Abstract
Despite advances in various chemotherapy regimens, current therapeutic options are limited for ovarian cancer patients. Oxidative stress-induced growth inhibitor 1 (OSGIN1), which is a tumor suppressor gene known to regulate the cellular stress response and apoptosis, is associated with ovarian cancer development. However, the underlying mechanisms involved in ferroptosis regulation have not been elucidated. Thus, this study aimed to investigate the effect and underlying regulatory mechanism of the OSGIN1 gene on ovarian cancer cells. Our results demonstrated that loss of the OSGIN1 gene promoted ovarian cancer growth and conferred resistance to drug-induced ferroptosis. Mechanistically, the loss of OSGIN1 activates AMPK signaling through ATM, leading to the upregulation of SLC2A3, which protects cells from ferroptosis and renders them insensitive to ferroptosis inducers. Notably, an SLC2A3-neutralizing antibody enhances the ferroptosis-inducing and anticancer effects of sorafenib on ovarian cancer patient-derived xenograft tumors. Overall, anti-SLC2A3 therapy is a promising method to improve ovarian cancer treatment by targeting ferroptosis.
Subject terms: Ovarian cancer, Drug development
Introduction
Ovarian cancer is a prevalent gynecological malignancy characterized by a high lethality rate1. Although platinum drugs are frequently applied in the treatment of advanced ovarian cancer, these drugs tend to induce tumor resistance and undesirable side effects, undermining their efficacy. Therefore, the long-term survival of patients, especially those with peritoneal metastases, is poor2,3. To improve the overall prognosis of ovarian cancer patients, it is imperative to develop new and effective therapeutic approaches.
Sorafenib, an orally administered multitarget, multikinase inhibitor, has been approved for treating several tumor types, including hepatocellular carcinoma and renal cell carcinoma4. While sorafenib has shown promise in some phase II trials for ovarian cancer5, it has failed to achieve a sufficient objective response or sustained disease stabilization as a third-line treatment6. Furthermore, sorafenib has not been recommended as a front-line maintenance therapy for ovarian cancer patients in complete remission7. Therefore, enhancing tumor responsiveness to sorafenib has become a key area of focus.
Notably, recent studies have reported that sorafenib can induce ferroptosis in liver cancer cell lines8. Ferroptosis is a type of cell death caused by oxidative stress9. Unlike other forms of cell death, such as apoptosis, necrosis, and autophagy, ferroptosis is characterized by the abnormal metabolism of iron, amino acids, and lipids. During ferroptosis, in the cell antioxidant system, glutathione (GSH) fails to neutralize harmful substances in the antioxidant systems of cells, which leads to the accumulation of iron ions and fatty acids, resulting in the generation of oxygen species (ROS) that cause lipid peroxidation10. There is a growing interest in investigating the role of ferroptosis in different types of cancer, including ovarian cancer.
However, most studies on ferroptosis regulators have been conducted in cell line models, which may not fully recapitulate the therapeutic response of autochthonous tumors and clinical cancers. As a result, the translation of ferroptosis regulators to clinical benefits remains limited, and several outstanding questions need to be addressed. Specifically, it is crucial to investigate the genetic or epigenetic alterations in human cancers that play a major role in vulnerability to ferroptosis in vivo. Additionally, identifying effective and safe strategies to sensitize therapy-resistant cancers to ferroptosis is of utmost importance.
Oxidative stress-induced growth inhibitor 1 (OSGIN1), also known as ovarian kidney and liver protein 38 (OKL38), has been reported to be highly expressed in the ovary, kidney, and liver11. OSGIN1 functions as a tumor suppressor gene that participates in the regulation of cellular stress and apoptosis, and loss of the OSGIN1 protein contributes to the development of liver cancer12. However, the functional characterization of OSGIN1 in ovarian cancer in vivo has not yet been performed. In this study, we demonstrated that the deletion of OSGIN1 promotes the development of ovarian cancer and confers resistance to drug-induced ferroptosis through the AMPK-mediated upregulation of SLC2A3.
Results
OSGIN1 is downregulated in ovarian cancer tissues
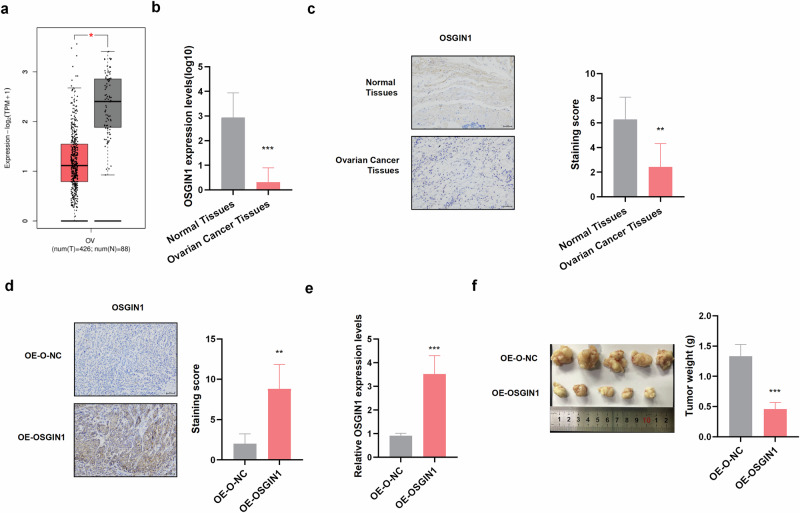
The expression of OSGIN1 in ovarian cancer was analyzed via The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases. The results revealed that OSGIN1 expression was significantly lower in ovarian cancer tissue (426 serous ovarian cancer tissues) than in normal ovarian tissue (n = 88) (Fig. 1a). Consistent with the RNA-seq data, qRT‒PCR and IHC assays of ovarian cancer tissues and normal tissues revealed that the expression of OSGIN1 was significantly lower in ovarian cancer tissues than in normal tissues (Fig. 1b‒c). These results showed that OSGIN1 was downregulated in ovarian cancer.
Fig. 1. OSGIN1 expression was reduced in human ovarian cancer tissues.
a The expression of OSGIN1 in normal ovary tissues (n = 88) (GTEx-OV) and ovarian cancer tissues in the TCGA-OV cohort (n = 426). b OSGIN1 mRNA expression in HGSOC tissues (n = 7) and normal ovarian tissues (n = 7). c IHC images showing OSGIN1 protein expression in ovarian cancer and normal ovarian tissues from seven ovarian cancer patients (n = 7). d IHC images showing OSGIN1 protein expression in tumor tissues from nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice bearing ovarian cancer (n = 5 mice/group). e Representative tumor images from specific populations (24 days) (n = 5 mice/group). f Tumor weights for the indicated cohorts (24 days). OSGIN1 mRNA expression levels in tumor tissues from nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice bearing ovarian cancer were determined by RT‒qPCR (n = 5 mice/group). All results are representative of at least three independent experiments. Statistical significance was determined by a two-tailed unpaired t-test (a–f). Error bars are s.e.m. *p < 0.05; **p < 0.01 and ***p < 0.001.
To explore the role of OSGIN1 in tumor growth, female NOD/SCID mice were intraperitoneally injected with OVCAR3 tumor cells (5 ×105 cells/mouse) overexpressing the OSGIN1 gene or the control vehicle. RT‒qPCR and IHC analysis revealed that OSGIN1 expression was upregulated in the OE-OSGIN1-treated group (Fig. 1d, e) and that OSGIN1 overexpression significantly decreased tumor volume and body weight (Fig. 1f). These results provide evidence that OSGIN1 has an inhibitory effect on ovarian cancer growth in vivo.
OSGIN1 was induced during erastin-induced ferroptosis of ovarian cancer cells
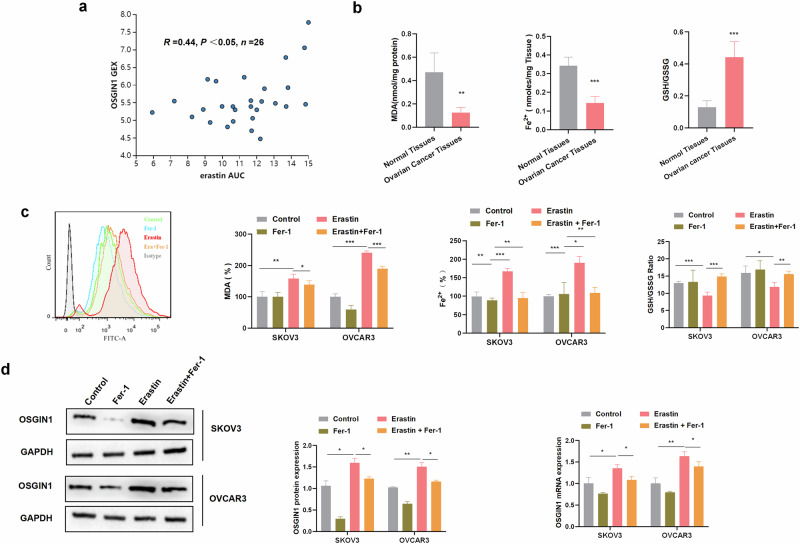
To assess whether OSGIN1 expression could influence drug response, we analyzed the Cancer Therapeutics Response Portal (CTRP), which is a platform allowing for the analysis of gene expression in response to 481 different compounds in various cancer cell lines13. We found that the OSGIN1 gene affected the sensitivity of ovarian cancer cells to erastin (Fig. 2a). Erastin is a well-known compound that is widely used as a tool compound in the study of ferroptosis14.
Fig. 2. OSGIN1 confers sensitivity to ferroptosis.
a Correlation between OSGIN1 expression and erastin sensitivity, based on the ovarian cancer cell lines (n = 26) from CTRP. Dose responses are normalized area under curve values. The linear relationship was determined by a two-tailed Pearson correlation analysis. b Comparison of the levels of MDA and iron and the GSH/GSSG ratio between ovarian cancer and normal ovarian tissues (n = 7). c SKOV3 and OVCAR3 cells were treated with DMSO (control) or erastin (10 μM) alone or in combination with ferrostatin-1 (fer-1, 10 μM). Levels of intracellular reactive oxygen species after the indicated treatments. The MDA/iron ratio and GSH/GSSG ratio after the indicated treatments. d Expression of OSGIN1 mRNA and protein in ovarian cancer cells after treatment with 10 μM erastin for 24 h. All results are representative of at least three independent experiments. Statistical significance in b was determined by a two-tailed unpaired t-test.The p values in c, d were determined by two-way ANOVA with multiple comparisons, Error bars are s.e.m. *p < 0.05; **p < 0.01 and ***p < 0.001.
