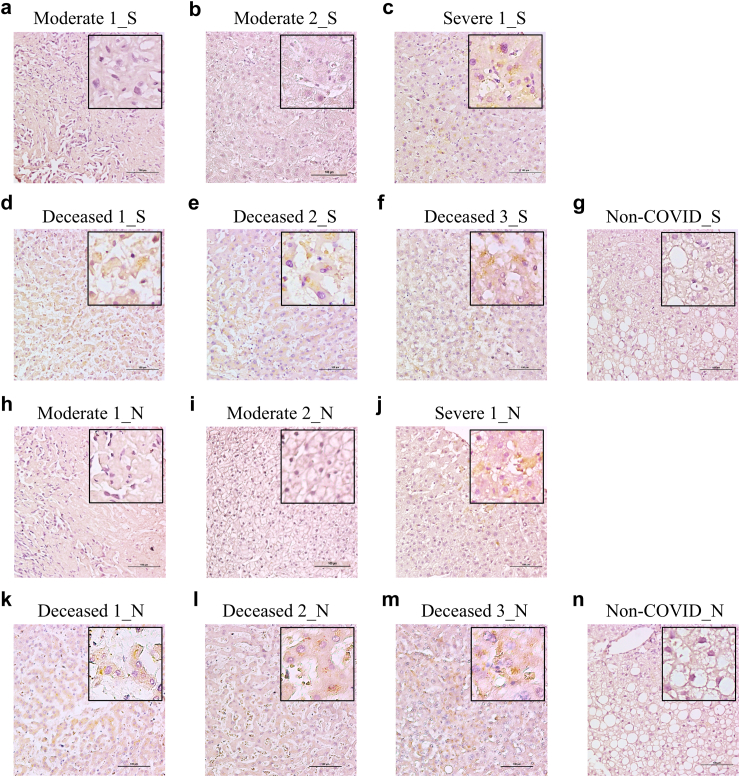
Thus, we obtained three autopsied liver tissue samples from deceased patients and three liver biopsy samples from patients with increased liver aminotransferases who ultimately recovered from COVID-19. Among the resolved patients, one remained positive for respiratory viral RNA for 31 days (severe), whereas the other two became negative within two weeks (moderate) (Supplementary Table S1). We stained the samples with anti-viral S or N protein antibodies. Interestingly, the S and N proteins were detected in the hepatocytes of patients with severe disease (Fig. 1c–j) and all autopsied liver tissues (Fig. 1d–f and k–n), whereas they were not detected in the livers of those with moderate disease or in the hepatitis B control tissue (Fig. 1a, b, g, h, i and n). These findings indicate that viral attack in the liver is related to COVID-19 severity.
Fig. 1.
Detection of viral spike (S) and nucleocapsid (N) proteins in liver tissues from patients. Immunohistochemistry (IHC) staining SARS-COV-2 S protein in livers from two moderate (a, b), a severe (c) and three deceased (d, e and f) patients with COVID-19 or in liver biopsy tissue from a non-COVID patient with hepatitis B (g). IHC staining SARS-COV-2 N protein in livers from two moderate (h, i), a severe (j) and three deceased (k, l and m) patients with COVID-19 or in liver biopsy tissue from a non-COVID patient with hepatitis B (n). Viral S and N proteins were visualized in yellow/brown.
Spatial landscape of SARS-CoV-2 in livers from patients with COVID-19
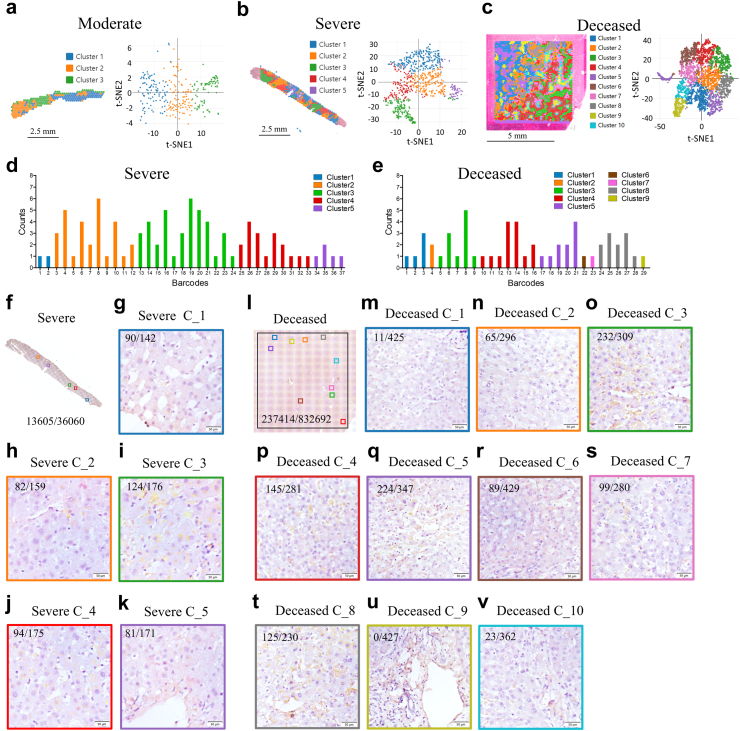
Notably, the levels of serum ALT, AST, and gamma-glutamyl transferase (GGT) were not elevated in the deceased patients examined in the last test before death (Supplementary Table S1). Therefore, to reveal the impacts of SARS-CoV-2 on the liver, we analyzed the spatial transcriptome in liver tissues from one moderate, one severe, and one deceased patient. Three pairs of probes targeting the viral nucleocapsid domain were designed (Supplementary Table S2). A total of 4981 spots were generated with a mean of 104,110 reads and a median of 5.720 genes or 30,899 transcripts unique molecular identifiers (UMIs) per spot in the deceased case; 751 spots were generated with a mean of 296,715 reads and a median of 5705 genes or 28,814 transcripts UMIs per spot in the severe case; and 271 spots were generated with a mean of 491,854 reads and a median of 3082 genes or 39,928 transcripts UMIs per spot in the moderate case (Supplementary Figure S2a, b and c). The graph-based algorithm partitioned the spots into three clusters in the moderately resolved case (Fig. 2a), five clusters in the severe case (Fig. 2b), or ten clusters in the deceased case (Fig. 2c), as shown in the section or in the t-stochastic neighbor embedding (t-SNE) visualization. Moreover, the sequencing saturation in all three tissues exceeded 60% (Supplementary Figure S2d, e and f), which means that most of converted probe in the kit from 10x Genomics ligation products have been sequenced.