To investigate the involvement of ferroptosis in ovarian cancer, we measured iron levels in tissues and observed relatively lower iron levels in ovarian cancer tissues (Fig. 2b). Excessive accumulation of free iron within cells can initiate ferroptosis15. Furthermore, our findings revealed lower expression of MDA, a biochemical marker associated with lipid peroxidation and oxidative stress16, in ovarian cancer tissues than in normal ovarian tissues. Additionally, we found a higher GSH/GSSG ratio in ovarian cancer tissues. Measuring the GSH/GSSG ratio allows indirect assessment of oxidative stress within cells and the degree of disruption in iron metabolism16. Low levels of MDA and a high GSH/GSSG ratio generally indicate decreased oxidative stress within tissues. These data suggest potential resistance to ferroptosis, indicating the presence of other regulatory factors or mechanisms that inhibit ferroptosis in ovarian cancer tissues.
To investigate the effect of ferroptosis inducers on ovarian cancer cells, we treated the cells with various concentrations of erastin (Supplementary Fig. 1a, b). Pretreatment with ferrostatin-1 (Fer-1), an effective antioxidant that can inhibit ferroptosis (10 μM), suppressed the decrease in cell viability that was induced by erastin (10 μM) at 12 hours, thereby confirming that the reduction in cell viability was due to ferroptosis (Supplementary Fig. 1a, b). Additionally, we observed that MDA/iron levels were significantly increased and that the GSH/GSSG ratio was decreased in cells treated with erastin (10 μM) for 12 h, and these changes could be reversed by pretreatment with Fer-1 (Fig. 2c). Considering the mutual promotion of intracellular reactive oxygen species (ROS) generation and ferroptosis17, we conducted flow cytometry to measure ROS production. Unsurprisingly, we observed that the increase in ROS levels induced by erastin treatment could be reversed by pretreatment with Fer-1 (Fig. 2c).
Remarkably, we observed an increase in OSGIN1 mRNA and protein levels in SKOV3 and OVCAR3 cells treated with erastin. However, pretreatment with Fer-1 alleviated the erastin-induced increase in OSGIN1 expression (Fig. 2d). These findings suggest that erastin promotes intracellular ferroptosis and upregulates OSGIN1 expression and that Fer-1 pretreatment suppresses OSGIN1 expression.
OSGIN1 accelerates ferroptosis in ovarian cancer cells
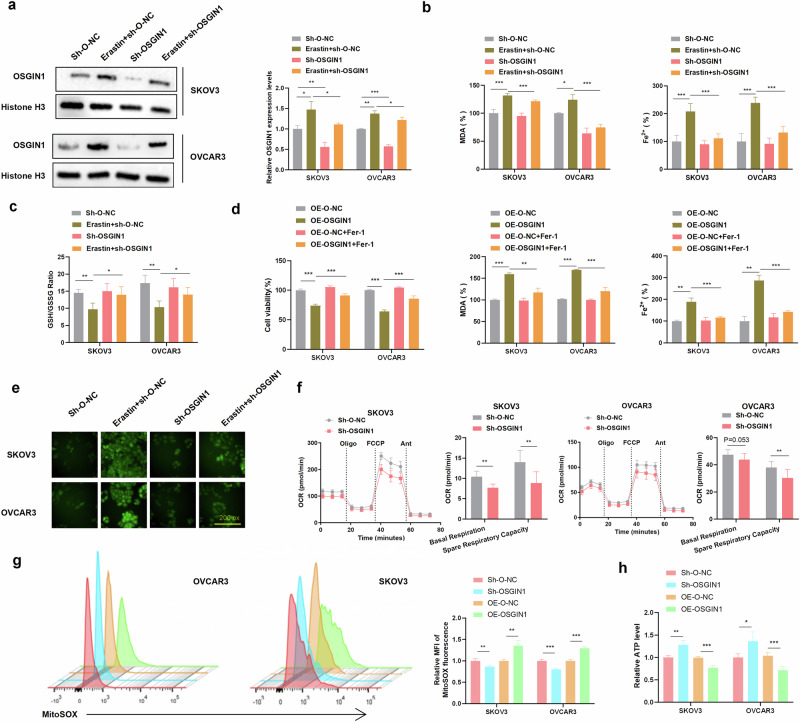
To investigate the role of OSGIN1 in erastin-induced ferroptosis, we manipulated the expression levels of OSGIN1 and evaluated their effect on ferroptosis. The knock-down or overexpression efficiency was verified through western blot and qRT-PCR analysis (Supplementary Fig. 2a–c). In OSGIN1-knockdown cells, the levels of intracellular ROS, MDA, and iron induced by erastin were effectively restored to near baseline levels (Fig. 3a–c, e). Furthermore, pretreating OSGIN1-overexpressing cells with Fer-1 for 24 h reversed the growth inhibition induced by OSGIN1 and mitigated the levels of MDA and iron (Fig. 3d). These findings indicate that OSGIN1 plays a role in modulating erastin-induced ferroptosis by influencing intracellular ROS, MDA, and iron levels.
Fig. 3. OSGIN1 gene downregulation inhibited erastin-induced ferroptosis.
a Relative mRNA and protein levels of OSGIN1 in sh-O-NC/sh-OSGIN1-transfected cells treated with or without erastin. b, c Effect of erastin treatment for 24 h on the intracellular levels of MDA, Fe2+ and GSH/GSSH in SKOV3 and OVCAR3 cells transfected with sh-O-NC or sh-OSGIN1 viruses. d Effect of Fer-1 pretreatment for 24 h on cell viability and MDA/Fe2+ levels in SKOV3 and OVCAR3 cells transfected with OE-O-NC or OE-OSGIN1 viruses. e ROS levels in sh-O-NC/sh-OSGIN1-transfected SKOV3 and OVCAR3 cells following treatment with 10 μM erastin. f OCR (oxygen consumption rate) results for SKOV3 and OVCAR3 cells transfected with sh-O-NC or sh-OSGIN1. g The rates of ROS generation by mitochondria in living cells from mutant and control cell lines were analyzed by FlowJo using the mitochondrial superoxide indicator MitoSOX Red (5 mM). Flow cytometry histogram showing MitoSOX Red fluorescence in various cell lines. Relative ratios of MitoSOX-Red fluorescence intensity. The average of three determinations for each cell line is shown. h Effect of sh-OSGIN1 on ATP levels in OVCAR3 cells. Each experiment was repeated three times. The p values in a–d, g, and h were determined by two-way ANOVA with multiple comparisons. The p value in f was obtained by two-tailed unpaired Student’s t test. Error bars are s.e.m. *p < 0.05; **p < 0.01 and ***p < 0.001.
During the process of ferroptosis, excessive accumulation of intracellular iron can lead to mitochondrial dysfunction, thereby affecting oxygen consumption rates (OCRs). OCR experiments can help us understand the cellular oxygen metabolism and energy metabolism status under iron-induced cell death conditions. We can evaluate changes in mitochondrial function and metabolic pathways within cells by measuring parameters such as the basal respiration rate and spare respiratory capacity18. Therefore, we quantitated the OCRs. The results showed that the basal respiratory OCR and alternate respiratory capacity OCR were decreased in sh-OSGIN1 cells, but the basal respiratory OCR remained unchanged in OVCAR3 cells (P = 0.053) (Fig. 3f). Additionally, as shown in Supplementary Fig. 3a, b, si-OSGIN1 treatment resulted in decreased ECAR levels in both SKOV3 and OVCAR3 cells. These findings suggest that the downregulation of OSGIN1 contributed to a reduction in both aerobic respiration and glycolysis.
Due to the importance of mitochondrial ROS in initiating ferroptosis19, mitochondrial ROS levels were quantified in ovarian cancer cells using MitoSOX. OVCAR3 and SKOV3 cells overexpressing OSGIN1 exhibited significantly higher levels of mitochondrial ROS compared to those of scrambled control cells (Fig. 3g). Conversely, after knocking down OSGIN1, a decrease in mitochondrial ROS levels was observed in both ovarian cancer cell lines.
Considering that mitochondrial oxygen consumption is coupled to ATP synthesis20, we assessed the cellular level of ATP in ovarian cancer cells. Notably, ATP levels were significantly higher in sh-OSGIN1 cells than in scrambled control cells (Fig. 3h). Conversely, ATP levels were significantly lower in OE-OSGIN1 cells than in scrambled control cells. These results strongly indicate that OSGIN1 significantly contributes to mitochondrial ROS, mitochondrial basal oxygen consumption and ATP production. Collectively, these findings highlight the critical involvement of OSGIN1 in ferroptosis and bioenergetics in ovarian cancer.
The OSGIN1 gene is a positive regulator of AMPK signaling and a negative regulator of SLC2A3 in ovarian cancer
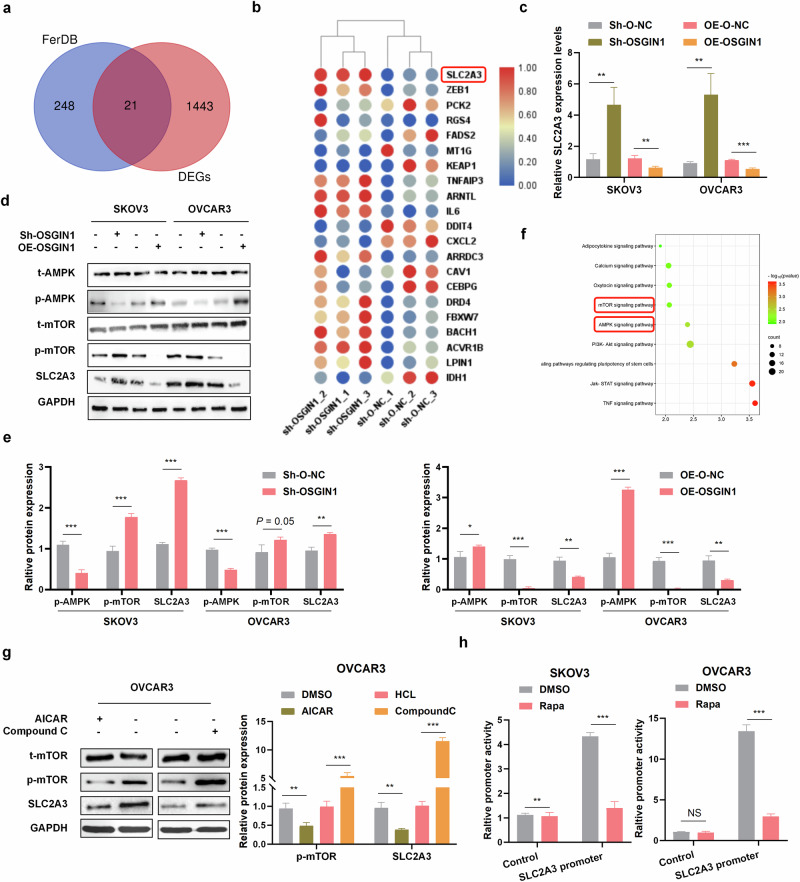
To explore the impact of OSGIN1 gene downregulation on the transcriptome of ovarian cancer cells, the RNA sequencing data of cells treated with sh-O-NC or sh-OSGIN1 were analyzed. Overall, 920 genes were upregulated and 547 genes were downregulated in sh-OSGIN1 cells compared to those in sh-O-NC cells. Differentially expressed genes (DEGs) were merged with ferroptosis-associated genes from the FERDB database to investigate the impact of OSGIN1 on ferroptosis. A total of 21 DEGs, including 13 upregulated and 8 downregulated genes, were found to be associated with ferroptosis following OSGIN1 knockdown (Fig. 4a, b).
Fig. 4. OSGIN1 inhibited SLC2A3 through the AMPK/mTOR pathway.
a A Venn diagram of the FerrDb database established by crossing related genes in the DEG and FerrDb datasets. b Volcano maps for the 21 genes. c SLC2A3 mRNA expression in SKOV3 and OVCAR3 cells with OSGIN1 knockdown or OSGIN1 overexpression. N = 3 samples. d, e The protein expression levels of t-AMPK, P-AMPK, t-mTOR, P-mTOR, and SLC2A3 were determined by Western blotting. f Dot plot of the KEGG pathway enrichment analysis. The horizontal axis represents the gene ratio, while the vertical axis represents the enriched pathway. The color scale indicates different thresholds of the p value, and the size of the dot indicates the number of genes corresponding to each pathway. g The expression of t-mTOR, P-mTOR, and SLC2A3 in ovarian cancer cells treated with 1 mmol/L AICAR (activator of AMPK) or 50 μmol/L compound C (inhibitor of AMPK) for 24 h. h Rapamycin stimulates SLC2A3 promoter activity. Activity of SLC2A3 promoter-driven luciferase reporter was measured in SKOV3 (Left) and OVCAR3 (Right) cells treated with or without 100 nM rapamycin for 24 h. Each experiment was repeated three times. The p values in c and g were determined by two-way ANOVA with multiple comparisons. The p values in e and h were determined by one-way ANOVA with multiple comparisons. Error bars are s.e.m. **p < 0.01; ***p < 0.001.
One of the highly upregulated genes in sh-OSGIN1 OVCAR3 cells was solute carrier family 2 member 3 (SLC2A3). It has been reported that SLC2A3 inhibits ferroptosis and is associated with poor prognosis in non-small cell lung cancer21. To confirm the regulatory effect of OSGIN1 on SLC2A3, we conducted qRT‒PCR and Western blot experiments. As expected, OSGIN1 knockdown resulted in the upregulation of SLC2A3 in ovarian cancer cells (Fig. 4c–e). Conversely, when OSGIN1 was overexpressed, SLC2A3 was significantly downregulated at both the protein and mRNA levels (Fig. 4c–e). This finding suggested that OSGIN1 may negatively regulate the expression of SLC2A3.
KEGG enrichment analysis revealed the involvement of differentially expressed genes in various pathways, such as mTOR signaling pathway, AMPK signaling pathway, PI3K-Akt signaling pathway, and TNF signaling pathway (Fig. 4f). The mTOR pathway has been demonstrated to activate SLC2A3 transcription22. Recent studies have shown that OSGIN1 activates the p-AMPKαT172/p-mTORS2448 signaling pathway in breast cancer23. Based on this information, we hypothesized that OSGIN1 may regulate SLC2A3 expression through the AMPK/mTOR signaling pathway in ovarian cancer. To investigate whether OSGIN1 regulates the AMPK/mTOR signaling pathway, we knocked down and overexpressed OSGIN1 in two ovarian cancer cell lines and evaluated the expression levels of p-AMPK and p-mTOR. OSGIN1 knockdown significantly inhibited αT172 phosphorylation in SKOV3 and OVCAR3 cells, and this effect was reversed by OSGIN1 overexpression (Fig. 4d, e). These findings indicate that OSGIN1 is involved in regulating the AMPK/mTOR signaling pathway.
To further investigate whether the AMPK/mTOR signaling pathway regulates SLC2A3, we treated OVCAR3 cells with AICAR (an AMPK activator) or Compound C (an AMPK inhibitor). Remarkably, treatment with AICAR (1 mmol/L), which activates AMPK, led to a significant downregulation of SLC2A3 expression (Fig. 4g). Conversely, treatment with Compound C (50 μmol/L), which inhibits AMPK, significantly upregulated SLC2A3 expression (Fig. 4g). To ask whether mTORC1 regulates SLC2A3 promoter activity, we constructed a SLC2A3 promoter-driven luciferase reporter and assayed its activity in SKOV3 or OVCAR3 cells. The luciferase activity decreased in response to rapamycin treatment (Fig. 4h). These findings suggest that OSGIN1 likely influences the AMPK/mTOR signaling pathway, which, in turn, regulates the expression of SLC2A3.
OSGN1 activates AMPK signaling through ATM, leading to ferroptosis
To understand how OSGIN1 regulates AMPK signaling, we searched BioGRID (https://thebiogrid.org/), a protein‒protein interaction database, for potential OSGIN1-interacting proteins. Among all candidate interactors, the DNA damage response serine/threonine kinase ATM (mutated in Ataxia-Telangiectasia) was reported to be an essential factor in ferroptosis, and silencing ATM suppressed erastin-induced ferroptosis in renal cell carcinoma24. Indeed, ATM, but not ATR, was pulled down with SFB-tagged OSGIN1 protein by S-protein beads but not with SFB-tagged GFP (Fig. 5a). ATM has been demonstrated to undergo autologous phosphorylation at Ser-1981, leading to the phosphorylation of AMPK and subsequently influencing cellular gene expression and energy metabolism25. In our study, we found that the overexpression of OSGIN1 activated AMPK signaling (Fig. 4d, e). This activation could be reversed by ATM knockdown or treatment with KU60019 (an ATM kinase inhibitor) at a concentration of 5 μM (Fig. 5b). These findings suggest that OSGIN1 facilitates the interaction between ATM and AMPK, leading to the activation of the AMPK signaling pathway through ATM. Collectively, these data indicate that OSGIN1 plays a role in modulating the ATM-AMPK interaction.
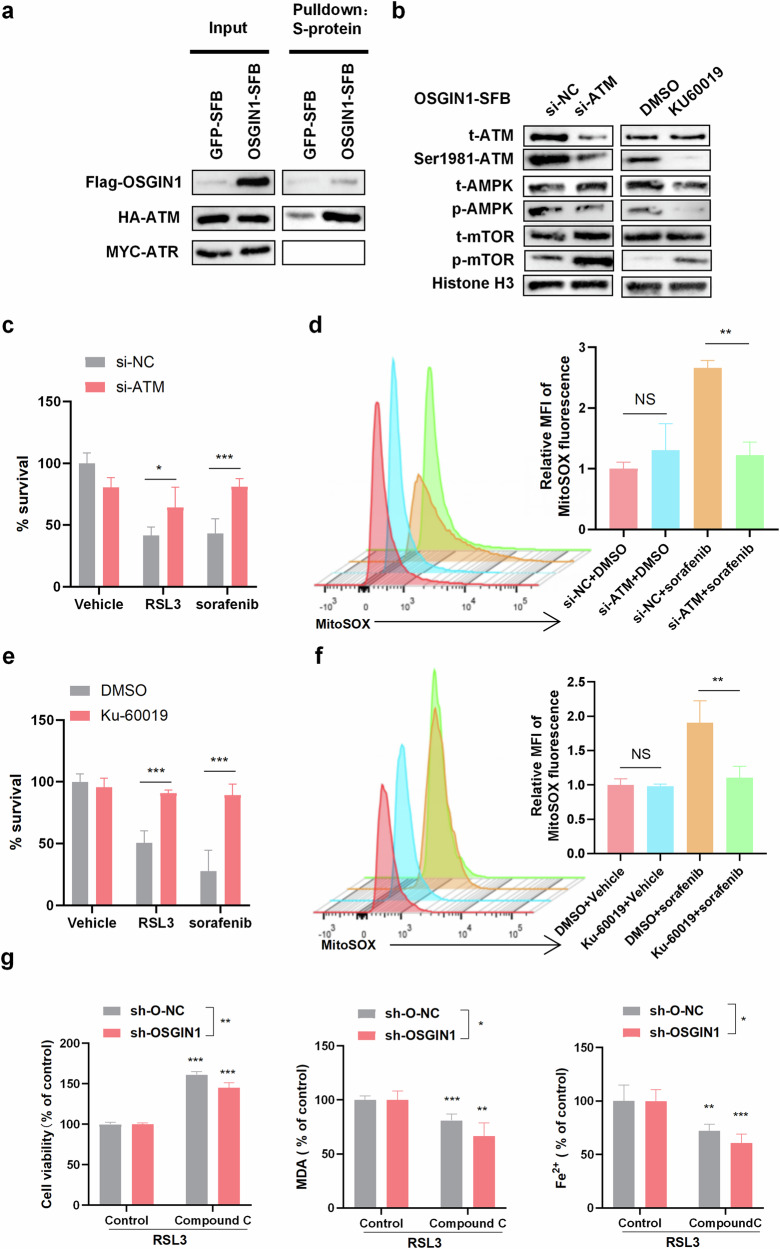
Fig. 5. OSGN1 activates AMPK signaling through ATM.
a HEK293T cells were transfected with HA-FLAG-ATM and SFB-tagged GFP or OSGIN1. The OSGIN1-SFB protein was pulled down with S-protein beads, followed by immunoblotting with antibodies against FLAG and HA. HEK293T cells were transfected with MYC-ARM and SFB-tagged GFP or OSGIN1. The OSGIN1-SFB protein was pulled down with S-protein beads, followed by immunoblotting with antibodies against FLAG and MYC. b Control and OSGIN1-overexpressing OVCAR3 cells were transduced with ATM siRNA and immunoblotted with the indicated antibodies. c OVCAR3 cells were transfected with si-NC or si-ATM for 48 h and then incubated with DMSO, RSL3 (10 μM) or sorafenib for 12 h. Cell viability or death was determined with a CCK8 kit. d Mitochondrial ROS were quantified in ovarian cancer cells that were transfected with siNC or siATM for 48 h and then incubated with DMSO or sorafenib (10 μM) for 12 h. e, f OVCAR3 cells were treated with Ku-60019 (3 μM) for 72 h and then treated with DMSO, RSL3 (10 μM) or sorafenib for 12 h before viability determination with a CCK8 kit. Mitochondrial ROS were quantified in ovarian cancer cells that were transfected with siNC or siATM for 48 h and then incubated with DMSO or sorafenib (10 μM) for 12 h. g OVCAR3 cells were transfected with sh-O-NC or sh-OSGIN1. The transfected cells were treated with 1 μM RSL3 combined with or without 5 μM compound C for 12 h. Cell viability, iron concentration, the GSH/GSSG ratio, and MDA levels were measured with a CCK8 kit, an iron assay kit, a GSH/GSSG ratio detection assay kit, and MDA assay reagent, respectively. The data are presented as the means ± SD. Each experiment was repeated three times. Statistical significance was determined by a two-tailed unpaired t-test (d and f). The p values in c, e and g were determined by one-way ANOVA with multiple comparisons. *p < 0.05; **p < 0.01 and ***p < 0.001; NS, p > 0.05.