Fig. 2.
Analyzing spatial feature of SARS-COV-2 RNA and proteins in livers from patients. Visium cell type classification showing distribution of clusters in liver tissue from moderate (a), severe (b) and deceased (c) subjects with COVID-19. (d) t-stochastic neighbor embedding (t-SNE) visualization of these clusters. (e) Distribution of SARS-COV-2 RNA in different cell clusters. (f) Whole scanned IHC image of S protein in a serial section from the transcript analyzed liver section of severe case (g–k) Magnified IHC views of S proteins in the corresponding areas of clusters from the severe patient. (l) Whole scanned IHC image of S protein in a serial section from the transcript analyzed liver section of the deceased patient (m–v) Magnified IHC views of S proteins in the corresponding areas of clusters from deceased patient. Viral S and N proteins were visualized in yellow/brown. Viral protein positive dots were counted using QuPath-0.5.1. The numbers depict viral positive dots/nucleuses (f–v).
Similar to the IHC results, SARS-CoV-2 RNA was detected only in those with severe disease and deceased patients but not in those with moderate disease (Fig. 2d and e). In severe cases, viral RNA was detected in all clusters and was higher in Clusters 2, 3, and 4 (Fig. 2d). In the deceased patients, viral RNA was detected in Clusters 1 to 9 but not in Cluster 10 (Fig. 2e), and viral RNA was high in Clusters 3, 4, 5, and 8 (Fig. 2e). Next, whole-scanned IHC images of S protein-stained samples from serial sections of the analyzed transcripts were obtained (Fig. 2f–l). The corresponding areas to the cluster were selected and zoomed. Consistently, in the severe case, Clusters 2, 3, and 4 (Fig. 2h–j), but not Clusters 1 and 5-matched areas (Fig. 2g–k), contained more SARS-CoV-2 proteins. In the deceased case, the viral protein expression was strong in Clusters 3, 4, 5, and 8 (Fig. 2o–q and t) but weak or not detected in Clusters 1, 2, 6, 7, 9, and 10 (Fig. 2m, n, r, s, u and v). These findings indicate that SARS-CoV-2 can infect particular areas of the liver.
ACE2 was almost undetectable in our liver sections (Supplementary Figure S2g, h). However, a hepatoma-derived line has been reported to support entry of SARS-CoV-2 via ACE2.22 Recently, one study revealed that ASGR1 is the receptor that mediates SARS-CoV-2 pseudovirus infection of primary hepatic parenchymal cells.23 Consistently, ASGR1 was most highly expressed then other reported entry factors including ACE2, KREMEN1, AXL, ATSL and TMPRSS2, in all clusters in the liver samples of deceased patients in our study (Supplementary Figure S2i). Nevertheless, level of ASGR1 was similar among the moderate, severe and deceased patients (Supplementary Figure S2i, j and k).