Based on the aforementioned findings, we further investigated the essential role of ATM in ferroptosis. We observed that silencing ATM rescued OVCAR3 cells from cell death triggered by sorafenib and RSL3 (Fig. 5c). This observation led us to focus on the functional role of ATM inhibition in protecting against ferroptosis. To confirm that the protective effect of ATM inhibition is specifically associated with ferroptosis, we quantified mitochondrial ROS levels in ovarian cancer cells that were transfected with siNC or siATM for 48 h and then treated with DMSO or sorafenib (10 μM) for 12 h. The si-ATM OVCAR3 cells exhibited significantly lower levels of mitochondrial ROS than did the scrambled control cells (Fig. 5d, e). Having confirmed that genetic silencing of ATM protected cells from erastin- or sorafenib-induced ferroptosis, we further investigated whether chemical inhibitors of ATM had similar effects. Treatment with the specific and structurally distinct ATM inhibitor Ku-60019 (3 μM) for 72 h significantly protected OVCAR3 cells from the ferroptosis phenotype induced by erastin (Fig. 5f, g). These findings highlight the critical role of ATM in ferroptosis.
To further explore the role of AMPK in OSGIN1-enhanced ferroptosis, cells were treated with Compound C to inhibit AMPK phosphorylation. As anticipated, Compound C not only inhibited AMPK phosphorylation but also counteracted the growth inhibitory effects of RSL3, which is known to inhibit glutathione peroxidase 4 (GPX4) and activate ferroptosis26. Moreover, Compound C reduced the accumulation of redox-active iron and the production of MDA in both sh-O-NC- and sh-OSGIN1 ovarian cancer cells (Fig. 5h). Collectively, these data indicate that OSGN1 activates AMPK signaling through ATM and that ATM depletion, or inhibition of its enzymatic activity, robustly confers ferroptosis resistance. These data indicate that AMPK is involved in OSGIN1-induced ferroptosis. Collectively, these findings indicate that OSGIN1 activates the AMPK signaling pathway through its interaction with ATM and that ATM depletion, or inhibition of its enzymatic activity, confers resistance to ferroptosis.
OSGIN1 promotes ferroptosis, whereas SLC2A3 inhibits ferroptosis
To understand the relationship between SLC2A3 and ferroptosis, lentiviruses carrying the SLC2A3 gene (OE-SLC2A3) or control lentiviruses (OE-S-NC) were administered to SKOV3 and OVCAR3 cells. The cells treated with erastin, RSL3 or FIN56 (an inducer of ferroptosis) were then subjected to flow cytometry analysis to measure the levels of 7-AAD (an indicator of DNA insertion and membrane integrity) and annexin V (a marker of phosphatidylserine accessibility). 7-AAD and annexin V are indicators of cell death but are not definitive markers of ferroptosis, apoptosis, or necroptosis. Therefore, we calculated the percentage of double-negative cells as a measure of 7-AAD and annexin V cell activity27.
The results demonstrated that SLC2A3 overexpression protected SKOV3 and OVCAR3 cells against cell death induced by erastin or RSL3 treatment (Fig. 6a, b). To better understand the relationship between ovarian cancer cell death and ferroptosis, the ferroptosis inhibitor liproxstatin-1 and the iron chelator desferrioxamine (DFO) were applied28. The results showed that SLC2A3 knockdown in OVCAR3 cells increased ferroptosis in response to erastin/RSL3/FIN56 treatment, which was reversed by simultaneous treatment with liproxstatin-1 or DFO (Fig. 6c). A similar effect on the levels of cellular MDA, ROS and iron was also observed (Fig. 6c–e). These findings suggest that SLC2A3 can inhibit ferroptosis.
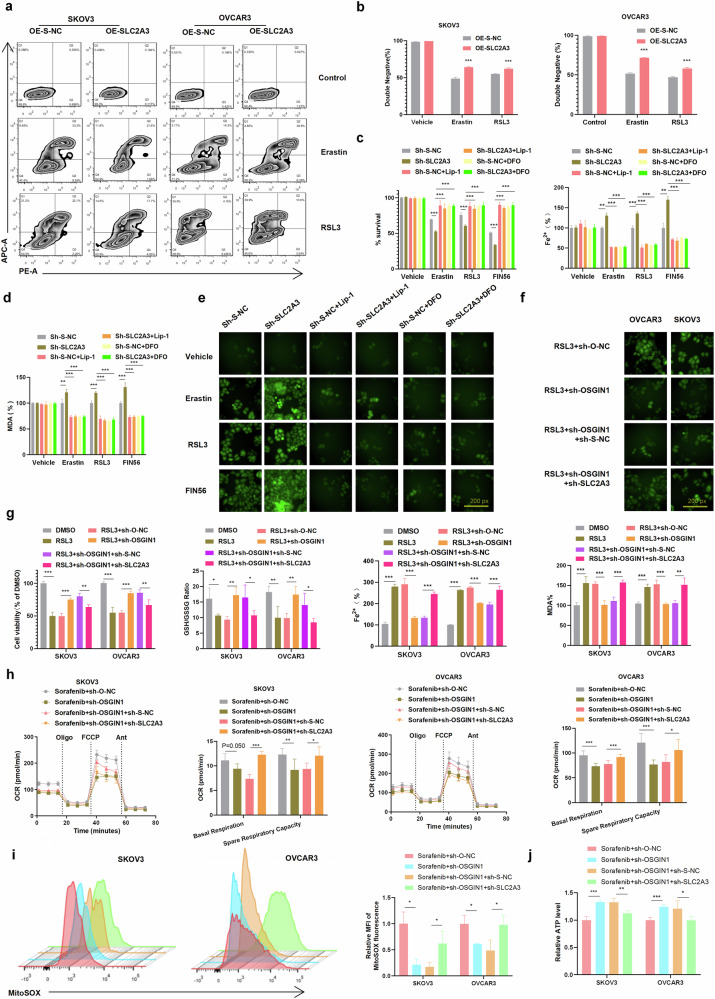
Fig. 6. OSGIN1 regulated ovarian cancer cell ferroptosis by suppressing SLC2A3 expression.
a, b SLC2A3 was overexpressed in SKOV3 and OVCAR3 cells after treatment with 10 μM erastin or 1 μM RSL3 for 12 h. G: 7-Aminoactinomycin (7-AAD) and annexin V staining results. H: The percentage of the annexin V and 7-AAD double-negative population. c, d The cell viability/Fe2 + /MDA level of OVCAR3 cells after SLC2A3 transfection and treatment with 10 μM erastin, 1 μM RSL3, and 50 μM FIN56, alone or with liproxstatin-1 (lip-1, 10 μM) or DFO (100 μM). e ROS levels in sh-S-NC/sh-SLC2A3-transfected SKOV3 cells following treatment with 10 μM erastin, 1 μM RSL3, or 50 μM FIN56 alone or with 10 μM liproxstatin-1 (lip-1) or 100 μM DFO. f ROS levels in sh-O-NC/sh-OSGIN1- or sh-S-NC/sh-SLC2A3-transfected SKOV3 cells after treatment with 1 μM RSL3. g The expression of OSGIN1 and/or SLC2A3 was downregulated in SKOV3 and OVCAR3 cells, alone or in combination with 1 μM RSL3. Cell viability, iron concentration, the GSH/GSSG ratio, and MDA levels were measured with a CCK8 assay, an iron assay kit, a GSH/GSSG ratio detection assay kit, and MDA assay reagent, respectively. h OCRs of SKOV3 and OVCAR3 cells treated with sorafenib+sh-O-NC, sorafenib+sh-OSGIN1, sorafenib+sh-OSGIN1+sh-S-NC, and sorafenib+sh-OSGIN1+sh-S-SLC2A3. i The rates of ROS generation by mitochondria in living cells from mutant and control cell lines were analyzed by FlowJo using the mitochondrial superoxide indicator MitoSOX Red (5 mM). Flow cytometry histogram showing MitoSOX Red fluorescence in various cell lines. Relative ratios of MitoSOX fluorescence intensity. The average of three determinations for each cell line is shown. j Effect of sh-OSGIN1 on ATP levels in OVCAR3 cells. Each experiment was repeated three times. The p values in c, d and g–j were determined by two-way ANOVA with multiple comparisons. The p values in b were determined by one-way ANOVA with multiple comparisons. Error bars are s.e.m. *p < 0.05; **p < 0.01 and ***p < 0.001.
To investigate the relationship between the OSGIN1-SLC2A3 axis and ferroptosis, we evaluated ferroptosis-related cellular markers, including MDA levels, ROS levels, the GSH/GSSG ratio, and iron levels, in sh-SLC2A3 and control cells infected with the sh-OSGIN1 virus after treatment with RSL3 (1 μM). As expected, knockdown of OSGIN1 protected SKOV3 and OVCAR3 cells against RSL3-induced cell death following 12 hours of RSL3 treatment. Moreover, compared to that in control cells, the cellular viability of OSGIN1-knockdown ovarian cancer cells was restored after SLC2A3 knockdown and RSL3 treatment. Consistently, SLC2A3 knockdown also reversed the effect of OSGIN1 downregulation on cellular MDA/iron levels and the GSH/GSSG ratio (Fig. 6f, g). These findings suggest that inhibiting SLC2A3 in OSGIN1-depleted ovarian cancer cells enhances ferroptosis.
To establish the biochemical mechanism responsible for the effect of SLC2A3 on sorafenib-induced ferroptosis in sh-OSGIN1 cells, we assessed mitochondrial function, including OCRs, ATP levels, and ROS levels, in control and sh-OSGIN1 cells (Fig. 6h–j). Remarkably, OSGIN1 silencing significantly inhibited the basal respiratory OCR, alternate respiratory capacity OCR, and mitochondrial ROS in SKOV3 and OVCAR3 cells following treatment with sorafenib (10 μM) for 12 h. This inhibitory effect was partially reversed by cotransfection with sh-SLC2A3, as depicted in Fig. 6h. Furthermore, we evaluated the cellular ATP level in sh-SLC2A3 and control cells infected with the sh-OSGIN1 virus after treatment with sorafenib (10 μM) for 12 h. While the control cells exhibited a significant increase in ATP levels, sh-SLC2A3 resulted in a substantial decrease in cellular ATP levels in OSGIN1-downregulated cells (Fig. 6j). These results suggest that when OSGIN1 is deficient, SLC2A3 significantly contributes to mitochondrial ROS, mitochondrial basal oxygen consumption and inhibitory control ATP production. Overall, these results demonstrate that OSGIN1 regulates ferroptosis in ovarian cancer cells by suppressing SLC2A3.