Intrahepatic immune characteristics in SARS-CoV-2-infected livers
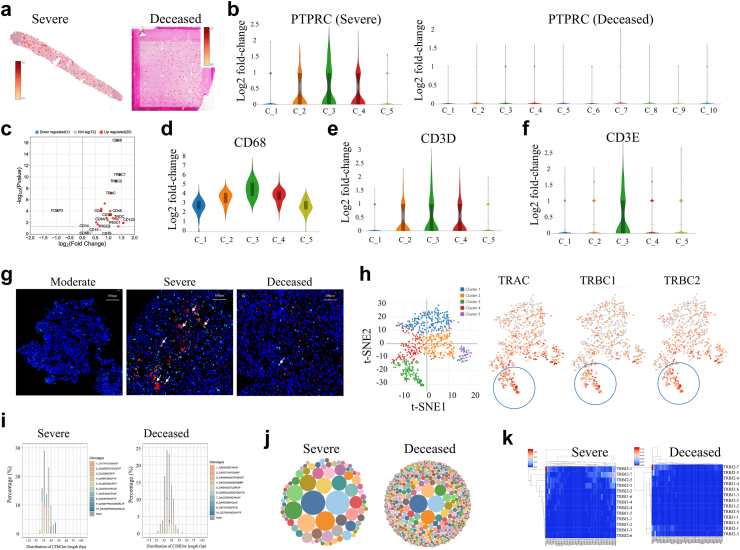
To investigate the immune features of SARS-CoV-2-infected livers, we checked the level of PTPRC, which encodes CD45, a panmarker of immune cells. Approximately 5% of the spots were PTPRC positive in the deceased patient (248 out of 4981), whereas approximately 25.3% of the spots had PTPRC expression in the patient with severe disease (190 out of 751) (Fig. 3a). PTPRC expression was not enriched in any cluster in the deceased patient but was enriched in Clusters 2, 3, and 4 in the surviving patient with severe disease (Fig. 3b). Interestingly, these clusters were enriched in viral RNA (Fig. 2d), indicating that immune cells participate in liver viral control.
Fig. 3.
Spatial immune features from intrahepatic SARS-COV-2 positive livers from patients. (a) Distributions of PTPRC/CD45 transcription level in liver sections from the deceased and severe patients. (b) Transcription level of PTPRC in clusters. (c) Levels of immune marker genes in Cluster 2, 3 and 4 from liver tissue of the severe. Transcription level of CD68 (d), CD3D (e) and CD3E (f) in clusters. The Y-axis is an estimated change of the log2 ratio of expression in a cluster to that in all other spots. (g) Immunofluorescence staining CD3 (Green) and viral N protein (Red) in liver tissue from moderate, severe and deceased patients. White arrows show CD3 positive cells adjacent to viral protein. (h) t-SNE visualization of TRAC, TRBC1 and TRBC2 gene levels in clusters. (i) Distribution of TCRα CDR3 (CDR3α) sequences in different length from deceased and severe cases (j) Bubble graph showing clonality and diversity of CDR3α in livers. (k) Recombination of CDR3β V and J genes in livers.
Furthermore, marker genes for macrophages, granulocytes, B cells, NK cells, and T cells were analyzed. In Cluster 3, which contained the most abundant viral proteins (Fig. 2i), CD68, TRAC, TRBC1, TRBC2, CD3D, CD3E, CD4, and CD8A expression levels were significantly increased (Fig. 3c). CD14 and CD15 expression levels were higher in Clusters 2, 3, and 4 (Supplementary Figure S3a, b), but CD19 and CD56 were almost undetectable in all clusters (Supplementary Figure S3c, d). Cluster analysis also confirmed that CD68 and CD3D were expressed mainly in Clusters 2, 3, and 4, which were positive for viral proteins (Fig. 3d and e). Strikingly, CD3E was increased only in Cluster 3 (Fig. 3f). We then conduced IF staining of CD3 and viral N protein. As expected, viral N protein was detected in livers from the severe and deceased, but not in liver from the moderate patient (Fig. 3g). Interestingly, more CD3+ cells are adjacent to viral positive cells in the surviving severe, but not in the deceased patient (Fig. 3g). Because the CD3 heterodimer composed of CD3E/CD3D or CD3E/CD3G is essential for T cell receptor (TCR) expression,24 CD3E in Cluster 3 indicates T-cell activation. TCR genes including, TRAC, TRAC1, and TRBC2, were examined. These genes tended to be highly expressed in Cluster 3 (Fig. 3h).
Considering the crucial roles of T cells in elimination of virus, we next conducted TCR sequencing on FFPE liver tissues to compared their features between surviving severe and deceased patients. Compared with those from deceased patients, liver tissue from resolved patients contained shorter CDR3s in the TCRα chain (CDR3α) (Fig. 3i), with highly expanded clones in resolved patients (Fig. 3j). Interestingly, we observed a new recombination of TRBV20-1 and TRBJ207 in the TCRβ chain in the resolved case (CDR3β) (Fig. 3k). The top sequences of CDR3α and CDR3β for each patient are shown in Supplementary Table S3 and Supplementary Table S4. These results suggest that T cells are crucial for the intrahepatic control of SARS-CoV-2.