OSGIN1 reduces resistance to sorafenib in ovarian cancer in vivo
In this study, the effect of OSGIN1 on ovarian tumors was evaluated using a NOD/SCID mouse xenograft model. Five types of human ovarian cancer cells, namely, OVCAR3 (noninfected, OE-O-NC, OE-OSGIN1, OE-OSGIN1 + OE-S-NC, and OE-OSGIN1 + OE-SLC2A3), were implanted into NOD/SCID mice. The mice were then intraperitoneally injected with either saline or sorafenib starting on Day 8 and at an interval of 2 days for 30 days. At the end of the experiment, the mice were sacrificed by CO2 inhalation, and the tumors were collected and weighed. The tumors in the mice transplanted with OVCAR3 (noninfected) cells and injected with saline continued to grow over time. In contrast, the tumors in the sorafenib-treated mice were smaller than those in the saline-treated mice, indicating that sorafenib inhibited the growth of ovarian tumors in vivo (Fig. 7b, c). OSGIN1 and SLC2A3 expression in the tumors was measured using IHC and qRT‒PCR assays. The results showed that OSGIN1 expression was upregulated and SLC2A3 expression was downregulated in the mice transplanted with OVCAR3 (OE-OSGIN1) cells (Fig. 7a). Furthermore, tumors in mice injected with OVCAR3 (OE-OSGIN1) cells and treated with sorafenib (sorafenib+OE-OSGIN1) were significantly smaller than those in mice injected with OVCAR3 (OE-O-NC) cells (sorafenib + OE-O-NC) (Fig. 7b, c). This result indicated that OSGIN1 overexpression promoted the antitumor effects of sorafenib. However, the tumors of sorafenib-injected mice implanted with OE-OSGIN1 + OE-SLC2A3-infected cells were larger than those in mice implanted with OE-OSGIN1 + OE-S-NC-infected cells (sorafenib + OE-OSGIN1 + OE-S-NC), which suggests that SLC2A3 overexpression decreased the antitumor effect of OSGIN1.
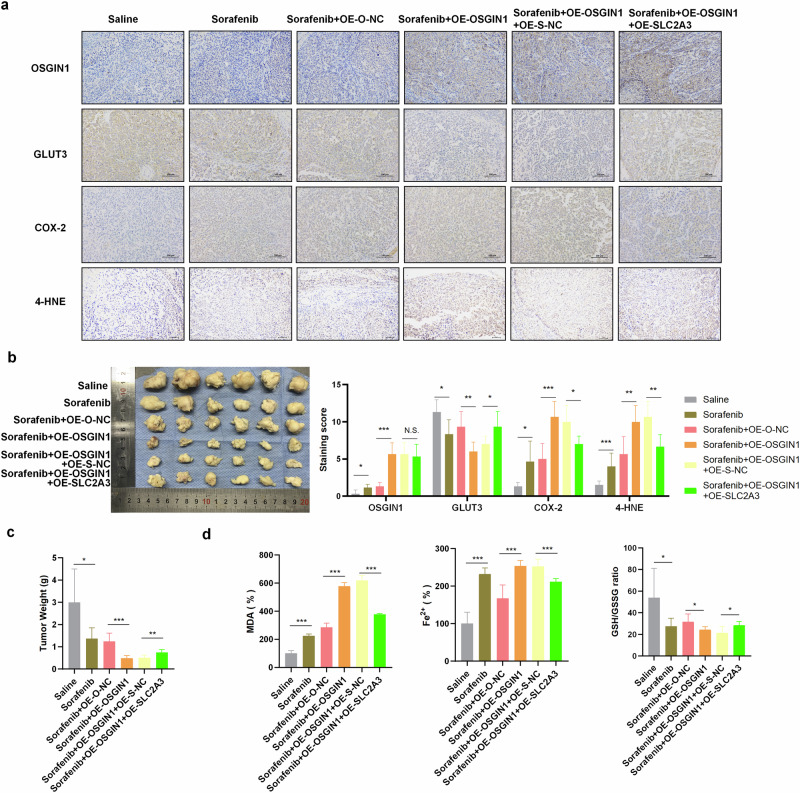
Fig. 7. OSGIN1 decreased the resistance of ovarian cancer to sorafenib in vivo.
a Relative protein levels of OSGIN1, GLUT3, COX-2 and 4-HNE in mouse tumor tissues. b Representative images of tumors from each group. n = 6 in each group. c Tumor weight in each experimental group. d MDA content in tumor tissues from each group. Cellular GSH/GSSG ratio in tumors from the indicated groups. Cellular iron levels in tumor tissues from the indicated groups. Each experiment was repeated three times. The p values in b were determined by two-way ANOVA with multiple comparisons. Statistical significance in c, d was determined by a two-tailed unpaired t-test. Error bars are s.e.m. *p < 0.05; **p < 0.01 and ***p < 0.001.
At the end of the experiment, the mice were euthanized, and the tumors were collected for further analysis. COX-2, 4-HNE, MDA, ROS, and iron levels were increased in the tumors following sorafenib treatment. Conversely, there was a decrease in the GSH/GSSG ratio (Fig. 7a, d). The overexpression of OSGIN1 exacerbated these changes. However, the coexpression of OSGIN1 and SLC2A3 reversed the anticarcinogenic effect induced by OSGIN1 overexpression (Fig. 7a, d).
SLC2A3 mediates ferroptosis resistance and is an actionable target for enhancing sorafenib efficacy
To evaluate the potential clinical implications of combining sorafenib with SLC2A3 neutralizing antibodies, we administered sorafenib at a dose of 30 mg/kg body weight via oral gavage once daily and SLC2A3-neutralizing antibodies at a dose of 100 μg per mouse via intraperitoneal injection once daily, both until day 21. This treatment regimen did not result in observable changes in body weight or clinical symptoms (Supplementary Fig. 4a). Subsequent post-mortem analyses revealed no significant differences in the weights of major organs (Supplementary Fig. 4b, c), and serum biochemical tests indicated the absence of hepatotoxicity, as evidenced by normal ALT and AST levels (Supplementary Fig. 4d). Furthermore, treatment did not impact colon length (Supplementary Fig. 4e), and hematological evaluations revealed no signs of anemia, leukocyte reduction, or leukocytosis among mice subjected to the combination therapy (Supplementary Fig. 4f). Additionally, platelet, lymphocyte, monocyte, and granulocyte counts remained unchanged (Supplementary Fig. 4f, g). Cardiovascular assessments, encompassing heart rate (Supplementary Fig. 4h) and blood pressure (Supplementary Fig. 4i), demonstrated no significant deviations between mice treated with the combination therapy and those receiving IgG + vehicle, thereby confirming the excellent safety profile of sorafenib and SLC2A3 neutralizing antibodies in combination.
To address whether SLC2A3-targeting agents can improve the therapeutic efficacy of sorafenib, we established ovarian cancer patient-derived xenograft (PDX) models in NOD-SCID-IL2Rγc–/– mice. In preclinical testing, we implanted PDX tumor tissues into NOD-SCID-IL2Rγc–/– mice and started treatment when the tumors reached 95–145 mm3. The mice were assigned to four treatment groups: (1) IgG + vehicle; (2) SLC2A3 antibody + vehicle; (3) IgG + sorafenib; and (4) SLC2A3 antibody + sorafenib. Sorafenib was administered at 30 mg/kg body weight by oral gavage once a day until the endpoint, and the SLC2A3-neutralizing antibody was administered at 100 μg per mouse by intraperitoneal injection once a day until the endpoint, which reduced SLC2A3 levels in the tumor tissues (Fig. 8a).
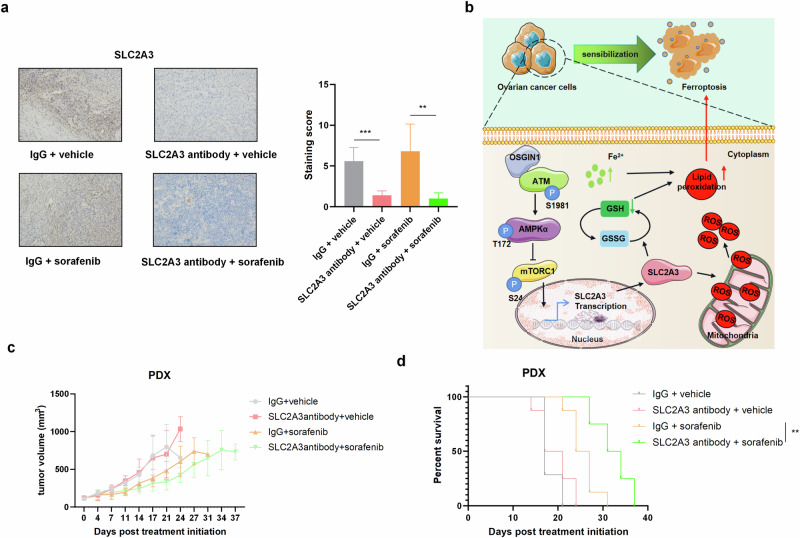
Fig. 8. An SLC2A3-neutralizing antibody enhances the antineoplastic effect of sorafenib on HGSOC patient-derived xenograft tumors with low OSGIN1 expression.
a Representative IHC staining images of tumor tissues collected from NSG mice bearing PDX models. Scale bar: 50 μm. All the data are expressed as the means ± SDs. b Model for the role of the OSGIN1−AMPK−SLC2A3 axis in ferroptosis. Loss of the OSGIN1 gene promoted the growth of ovarian cancer and induced resistance to drug-induced ferroptosis. Mechanistically, the loss of OSGIN1 activates AMPK signaling through ATM, leading to the upregulation of SLC2A3, which protects cells from ferroptosis and renders them insensitive to ferroptosis inducers. c, d Tumor growth curves and Kaplan−Meier survival curves of NSG mice bearing PDX models. Mice were treated with anti-SLC2A3 antibodies and sorafenib, either alone or in combination (n = 8). The survival time of the tumor-bearing mice was determined. Statistical significance was determined by a two-tailed unpaired t-test a and log-rank test c. Error bars are s.e.m. *p < 0.05; **p < 0.01 and ***p < 0.001.
In the PDX mouse model, treatment with the SLC2A3 neutralizing antibody alone had minimal effect on tumor growth, whereas the combination treatment achieved a greater antitumor effect than sorafenib treatment alone (Fig. 8c). Moreover, the survival time of the mice was significantly prolonged after combination treatment (Fig. 8d). We considered whether the combined effect on the PDX model was associated with ferroptosis. Collectively, these data suggested that the SLC2A3 neutralizing antibody enhanced the ferroptosis-inducing effect of sorafenib on ovarian cancer patient-derived xenograft tumors with low OSGIN1 expression and high SLC2A3 expression.