Distribution of and changes in the liver after SARS-CoV-2 infection
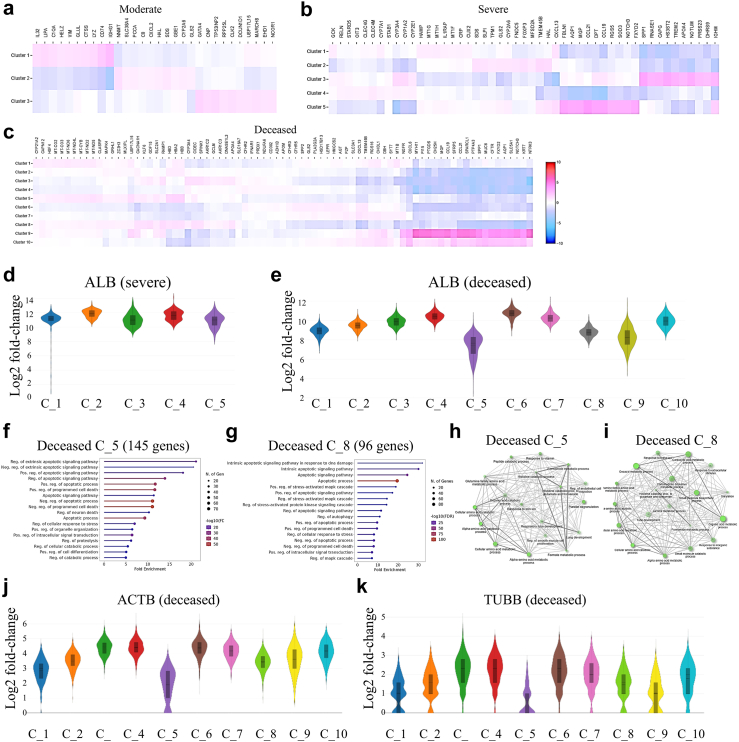
Spatial transcriptomics analysis revealed enriched characteristic genes in clusters in the samples from moderate, severe, and deceased cases (Fig. 4a–c). We investigated well-known liver nonparenchymal cell-related genes. Interestingly, erythroid genes (AHSP, CA1, and SLC4A1) were found in viral RNA-enriched Clusters 2 and 4 from the severe case (Supplementary Figure S4a) and Clusters 3, 4, 5, and 8 from the deceased case (Supplementary Figure S4b). The AHSP (erythroid cells), ISLR (hepatic stellate cells), ISGF6 (Kupffer cells), JCHAIN (Plasma cells) and CD3D (T cells) are also showed in Violin spots (Supplementary Figure S4c-l). This finding is supported by reports that SARS-CoV-2 can infect red blood cells and cause hemagglutination.25
Fig. 4.
Features of livers from patients with COVID-19. Top up and down-regulated genes in clusters from moderate (a), severe (b) and deceased (c) cases. Transcription level of albumin (ALB) in different clusters from severe (d) and deceased (e) patients. GO Biological Processes enrichment of apoptotic process (GO: 0006915) in top 1000 up-regulated genes from Cluster 5 (f) and Cluster 8 (g). GO Biological Processes enrichment of pathway in down-regulated genes from Cluster 5 (h) and Cluster 8 (i). Levels of housekeeping genes, ACTB (j) and TUBB (k), in clusters from the deceased patient. In d, e, j and k, the Y-axis is an estimated change of the log2 ratio of expression in a cluster to that in all other spots.
We then investigated whether the liver parenchyma was affected by viruses and analyzed the transcription of marker genes for hepatocyte-1 (ADH6, CMBL, and SLC10A1), hepatocyte-2 (APOF, AOX1, and C8A), cholangiocyte (APINT1, EPCAM, and KRT71), vascular endothelial cells (ECSCR, ARAP3, and RHOJ) and sinusoid endothelial cells (CLEC1B, EHD3, and GPR182). In the severe case, Clusters 1 and 4 consisted of hepatocytes with abundant sinusoids, Cluster 2 comprised hepatocytes, and Clusters 3 and 5 highly expressed cholangiocyte- and vascular-specific genes (Supplementary Figure S4m). In the deceased patient, Clusters 9 and 10 were enriched for cholangiocyte- and vascular endothelial cell-specific genes, and the number of sinusoid-related genes was high in Clusters 2, 3, 4, 5, and 8 (Supplementary Figure S4n). Clusters 1, 2, 3, 4, 6, and 7 also highly expressed hepatocyte-specific genes (Supplementary Figure S4n). The EPCAM, SLC10A1, C8A, EHD3 and ECSCR are also showed in Violin spots (Supplementary Figure S4o-x). Interestingly, SARS-CoV-2 RNA was enriched in Clusters 3, 4, 5, and 8 from the deceased patient (Fig. 2e). We then suspected that the clusters with high levels of sinusoid genes were dysfunctional hepatocytes. The transcription of ALB was evaluated to investigate this. As expected, Clusters 5 and 8 from the deceased patient had obviously decreased ALB levels (Fig. 4e), whereas ALB expression in clusters was unchanged in the severe case (Fig. 4d).
We subsequently conducted GO biological process enrichment for the top 1000 upregulated genes in Clusters 5 and 8 with decreased ALB. A total of 145 genes from Cluster 5 and 96 genes from Cluster 8 were enriched in the apoptotic process (GO: 0006915) (Fig. 4f and g). In addition, the extrinsic apoptotic pathway was identified in Cluster 5 (Fig. 4f), and the intrinsic apoptotic pathway tended to be activated in Cluster 8 (Fig. 4g). Moreover, the expression of genes related to glutamine family amino acid metabolism, cellular amino acid catabolism and metabolism, and alpha-amino acid catabolism and metabolic processes significantly decreased in both Clusters 5 and 8 (Fig. 4h and i). These data suggested that there might be a general drop across all transcripts due to cell death. We therefore checked two housekeeping genes, ACTB (ENSG00000163631, encoding beta-actin) and TUBB (ENSG00000075624, encoding beta-tubulin), and found both of them are decreased in the apoptotic clusters, especially in the Cluster-5 (Fig. 4j and k). We also stained cleaved caspase-3 to view the apoptotic in liver sections, and found that it is unexpectedly less in the deceased than in the severe patient (Supplementary Figure S5a-d). Considering the general decrease of all transcripts due to cell death, we checked transcription level of caspase-3 gene (CASP3). In the apoptotic Cluster-5, it is much lower than other clusters (Supplementary Figure S5e), which should be the reason for less presentation of its cleaved form in the deceased patient. Interestingly, we found viral N protein positive cells tend to have weak nucleuses (Supplementary Figure S5b), which might be caused by death of hepatocytes. These findings suggested that the protein synthesis function of hepatocytes was greatly impaired by SARS-CoV-2, promoting their apoptosis in deceased patients.
Dynamic fluctuations in the ALB concentration are associated with outcomes of patient with COVID-19
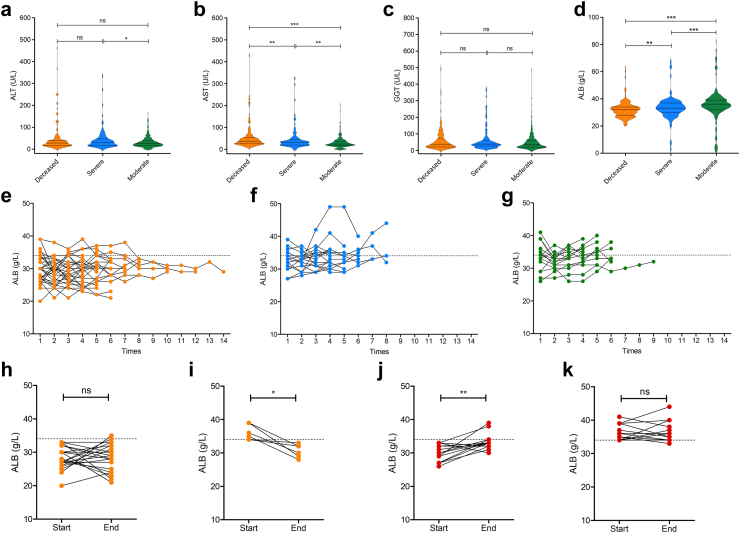
The spatial analysis suggested that in the deceased patient, ALB synthesis was significantly impaired, whereas ALT, AST, and GGT levels were elevated, accompanied by immune cell infiltration, in the resolved severe patient. Therefore, we retrospectively studied hospitalized patients with COVID-19, including 165 deceased (median age was 87 years, range from 51 to 104; 48 females and 117 males), 122 severely ill (median age was 75 years, range from 32 to 94; 40 females and 82 males), and 363 moderately ill patients (median age was 75 years, range from 18 to 99; 140 females and 223 males). We did not observe any difference in baseline ALT or GGT levels among the deceased, severe, and moderate cohorts (Fig. 5a–c). However, AST levels increased with increasing disease severity (Fig. 5b). In contrast, the baseline ALB concentration decreased with severity (Fig. 5d).
Fig. 5.
Characteristics of liver function indicators in COVID-19 clinical cohorts. (a) Serum ALT levels in deceased (Orange, n = 165), severe (Blue, n = 122) and moderate (Green, n = 363) cohorts. (b) Serum AST levels in deceased, severe and moderate cohorts. (c) Serum GGT levels in deceased, severe and moderate cohorts. (d) Serum ALB levels in deceased, severe and moderate cohorts. (e) Dynamic monitor of ALB in 30 deceased cases. (f) Dynamic monitor of ALB in 17 severe cases. (g) Dynamic monitor of ALB in 15 moderate cases. Change of ALB between start and end points in deceased cases, with declined ALB (h) or normal ALB (i) at the beginning. Change of ALB between start and end points in survived severe and moderate cases (Red), with declined ALB (j) or normal ALB (k) at the beginning. Statistical significance was assessed by Mann–Whitney test (a–d) or paired two-tailed t tests (h–k). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Moreover, among them, 30 deceased, 17 severe, and 15 moderate cases had dynamic monitored data (≥4 times). In the deceased patients, 12 initially and 10 ultimately had increased AST levels (Supplementary Figure S6a). Among the severe cases, five patients initially and three ultimately had increased AST levels (Supplementary Figure S6b). Among the moderate cases, six initially and four ultimately had increased AST levels (Supplementary Figure S6c). Although AST slightly decreased in these patients, we did not observe a clear trend. Thus, we directly compared the data between the start and end points. There was no significant change in AST levels in patients without enhanced baseline AST, regardless of their outcome (Supplementary Figure S6d, f). In the survival cohorts, which included the moderate and severe cohorts, increased AST levels also did not significantly change (Supplementary Figure S6g). Surprisingly, in the deceased patients, the increase in AST initially returned to normal (Supplementary Figure S6e). This finding indicates that the baseline AST level could be used as a prognostic marker for mortality risk in patients with COVID-19, while its regression does not indicate a decrease in risk.