Discussion
Homologous recombination deficiency (HRD) occurs in approximately 50% of patients, including mutations in various proteins associated with HR-mediated DNA repair29. Unfortunately, 50% of high-grade serous ovarian carcinoma (HGSOC) patients typically exhibit poor responses to current treatments and worse overall survival30. ATM, a serine/threonine kinase, is crucial for HR-mediated DNA double-strand break (DSB) repair31. Germline mutations in ATM lead to ataxia-telangiectasia (A-T), which has numerous pathological consequences, including cancer predisposition and metabolic dysfunctions31. Given the heightened cancer risk in A-T patients and the development of malignancies in ATM-/- mice31,32, ATM is considered a tumor suppressor. However, many tumors rely on elevated DNA repair pathways, and recent literature suggests that tumorigenesis requires ATM33. The majority of HGSOC patients carry wild-type ATM alleles, and ATM kinase activity is upregulated in HGSOC samples compared to that in fallopian tubes34. Collectively, these studies suggest that ATM may be an actionable target in both wild-type and elevated tumor subgroups. However, ATM inhibitors have remained in clinical development for the past two decades35. Preclinical studies have indicated that ATM inhibitor monotherapy is unlikely to be effective36. Recent studies have highlighted the tumor-suppressive and metastasis-inhibiting effects of ferroptosis37. Cancer proteins and tumor suppressors, such as CSPP1, EGFR, and IDH1, have been shown to regulate ferroptosis in cancer cells38. Chen and colleagues recently conducted a genetic-based forward kinase screen for ferroptosis and identified ATM as a necessary kinase for ferroptosis induction39. Chemical inhibition or siRNA-mediated knockdown of ATM leads to primary resistance to cysteine deprivation and erastin-induced ferroptosis40. PARP inhibitors and radiotherapy sensitize ovarian cancer cells to ferroptosis through synergistic activation of ATM40. We report that OSGIN1 interacts with ATM to activate AMPK signaling, resulting in downregulation of the target gene mTOR and subsequent downregulation of SLC2A3 expression, leading to ferroptosis sensitivity (Figs. 4 and 5). Furthermore, our study extends the role of ATM kinase in ferroptosis in ovarian cancer.
In this study, we used erastin, a ferroptotic inducer, to kill ovarian cancer cells by modulating ferroptosis in vivo. To our surprise, we found that erastin treatment significantly increased the expression of OSGIN1, a gene that is weakly expressed in ovarian cancer. OSGIN1 is known as a tumor suppressor gene that is linked to liver cancer and poor patient survival11. However, the precise mechanism by which OSGIN1 expression is regulated in the context of ferroptosis remains unclear. We speculate that OSGIN1 expression may be influenced by signaling pathways involved in ferroptosis, such as the NRF2-KEAP1 pathway or the lipid peroxidation pathway8,40. These pathways have been implicated in the regulation of cellular responses to oxidative stress and could potentially modulate OSGIN1 expression in response to ferroptotic stimuli.
Our results showed that inhibiting OSGIN1 in erastin-treated ovarian cancer cells restored intracellular ROS in erastin-treated cells. There is conflicting evidence regarding the relationship between OSGIN1 and ROS, with some studies suggesting that OSGIN1 overexpression improves ROS12,23, while others have reported that OSGIN1 overexpression does not significantly increase ROS production41. Our study indicated that OSGIN1 downregulation decreases ROS levels in ovarian cancer cells and may affect mitochondrial oxidative phosphorylation. The relationship between OSGIN1 and ferroptosis is still unclear. Recent studies have indicated that OSGIN1 may have an impact on apoptosis and autophagy42. Our repeated experiments showed that Fer1 was unable to stimulate cell growth in cells treated with OE-O-NC to the same extent as that in OE-OSGIN1-treated cells. These findings suggest that OSGIN1 may not only play a role in promoting ferroptosis but also inhibits tumor growth through other pathways. Here, using human ovarian cancer cells overexpressing OSGIN1 in NOD/SCID mice, we demonstrated that OSGIN1 is a bona fide ovarian cancer suppressor and that its loss confers resistance to drug-induced ferroptosis (Fig. 2). Thus, OSGIN1 loss not only promotes ovarian cancer progression but also renders ovarian cancer cells resistant to ferroptosis.
In this study, both 2D and 3D models were utilized to investigate the differences in sensitivity to ferroptosis. Although 2D in vitro cell culture models are more practical for analyzing cell death signaling, there are significant differences in ferroptosis sensitivity between 2D and 3D models. In 3D models, such as organoids, xenografts, or human cancers, the natural tumor microenvironment inhibits ferroptosis, and the expression of antioxidant genes, such as GPX4, is upregulated to counteract oxidative stress. On the other hand, the 2D monolayer culture model does not exhibit ferroptosis without artificial intervention, and GPX4 expression is relatively low.
Notably, when xenograft tumors derived from ovarian cancer patients were treated with a combination of sorafenib and SLC2A3 neutralizing antibodies, better treatment responses were achieved compared to those with sorafenib treatment alone. This finding highlights the potential of combining targeted therapies to improve treatment outcomes in patients with ovarian cancer.
Furthermore, considering that radiotherapy and immunotherapy trigger lipid oxidation and ferroptosis by inhibiting system Xc-40, we hypothesize that neutralizing antibodies against SLC2A3 may have the potential to sensitize tumors to radiation and immunotherapy, which warrants further investigation. In summary, this study revealed an actionable axis that controls ovarian cancer iron homeostasis, tumor progression, and vulnerability to ferroptosis, which may pave the way for targeted ferroptosis strategy to improve cancer therapy.
Methods
Collection of human ovarian cancer specimens
Human ovarian tumor specimens (N = 15) and normal ovarian tissues (N = 14) were collected from patients with ovarian tumors at Beijing Obstetrics and Gynecology Hospital Affiliated Capital Medical University. Fifteen ovarian tissue samples from serous ovarian cancer patients, including four low-stage (FIGO stage I and II) and eleven advanced-stage (FIGO stages III and IV) patients, were collected at the time of primary ovarian cancer surgery (patient details are summarized in Supplementary Table 1). Fourteen ovarian tissue samples were obtained from nonovarian carcinoma patients who underwent hysterectomy or bilateral salpingo-oophorectomy due to prediagnosed medical conditions (patient details are summarized in Supplementary Table 2). This study was approved by the ethics committee of Beijing Obstetrics and Gynecology Hospital Affiliated Capital Medical University (2024-KY-101-01) and written informed consent was obtained from each participant. The study complied with the Declaration of Helsinki.
Cell culture and reagents
The ovarian cancer cell lines SKOV3 (ATCC, Cat # HTB-77) and ES-2 (ATCC, Cat # CRL-1978) were cultured in modified medium (McCoy’s 5A, Procell Life, PM150710, China). The ovarian cancer cell lines A2780 (ECACC, Cat # 93112519), COC1 (CCTCC, Cat # CL-0064) and OVCAR3 (ATCC, Cat # HTB-161) were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Procell Life, PM150110, China). As we described previously43, the culture media were supplemented with 10% fetal bovine serum (Gibco, 16000044, USA) and 100 units/ml penicillin‒streptomycin solution (Procell Life, PB180120, China). The cell lines selected for this study represent diverse histological subtypes of ovarian cancer. SKOV3 cells are commonly associated with endometrioid histology, whereas OVCAR3 cells are typically representative of high-grade serous ovarian cancer. Additionally, ES-2 cells exhibit characteristics of clear cell histology, while A2780 cells are not of likely ovarian cancer origin. The cells were cultured at 37 °C and 5% CO2. These ovarian cancer cell lines were donated by Procell Life. All cell lines were authenticated and tested negative for mycoplasma contamination.
Cell grouping and transfection
Prior to transfection, the cells were seeded onto six-well plates and incubated for 24 h. Once the cells reached 75% confluence, transfection was performed using a Lipofectamine 3000 kit (Thermo Fisher Scientific, L3000015, UK). For transfection, the following constructs were used: OSGIN1 (sh-OSGIN1), OSGIN1 (OE-OSGIN1), and SLC2A3 (sh-SLC2A3). The control vectors used were sh-O-NC, sh-S-NC, and oe-S-NC. We used reverse transcription (RT) to determine the knockdown efficiency of sh-OSGIN1#1, sh-OSGIN1#2, sh-OSGIN1#3, sh-SLC2A3#1, sh-SLC2A3#2, and sh-SLC2A3#3. The levels of OSGIN1 and SLC2A3 knockdown met the minimum threshold required for further research. All plasmids, vectors, sequences, viral packet, and titer testing were performed by the SyngenTech Company (Beijing, China).
Immunohistochemistry (IHC)
Cells were fixed with 4% paraformaldehyde (PFA), wrapped in paraffin, and sectioned into 5-μm thick slices. The sections were incubated in sodium citrate buffer (10 mM, pH 6.0) at 95 °C for 20 min to retrieve the antigen. The sections were blocked with 5% goat serum diluted in PBS and incubated with antibodies against the following target proteins overnight at 4 °C: OSGIN1 (1:200, Abcam, Ab68793, USA), GLUT3 (1:200, Abcam, Ab41525, USA), COX-2 (1:100, Abcam, Ab179800, USA), and 4-HNE (1:50, Abcam, Ab46545, USA). The sections were then stained with the SPlink assay reagent (ZSGB-BIO) (Biotin-Streptavidin HRP assay system) and developed using a DAB kit (ZSGB-Bio) following the manufacturer’s instructions. Immunohistochemical staining was semiquantitatively analyzed using the immunoreactive score (IRS) system. The percentage of positive cells was scored on a scale of 0–4: 0 if 0% of tumor cells were positive, 1 if 1–10% were positive, 2 if 11–50% were positive, 3 if 51–80% were positive, and 4 if 81–100% were positive. The staining intensity was scored on a scale of 0–3 (3 is the strongest). The final IRS was calculated as (score of the staining intensity) × (score of the percentage of positive cells).
Cell viability assay
Ovarian cancer cells were seeded in a 96-well plate at a density of 1 × 105 cells/well in 100 ml of culture medium. After preincubation for 36 h, 10 μL of Cell Counting Kit-8 (CCK8) solution was added to each well, and the plates were incubated at 37 °C in a 5% CO2 incubator for 1 hour. Finally, the absorbance of the solution was measured using a SpectraMax M5 spectrophotometer at a wavelength of 450 nm.