We next examined the fluctuation of ALB levels. Among the deceased patients, 24 initially 28 ultimately had decreased ALB levels (Fig. 5e). Among the severe cases, 10 initially and 8 ultimately had decreased ALB levels (Fig. 5f). Among the moderate cases, seven initially and five ultimately had decreased ALB levels (Fig. 5g). In the deceased group, the ALB concentration decreased below the normal level (Fig. 5e), whereas in the surviving severe and moderate cohorts, the ALB concentration tended to increase (Fig. 5f and g). A comparison of the ALB levels between the start and end points revealed that the patients whose serum ALB level decreased at the beginning of the study did not recover at the end of the study (Fig. 5h). Even when the serum ALB concentration initially returned to normal, it decreased by the end of the study (Fig. 5i). In contrast, in surviving patients, patients whose ALB levels initially decreased tended to recover (Fig. 5j), and those with normal ALB levels maintained this trend until the end of the study (Fig. 5k). As hepatocytes specifically synthesize ALB, its decrease reflects the impact of SARS-CoV-2 on the liver. Importantly, fluctuations in the serum ALB concentration could be used to assess the mortality risk of patients with COVID-19.
Discussion
Liver injury is commonly reported in patients with COVID-19, but whether SARS-CoV-2 can infect the liver remains controversial. In this study, we detected viral proteins and RNA in the livers of severely ill deceased patients. Although our findings do not provide direct evidence that the liver supports the entire life cycle of SARS-CoV-2, we indicated that viral entry and translation were accessible. The spatial transcriptomics analysis of tissues partitioned liver spots into 3 clusters in moderate cases, 5 in severe cases, and 10 in deceased cases. This finding suggested that, as the severity of the disease increased, the impact of the virus on the liver became more severe. As no viral proteins were detected in the livers of moderate cases, it is possible that SARS-CoV-2 was already controlled before entering liver in these patients. Therefore, invasion of the liver potentially occurs after long-term viral exposure in patients with moderate and severe COVID-19.
Many studies have shown that ACE2 is detected in the bile duct, sinusoidal, and capillary endothelial cells and that the level of ACE2 is 20 times greater in these cells than that in hepatocytes.26 However, we detected few viral proteins or RNAs in cholangiocytes, mainly observing them in hepatocytes and hepatic sinusoids. ACE2 expression is upregulated in hepatocytes from livers with cirrhosis27; however, ACE2 transcription is almost undetectable, indicating a limited role for ACE2 in the liver and suggesting that other intracellular restriction factors or receptors might contribute to SARS-CoV-2 infection in hepatocytes. Thus, we preliminarily examined the transcription level of ASGR1 and found that it was dramatically high. Notably, ASGR1 can mediate the entry of SARS-CoV-2 in an ACE2-independent manner.28 Recently, ASGR1 was shown to mediate SARS-CoV-2 pseudovirus entry into primary hepatic parenchymal cells,23 and our work herein provides clinical evidence for its role in the liver. Moreover, we found that the ASRG1 level in different patients are almost equal, suggesting it is not a distinct factor for liver infection, and whether the virus can reach the liver through numerous obstacles should be the key issue.
Consistent with our finding that increased liver T cells were observed only in patients with finally resolved virus-infected livers but not in deceased patients, a decreased magnitude of the T-cell response was associated with severe COVID-19.29 Surprisingly, one study demonstrated that normal hepatocytes, which present ABCD3 peptides, can be cross-recognized by SARS-CoV-2-specific T cells in patients with COVID-19.30 The ABCD3-specific and SARS-CoV-2-specific TCRs share the same CDR3β and a single amino acid-substituted CDR3α.30 These findings indicate that T-cell responses triggered by SARS-CoV-2 may lead to liver damage. We also analyzed CDR3αβ in the liver. No similar CDR3 sequence was identified, but clonal expansion of CDR3α and novel recombination of CDR3β were characterized in SARS-CoV-2-positive liver samples, which were ultimately resolved, indicating a crucial role of T cells in viral control.
A systematic summary and meta-analysis of investigations from the first four months of 2020 reported that 19% of patients had increased ALT and/or AST levels.31 Moreover, 50% of hospitalized patients were demonstrated to have elevated liver biochemistry.32 Although we detected SARS-CoV-2 proteins and RNA in liver tissues from deceased patients with COVID-19, none presented increased ALT, AST, or GGT levels. Conversely, there were increased ALT, AST, and GGT levels in survived patients, who also had expanded T cells colocalized with the virus. It is possible that cytotoxic T cells rather than viruses cause the increases in liver aminotransferases, which needs further investigation.