Cellular ROS, MDA, GSH/GSSG and iron detection
Intracellular ROS were measured using the DCFDA/H2DCFDA-Cellular ROS Assay (Abcam, ab113851, UK). Briefly, PBS was used to wash each cell twice. Then, 1 mL of DCFH-DA solution (at a final concentration of 10 μM) was added to each well and incubated in a CO2 incubator at 37 °C for 30 min. The cells were then washed with PBS three times to remove the DCFH-DA that did not enter the cells. Cellular ROS content assessment was performed using a fluorescence microscope. MDA (Abcam, ab118970, UK), the GSH/GSSG ratio (Abcam, ab138881, UK) and iron levels (Abcam, ab83366, UK) were estimated using commercial kits. Ovarian tissues or cells were washed with PBS, homogenized in lysis buffer and sonicated. After sonication, the lysed tissue was centrifuged (10,000 × g, 10 min) to remove debris, and the supernatant was retained. A microplate reader was used to measure the levels of MDA, GSH, GSSG and iron in the supernatant. In addition, the MDA, GSH, GSSG, and iron levels were normalized according to the protein concentration.
RNA extraction and qRT‒PCR
Total RNA was extracted using TRIzol Reagent containing DNaseI to prevent DNA contamination (CWBIO, CW0580, China). The extracted RNA was reverse-transcribed to cDNA using a commercially available UltraSYBR One Step RT‒qPCR Kit (CWBIO, CW0659, China). The samples were mixed with SYBR Green Principal Component Mix and subjected to PCR. The mRNA expression of OSGIN1 and SLC2A3 was normalized to that of GAPDH, which was used as an internal control. The relative expression was calculated using the 2-ΔΔCt method. The primers for the target genes were designed by GenePharma and are listed below:
OSGIN1 forward primer: 5′-GCAGAAGAAGCGAAGAGGTC-3′;
OSGIN1 reverse primer: 5′-CTACAGCACCGGACACAAAG-3′;
SLC2A3 forward primer: 5′-ATTGTGCTCCAGCTCTCTCA-3’′;
SLC2A3 reverse primer: 5′-CTCTGGGTTCTCTGCCGTAG-5′;
GAPDH forward primer: 5′-GAGTCCTTCTTCCTGCCCTT-3′;
GAPDH reverse primer: 5′-TCAGCTCAGGGATGACCTTG-3′.
Western blotting
The cells were lysed using RIPA lysis buffer (Beyotime, P0013B, China) containing protease and phosphatase inhibitors (GenDEPOT). The extracted proteins were separated using 4–20% SDS‒PAGE and transferred to nitrocellulose membranes. After transfer, the membranes were blocked with 5% skim milk mixed with 0.05% Tween-20 (TBST) diluted in Tris buffer. The membranes were then incubated with primary antibody overnight at 4 °C, followed by incubation with secondary antibody. The protein bands were visualized with an enhanced ECL chemiluminescence kit (Beyotime, P0018M, China). The primary antibodies used were OSGIN1 (1:1000, Abcam, Ab68793, USA), GLUT3 (1:1000, Abcam, Ab41525, USA), mTOR (1:1000, Abcam, Ab134903, USA), phospho-mTOR (1:1000, Abcam, Ab109268, USA), AMPKα (1:1000, Abcam, Ab207442, USA), phospho-AMPKα (1:1000, Abcam, Ab133448, USA), ATR (1:15000, Abcam, Ab10312, USA), ATM (1:1000, Abcam, Ab32420, USA), phospho-Ser1981-ATM (1:1000, Abcam, Ab308338, USA), alpha-Tubulin (1:5000, Abcam, Ab7291, USA) and GAPDH (1:1000, Abcam, Ab8245, USA). All original images are provided (Supplementary Figs. 5–12). All blots within each relevant panel were derived from same experiment and processed in parallel.
Flow cytometry
The harvested cells were washed twice with a freezing solution of 2% FBS in PBS and then resuspended in 5 µL of APC Annexin V and 5 µL of 7-AAD Viability Staining Solution (BioLegend, 640930) at a concentration of 1 ×106 cells. The cells were gently shaken at room temperature (25°C) and incubated in the dark for 15 min for subsequent 7-AAD and annexin V staining. Then, 400 μL of Annexin V-Buffer was added to each tube, and the samples were subjected to flow cytometry analysis. To evaluate lipid peroxidation, the cells were incubated at 37 °C in medium supplemented with 2 μM BODIPY 581/591C11 (Thermo Fisher, D3861) for 30 min. The cells were resuspended in fresh cell-staining buffer containing 2% FBS and analyzed with an Attune NxT Flow Cytometer (Invitrogen) and FlowJo software. Gating strategies used to determine cell percentages are detailed in Supplementary Figs. 13 and 14.
RNA-seq analysis
To assess the quality of the FASTQ files, mRNA sequencing was conducted on replicate RNA samples transfected with sh-O-NC (n = 3) and sh-OSGIN1 (n = 3) using sequencing services provided by Wekemo Tech Group Ltd. Total RNA was then extracted with a TRIzol Plus RNA Purification Kit (Invitrogen, #12183555) following the manufacturer’s protocol. RNA-seq was then performed by a NovaSeq 6000 (Illumina), and the results were analyzed by the HISAT2 – StringTie – Ballgown pipeline. Gene Ontology enrichment analysis was performed through the Gene Ontology Resource (http://geneontology.org/). An unbiased representation of the RNA-seq results is provided in the Supplementary Dataset 1.
TCGA analysis
In the TCGA analysis, we analyzed the expression of OSGIN1 in serous ovarian cancer tissues (from TCGA, n = 426) and normal ovarian tissues (from GTEx, n = 88) using Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/). This analysis included all subtypes of ovarian cancer available in the TCGA dataset.
Splicing reporter assay
5 × 104 OVCAR3 cells were plated in a 12-well plate. After culturing overnight, cells were co-transfected with SLC2A3_2-specific siRNA and the pTN24 splicing reporter plasmid, which expresses a functional luciferase upon removal of a translation termination signal sequence by splicing, and expresses constitutive β-galactosidase reporter for normalization. 48 h after transfection and rapamycin treatment, cells were harvested for detection of reporter expression using the Dual Light Reporter System (Applied Biosystems). Data was analyzed by calculating the ratio of luciferase to β-galactosidase signals. Statistical analysis was performed with Student’s t test.
In vivo studies
NOD/SCID mice (5–6 weeks old, weighing 20–22 g) were housed at Beijing Weitahe Experimental Animal Technology Co., Ltd. (JING) (production license number: SCXK (JING) 2021-0006; license number: SYXK (JING) 2017-0033). The mice were handled following the ethical standards for experimental animals provided by the Chinese Ministry of Science and Technology GB/T35892-2018 “Guiding Opinions on Ethical Review of Laboratory Animal Welfare”. All animal experiments were approved by the National Center for Protein Science (JUN) 2020-0002. During intraperitoneal injections, gavage administration, and euthanasia procedures, the mice were not subjected to anesthesia and no analgesics were administered to alleviate pain.
Mouse tumor growth models
Ovarian cancer cells were infected with oe-O-NC, oe-OSGIN1, oe-OSGIN1 + oe-S-NC, or oe-OSGIN1 + oe-SLC2A3 lentivirus for 48 h. Subsequently, NOD/SCID mice were intraperitoneally injected in the lower left abdominal quadrant (5 × 105 cells per mouse) to induce tumor formation. The saline-treated group and sorafenib-treated group also included NOD/SCID mice inoculated with noninfected OVCAR3 cells. OVCAR3 tumor cells were intraperitoneally injected into mice (5 ×105 cells/mouse). Each group comprised 5 mice. For sorafenib treatment, 10 mg/kg sorafenib was intraperitoneally injected into the mice every 2 days beginning on the 8th day. Mice in the saline group received an intraperitoneal injection of saline every 2 days beginning on the 8th day. The mice were then intraperitoneally injected with either saline or sorafenib starting on Day 8 and at an interval of 2 days for 30 days. At the end of the experiment, the mice were sacrificed by CO2 inhalation, and the tumors were collected and weighed. Fresh tumor tissues were processed and subjected to real-time qPCR or IHC as previously described44.
Blood pressure and heart rate measurements
Five-week-old female NOD-SCID-IL2Rγc–/– mice were intraperitoneally administered sorafenib orally at a dose of 30 mg/kg body weight once daily until day 21 in NOD-SCID-IL2Rγc–/– mice. Additionally, SLC2A3-neutralizing antibodies were administered via intraperitoneal injection once daily at a dose of 100 μg per mouse until day 21. Blood pressure (including systolic, diastolic, and mean blood pressure) and heart rate were measured one day after the final administration.
Blood and organ collection
Blood was collected from surviving mice and used for complete blood cell counts (CBCs) and plasma collection, as we described previously45. The CBC was measured with an automated BC-2800 Vet cell counter (Mindray, China). The body and organ weights of the heart, liver, lung, kidney and spleen were determined for each euthanized mouse.
Patient-derived xenograft (PDX) models
Human ovarian cancer tissue was obtained from Beijing Obstetrics and Gynecology Hospital Affiliated Capital Medical University. PDX tumors in cold Dulbecco’s modified Eagle’s medium (DMEM) were minced into 1–2 mm3 fragments, and each fragment was subcutaneously transplanted into the dorsal flank of 5-week-old female NOD-SCID-IL2Rγc–/– mice. Tumors were measured twice weekly with calipers. The tumor volume was calculated according to the formula: volume = 0.5 × length × width2. Treatment started when the tumors were between 95 and 145 mm3. Mice were assigned to treatment groups based on the average tumor volume per group. Mice were culled when the tumors reached a volume of 600 mm3.
Ethics approval and consent to participate
The study was carried out in accordance with the Declaration of Helsinki and was approved by the ethics committee of Beijing Obstetrics and Gynecology Hospital Affiliated Capital Medical University (Approval No. IEC-B-03-V01-FJ1). All animal experiments were conducted following the principles outlined in the ARRIVE guidelines and the Basel Declaration. All animal experiments were approved by the National Center for Protein Science (JUN) 2020-0002 (Approval No. IACUC-20211116-37MT).
Statistical analysis
The data are presented as the means ± SD or means ± SEM from at least three biological replicate experiments, as indicated in the corresponding figure legends. Statistical analysis was performed by t tests, one-way ANOVA and two-way ANOVA as indicated in the corresponding figure legends using GraphPad Prism (version 8.1.0). The statistical significance is annotated as follows: ns, nonsignificant; *p < 0.05; **p < 0.01 and ***p < 0.001 Differences with p < 0.05 were considered to indicate statistical significance.
Supplementary information
Supplementary Table 1-2 and Supplementary Figure 1-14
Acknowledgements
We thank the Home for Researchers editorial team (www.home-for-researchers.com) for language editing services. Special thanks to Zhang Jinxu, Zhang Wenjuan, Jin Zhaoyu, Liu Hailong and Wu Ming from the National center for protein sciences (Beijing) for their contributions to cellular experiments procedures. We appreciate Qiu Chen’s guidance on animal experimental procedures. This study was funded by the Beijing Hospitals Authority’s Ascent Plan (Grant number: DFL20221201), National Natural Science Foundation of China (Grant number: 82271677), Beijing Hospitals Authority Clinical Medicine Development of Special Funding Support (Grant number: ZYLX202120), Beijing Natural Science Foundation (Grant number: 7162063), Capital Medical University Laboratory for Clinical Medicine and Gynecological Tumor Precise Diagnosis and Treatment Innovation Studio.