This finding raises the question of the impact of viruses on the liver. We demonstrated that there is a general drop across all transcripts due to cell death in Cluster-5 from deceased patient. Nevertheless, one limitation of this study is the mechanism by which hepatocytes die is unclear and deserves further investigation. In the deceased patient, we observed that ALB expression was dramatically impaired in clusters with high levels of SARS-CoV-2 RNA. Consistently, the serum ALB concentration was markedly decreased in patients with severe SARS-CoV-2-positive hepatic disease, which was confirmed in clinical cohorts, with 165 deceased, 122 severely ill, and 363 moderately ill patients. This finding is supported by a study reporting that patients with low ALB levels at baseline have an increased risk of death versus those with high ALB levels at baseline.33 We found that the recovery of serum ALB, but not AST, indicated patients’ clinical improvement. These findings suggest that downregulated serum ALB expression, but not upregulated liver aminotransferase expression, could serve as a potential indicator of liver involvement in SARS-CoV-2 infection.
Overall, we profiled the liver tropism of SARS-CoV-2 via the detection of viral RNA and S and N proteins, revealed the spatial distribution and immune features of the virus in the liver, and suggested that the clonality and recombination of TCR CDR3s from T cells are crucial for controlling intrahepatic viruses. We also demonstrated that SARS-CoV-2 promotes the apoptosis of hepatocytes in deceased patients, which results in impaired ALB expression. Furthermore, the decrease in serum ALB expression rather than that of aminotransferases can be used to evaluate a patients mortality risk. Although we have not provided detail evidences on the liver tropism, T cell activation and dysfunction of liver in SARS-CoV-2 infection, this clinical-based study may help elucidate the viral pathogenic mechanism and immune responses against SARS-CoV-2.
Contributors
ZF, SC and QY conceived the study. ZY, ZF and SC designed the Study. SC collected the patients’ clinical information. YZ, SC, SS, WW, ZD and JZ collected liver tissue samples. AA, YL, JB and JC performed immunohistochemistry staining. HG, ZH, LY, YW, YG, QY and NX conducted the infection of golden (Syrian) hamsters. CW, DT and CH analyzed data. YL and ZF prepared the figures and/or wrote the manuscript. SC and ZF had verified the underlying data. All authors read and approved the final version of the manuscript, and ensure it is the case.
Data sharing statement
The data that support the findings of this study are available from the corresponding author upon request.
Declaration of interests
The authors declared no conflict of interest.
Acknowledgements
We thank Yiwei Zhong at School of Basic Medical Sciences, Fudan University for excellent technical support.
Funding: National Key R&D Program of China (2021YFC2300602), National Natural Science Foundation of China (92369110), National Natural Science Foundation of China (U23A20474), Shanghai Municipal Science and Technology Major Project (ZD2021CY001), Shanghai Jinshan District Medical and Health Technology Innovation Fund Project (2023-WS-31).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105517.
Contributor Information
Ningshao Xia, Email: nsxia@xmu.edu.cn.
Zhenghong Yuan, Email: zhyuan@shmu.edu.cn.
Jiming Zhang, Email: jmzhang@fudan.edu.cn.
Quan Yuan, Email: yuanquan@xmu.edu.cn.
Zhong Fang, Email: zhongfang13@fudan.edu.cn.
Appendix A. Supplementary data
References
- 1.Vinayagam S., Sattu K. SARS-CoV-2 and coagulation disorders in different organs. Life Sci. 2020;260 doi: 10.1016/j.lfs.2020.118431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y., Geng X., Tan Y., et al. New understanding of the damage of SARS-CoV-2 infection outside the respiratory system. Biomed Pharmacother. 2020;127 doi: 10.1016/j.biopha.2020.110195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loganathan S., Kuppusamy M., Wankhar W., et al. Angiotensin-converting enzyme 2 (ACE2): COVID 19 gate way to multiple organ failure syndromes. Respir Physiolo Neurobiol. 2021;283 doi: 10.1016/j.resp.2020.103548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang C., Shi L., Wang F.S. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol Hepatol. 2020;5:428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wanner N., Andrieux G., Badia I.M.P., et al. Molecular consequences of SARS-CoV-2 liver tropism. Nat Metab. 2022;4:310–319. doi: 10.1038/s42255-022-00552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes E. Infection of liver hepatocytes with SARS-CoV-2. Nat Metab. 2022;4:301–302. doi: 10.1038/s42255-022-00554-4. [DOI] [PubMed] [Google Scholar]
- 7.Heinen N., Khanal R., Westhoven S., et al. Productive infection of primary human hepatocytes with SARS-CoV-2 induces antiviral and proinflammatory responses. Gut. 2024;73:e14. doi: 10.1136/gutjnl-2023-330961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinen N., Klöhn M., Westhoven S., et al. Host determinants and responses underlying SARS-CoV-2 liver tropism. Curr Opin Microbiol. 2024;79 doi: 10.1016/j.mib.2024.102455. [DOI] [PubMed] [Google Scholar]