Author contributions
D.M.: Conceptualization, data curation, formal analysis, investigation, methodology, project administration, software, validation, visualization, writing the original draft, and reviewing and editing the draft. T.F., J.X. and Z.Y.: Methodology, formal analysis, investigation, resources, data curation, writing of the original draft, and visualization. C.X., Z.Y. and Y.R.: Conceptualization, supervision, reviewing and editing the draft. L.P.: Conceptualization, data curation and formal analysis. H.J. and J.J.: Project supervision and initial design. M.J.: Funding acquisition, project administration, resources, and reviewing and editing the draft. All the authors have read and approved the final manuscript.
Data availability
The RNA-seq data generated in this study are publicly available in the ERA at PRJNA1094389. The TCGA database for the ovarian cancer cohort can be accessed and analyzed by Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/). Qualified researchers may request the rest of the data, materials, and/or methods directly from Jinwei Miao.
Code availability
The quality of the FASTQ files was evaluated using FastQC (v0.11.5, https://www.babraham.ac.uk/projects/fastqc/), and annotated documentation of reference genomes and gene patterns was obtained directly from the genome site. The index of the reference genome was established and matched with the reference genome using Hisat2v2.0.5. Using a reference-based approach, Stringtie was used to count the genes (v1.3.3b). We analyzed two groups, each consisting of three biological replicates, for differential gene expression using the DESeq2R package (v1.16.2). DESeq2R uses the negative binomial distribution model to detect differential expression in digital gene expression data. To control for false discovery rates, we adjusted P values using the Benjamini and Hochberg method. We identified genes as differentially expressed when their adjusted P values were < 0.05, as determined by DESeq2. The enrichment of DEGs in KEGG pathways was analyzed by KOBAS (http://kobas.cbi.pku.edu.cn/).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-024-00791-8.
References
- 1.Chandrasekaran, A. et al. Synthetic Lethality in ovarian cancer. Mol. Cancer Ther.20, 2117–2128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caruso, G. et al. Poly (ADP-ribose) polymerase inhibitors (PARPi) in ovarian cancer: lessons learned and future directions. Int. J. Gynecol. Cancer33, 431–443 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Deng, M. et al. Immunotherapy for Ovarian Cancer: Disappointing or Promising? Mol. Pharm.21, 454–466 (2024). [DOI] [PubMed] [Google Scholar]
- 4.Abdalla, A. M. E. et al. Current Challenges of Cancer Anti-angiogenic Therapy and the Promise of Nanotherapeutics. Theranostics8, 533–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chekerov, R. et al. Sorafenib plus topotecan versus placebo plus topotecan for platinum-resistant ovarian cancer (TRIAS): a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol.19, 1247–1258 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Bodnar, L. et al. Sorafenib as a third line therapy in patients with epithelial ovarian cancer or primary peritoneal cancer: a phase II study. Gynecol. Oncol.123, 33–36 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Herzog, T. J. et al. A randomized phase II trial of maintenance therapy with Sorafenib in front-line ovarian carcinoma. Gynecol. Oncol.130, 25–30 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Ren, X. et al. Overcoming the compensatory elevation of NRF2 renders hepatocellular carcinoma cells more vulnerable to disulfiram/copper-induced ferroptosis. Redox Biol.46, 102122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu, L. et al. Ferroptosis in life: To be or not to be. Biomed. Pharmacother.159, 114241 (2023). [DOI] [PubMed] [Google Scholar]
- 10.Wang, F. et al. miR-672-3p Promotes Functional Recovery in Rats with Contusive Spinal Cord Injury by Inhibiting Ferroptosis Suppressor Protein 1. Oxid. Med. Cell Longev.2022, 6041612 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ong, C. K. et al. Genomic structure of human OKL38 gene and its differential expression in kidney carcinogenesis. J. Biol. Chem.279, 743–754 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Liu, M. et al. Allele-specific imbalance of oxidative stress-induced growth inhibitor 1 associates with progression of hepatocellular carcinoma. Gastroenterology146, 1084–1096 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Zhao, T. et al. Assessment of alterations in histone modification function and guidance for death risk prediction in cervical cancer patients. Front. Genet.13, 1013571 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morales, M. et al. Targeting iron metabolism in cancer therapy. Theranostics11, 8412–8429 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, L. et al. Attenuation of Sepsis-Induced Acute Kidney Injury by Exogenous H2S via Inhibition of Ferroptosis. Molecules28, 4770 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jungsuwadee, P. et al. The G671V variant of MRP1/ABCC1 links doxorubicin-induced acute cardiac toxicity to disposition of the glutathione conjugate of 4-hydroxy-2-trans-nonenal. Pharmacogenet Genomics22, 273–284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou, J. et al. The ferroptosis signature predicts the prognosis and immune microenvironment of nasopharyngeal carcinoma. Sci. Rep.13, 1861 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boehning, A. L. et al. Cell type-dependent effects of ellagic acid on cellular metabolism. Biomed. Pharmacother.106, 411–418 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh, S. J. et al. Mitochondrial event as an ultimate step in ferroptosis. Cell Death Discov.8, 414 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira, T. et al. Detachment of Hexokinase II From Mitochondria Promotes Collateral Sensitivity in Multidrug Resistant Chronic Myeloid Leukemia Cells. Front. Oncol.12, 852985 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneko, N. et al. siRNA-mediated knockdown against CDCA1 and KNTC2, both frequently overexpressed in colorectal and gastric cancers, suppresses cell proliferation and induces apoptosis. Biochem. Biophys. Res. Commun.390, 1235–1240 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Koutsioumpa, M. et al. Lysine methyltransferase 2D regulates pancreatic carcinogenesis through metabolic reprogramming. Gut68, 1271–1286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai, C. H. et al. Docosahexaenoic acid promotes the formation of autophagosomes in MCF-7 breast cancer cells through oxidative stress-induced growth inhibitor 1 mediated activation of AMPK/mTOR pathway. Food Chem. Toxicol.154, 112318 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Teng, P. N. et al. Pharmacologic inhibition of ATR and ATM offers clinically important distinctions to enhancing platinum or radiation response in ovarian, endometrial, and cervical cancer cells. Gynecol. Oncol.136, 554–561 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, P. H. et al. Kinome screen of ferroptosis reveals a novel role of ATM in regulating iron metabolism. Cell Death Differ.27, 1008–1022 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong, T. et al. PARP inhibition promotes ferroptosis via repressing SLC7A11 and synergizes with ferroptosis inducers in BRCA-proficient ovarian cancer. Redox Biol.42, 101928 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou, J. et al. m6A demethylase ALKBH5 controls CD4+T cell pathogenicity and promotes autoimmunity. Sci. Adv.7, eabg0470 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y. et al. Erastin induces ferroptosis via ferroportin-mediated iron accumulation in endometriosis. Hum. Reprod.36, 951–964 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Bitler, B. G. PARP inhibitors: clinical utility and possibilities of overcoming resistance. Gynecol. Oncol.147, 695–704 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tumiati, M. A functional homologous recombination assay predicts primary chemotherapy response and long-term survival in ovarian cancer patients. Clin. Cancer Res.24, 4482–4493 (2018). [DOI] [PubMed] [Google Scholar]
- 31.McKinnon, P. J. ATM and the molecular pathogenesis of ataxia telangiectasia. Annu. Rev. Pathol.7, 303–321 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Barlow, C. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell86, 159–171 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Liu, X. ATM paradoxically promotes oncogenic transformation via transcriptional reprogramming. Cancer Res.80, 1669–1680 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdel-Fatah, T. M. ATM, ATR and DNA-PKcs expressions correlate to adverse clinical outcomes in epithelial ovarian cancers. BBA Clin.2, 10–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin, M. H. & Oh, D. Y. ATM in DNA repair in cancer. Pharmacol. Ther.203, 107391 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Riches, L. C. Pharmacology of the ATM inhibitor AZD0156: potentiation of irradiation and olaparib responses preclinically. Mol. Cancer Therapeut.19, 13–25 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Kurman, R. J. & Shih Ie, M. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer-shifting the paradigm. Hum. Pathol.42, 918–931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDermott, J. E. Proteogenomic characterization of ovarian HGSC implicates mitotic kinases, replication stress in observed chromosomal instability. Cell Rep. Med.1, 100004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guleria, A. & Chandna, S. ATM kinase: much more than a DNA damage responsive protein. DNA Repair39, 1–20 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Lei, G. et al. Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer22, 381–396 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khoi, C. S. et al. Oxidative stress-induced growth inhibitor (OSGIN1), a target of X-Box-Binding Protein 1, protects palmitic acid-induced vascular lipotoxicity through maintaining autophagy. Biomedicines10, 992 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu, R. et al. A shortage of FTH induces ROS and sensitizes RAS-proficient neuroblastoma N2A cells to ferroptosis. Int. J. Mol. Sci.22, 8898 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng, M. et al. MiR-29c downregulates tumor-expressed B7-H3 to mediate the antitumor NK-cell functions in ovarian cancer. Gynecol. Oncol.162, 190–199 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Gao, A. et al. Tumor-derived ILT4 induces T cell senescence and suppresses tumor immunity. J. Immunother. Cancer9, e001536 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng, M. et al. Small-molecule inhibitor HI-TOPK-032 improves NK-92MI cell infiltration into ovarian tumours. Basic Clin. Pharmacol. Toxicol. 19, 629–642 (2024). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1-2 and Supplementary Figure 1-14
Data Availability Statement
The RNA-seq data generated in this study are publicly available in the ERA at PRJNA1094389. The TCGA database for the ovarian cancer cohort can be accessed and analyzed by Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/). Qualified researchers may request the rest of the data, materials, and/or methods directly from Jinwei Miao.
The quality of the FASTQ files was evaluated using FastQC (v0.11.5, https://www.babraham.ac.uk/projects/fastqc/), and annotated documentation of reference genomes and gene patterns was obtained directly from the genome site. The index of the reference genome was established and matched with the reference genome using Hisat2v2.0.5. Using a reference-based approach, Stringtie was used to count the genes (v1.3.3b). We analyzed two groups, each consisting of three biological replicates, for differential gene expression using the DESeq2R package (v1.16.2). DESeq2R uses the negative binomial distribution model to detect differential expression in digital gene expression data. To control for false discovery rates, we adjusted P values using the Benjamini and Hochberg method. We identified genes as differentially expressed when their adjusted P values were < 0.05, as determined by DESeq2. The enrichment of DEGs in KEGG pathways was analyzed by KOBAS (http://kobas.cbi.pku.edu.cn/).