- 9.Zanon M., Neri M., Pizzolitto S., et al. Liver pathology in COVID-19 related death and leading role of autopsy in the pandemic. World J Gastroenterol. 2023;29:200–220. doi: 10.3748/wjg.v29.i1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yadav D.K., Singh A., Zhang Q., et al. Involvement of liver in COVID-19: systematic review and meta-analysis. Gut. 2021;70:807–809. doi: 10.1136/gutjnl-2020-322072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulkarni A.V., Kumar P., Tevethia H.V., et al. Systematic review with meta-analysis: liver manifestations and outcomes in COVID-19. Aliment Pharmacol Ther. 2020;52:584–599. doi: 10.1111/apt.15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang R.X., Zheng R.D., Fan J.G. Etiology and management of liver injury in patients with COVID-19. World J Gastroenterol. 2020;26:4753–4762. doi: 10.3748/wjg.v26.i32.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nardo A.D., Schneeweiss-Gleixner M., Bakail M., et al. Pathophysiological mechanisms of liver injury in COVID-19. Liver Int. 2021;41:20–32. doi: 10.1111/liv.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X.T., Yu Y.H., Zhang C., et al. Mechanism of SARS-CoV-2 invasion into the liver and hepatic injury in patients with COVID-19. Mediterr J Hematol Infect Dis. 2022;14 doi: 10.4084/MJHID.2022.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonzogni A., Previtali G., Seghezzi M., et al. Liver histopathology in severe COVID 19 respiratory failure is suggestive of vascular alterations. Liver Int. 2020;40:2110–2116. doi: 10.1111/liv.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y., Huang X., Yuan L., et al. A recombinant spike protein subunit vaccine confers protective immunity against SARS-CoV-2 infection and transmission in hamsters. Sci Transl Med. 2021;13 doi: 10.1126/scitranslmed.abg1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan J.F., Zhang A.J., Yuan S., et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sia S.F., Yan L.M., Chin A.W.H., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitsui Y., Suzuki T., Kuniyoshi K., et al. Expression of the readthrough transcript CiDRE in alveolar macrophages boosts SARS-CoV-2 susceptibility and promotes COVID-19 severity. Immunity. 2023;56:1939–1954.e12. doi: 10.1016/j.immuni.2023.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Brevini T., Maes M., Webb G.J., et al. FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2. Nature. 2023;615:134–142. doi: 10.1038/s41586-022-05594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T., Xue W., Zheng Q., et al. Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants. Nat Commun. 2021;12:5652. doi: 10.1038/s41467-021-25997-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia S., Liu M., Wang C., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X.Y., Zheng X., Zhu Y.Q., et al. Asialoglycoprotein receptor 1 promotes SARS-CoV-2 infection of human normal hepatocytes. Signal Transduct Targeted Ther. 2024;9 doi: 10.1038/s41392-024-01754-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kjer-Nielsen L., Dunstone M.A., Kostenko L., et al. Crystal structure of the human T cell receptor CD3 epsilon gamma heterodimer complexed to the therapeutic mAb OKT3. Proc Natl Acad Sci U S A. 2004;101:7675–7680. doi: 10.1073/pnas.0402295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kronstein-Wiedemann R., Stadtmuller M., Traikov S., et al. SARS-CoV-2 infects red blood cell progenitors and dysregulates hemoglobin and iron metabolism. Stem Cell Rev Rep. 2022;18:1809–1821. doi: 10.1007/s12015-021-10322-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong P., Xu J., Yang D., et al. COVID-19-associated gastrointestinal and liver injury: clinical features and potential mechanisms. Signal Transduct Target Ther. 2020;5:256. doi: 10.1038/s41392-020-00373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Q., Xie Q., Shi C.C., et al. Expression of angiotensin-converting enzyme 2 in CCL4-induced rat liver fibrosis. Int J Mol Med. 2009;23:717–723. doi: 10.3892/ijmm_00000185. [DOI] [PubMed] [Google Scholar]
- 28.Gu Y., Cao J., Zhang X., et al. Receptome profiling identifies KREMEN1 and ASGR1 as alternative functional receptors of SARS-CoV-2. Cell Res. 2022;32:24–37. doi: 10.1038/s41422-021-00595-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnes E., Goodyear C.S., Willicombe M., et al. SARS-CoV-2-specific immune responses and clinical outcomes after COVID-19 vaccination in patients with immune-suppressive disease. Nat Med. 2023;29:1760-+. doi: 10.1038/s41591-023-02414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Wang Y., Peng Z., Li G., Wang J. T cell cross-reactivity in autoimmune-like hepatitis triggered by COVID-19. hLife. 2023;1:57–61. [Google Scholar]
- 31.Mao R., Qiu Y., He J.S., et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2020;5:667–678. doi: 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bloom P.P., Pasricha T.S., Andersson K.L., et al. Hepatology consultants often disagree on etiology of abnormal liver biochemistries in COVID-19 but agree on management. Dig Dis Sci. 2021;66:1852–1854. doi: 10.1007/s10620-020-06495-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang T.Y., Smith D.A., Campbell C., et al. Longitudinal analysis of the utility of liver biochemistry as prognostic markers in hospitalized patients with corona virus disease 2019. Hepatol Commun. 2021;5:1586–1604. doi: 10.1002/hep4.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.