Abstract
Background
Advanced gastric cancer (GC) exhibits a high recurrence rate and a dismal prognosis. Myocyte enhancer factor 2c (MEF2C) was found to contribute to the development of various types of cancer. Therefore, our aim is to develop a prognostic model that predicts the prognosis of GC patients and initially explore the role of MEF2C in immunotherapy for GC.
Methods
Transcriptome sequence data of GC was obtained from The Cancer Genome Atlas (TCGA), the Gene Expression Omnibus (GEO) and PRJEB25780 cohort for subsequent immune infiltration analysis, immune microenvironment analysis, consensus clustering analysis and feature selection for definition and classification of gene M and N. Principal component analysis (PCA) modeling was performed based on gene M and N for the calculation of immune checkpoint inhibitor (ICI) Score. Then, a Nomogram was constructed and evaluated for predicting the prognosis of GC patients, based on univariate and multivariate Cox regression. Functional enrichment analysis was performed to initially investigate the potential biological mechanisms. Through Genomics of Drug Sensitivity in Cancer (GDSC) dataset, the estimated IC50 values of several chemotherapeutic drugs were calculated. Tumor-related transcription factors (TFs) were retrieved from the Cistrome Cancer database and utilized our model to screen these TFs, and weighted correlation network analysis (WGCNA) was performed to identify transcription factors strongly associated with immunotherapy in GC. Finally, 10 patients with advanced GC were enrolled from Sun Yat-sen University Cancer Center, including paired tumor tissues, paracancerous tissues and peritoneal metastases, for preparing sequencing library, in order to perform external validation.
Results
Lower ICI Score was correlated with improved prognosis in both the training and validation cohorts. First, lower mutant-allele tumor heterogeneity (MATH) was associated with lower ICI Score, and those GC patients with lower MATH and lower ICI Score had the best prognosis. Second, regardless of the T or N staging, the low ICI Score group had significantly higher overall survival (OS) compared to the high ICI Score group. For its mechanisms, consistently, for Camptothecin, Doxorubicin, Mitomycin, Docetaxel, Cisplatin, Vinblastine, Sorafenib and Paclitaxel, all of the IC50 values were significantly lower in the low ICI Score group compared to the high ICI Score group. As a result, based on univariate and multivariate Cox regression, ICI Score was considered to be an independent prognostic factor for GC. And our Nomogram showed good agreement between predicted and actual probabilities. Based on CIBERSORT deconvolution analysis, there was difference of immune cell composition found between high and low ICI Score groups, probably affecting the efficacy of immunotherapy. Then, MEF2C, a tumor-related transcription factor, was screened out by WGCNA analysis. Higher MEF2C expression is significantly correlated with a worse OS. Moreover, its higher expression is also negatively correlated with tumor mutation burden (TMB) and microsatellite instability (MSI), but positively correlated with several immunosuppressive molecules, indicating MEF2C may exert its influence on tumor development by upregulating immunosuppressive molecules. Finally, based on transcriptome sequencing data on 10 paired tumor tissues from Sun Yat-sen University Cancer Center, MEF2C expression was significantly lower in paracancerous tissues compared to tumor tissues and peritoneal metastases, and it was also lower in tumor tissues compared to peritoneal metastases, indicating a potential positive association between MEF2C expression and tumor invasiveness.
Conclusions
Our prognostic model can effectively predict outcomes and facilitate stratification GC patients, offering valuable insights for clinical decision-making. The identified transcription factor MEF2C can serve as a biomarker for assessing the efficacy of immunotherapy for GC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-024-02082-4.
Keywords: Gastric cancer (GC), Prognostic model, Transcriptome sequencing, Immune microenvironment, Immunotherapy, Transcription factor (TF), Myocyte enhancer factor 2c (MEF2C), Biomarker
Background
Gastric cancer (GC) is a heterogeneous disease influenced by various factors, including genetic and environmental factors, with its exact etiology unclear. The Lauren classification system is commonly used to categorize GC into two main histological subtypes: intestinal and diffuse [1]. According to the 2020 data released by the International Agency for Research on Cancer, GC constitutes 5.6% of all new cancer cases and accounts for 7.7% of total cancer-related deaths worldwide [2]. Despite a declining trend in mortality rates in most regions, the annual incidence of GC still exceeds one million cases [3]. The prognosis for early-stage and advanced GC differs significantly; the 5-year survival rate after early-stage surgery can exceed 90%, whereas advanced GC exhibits a high recurrence rate [4] and a dismal prognosis with the median overall survival (OS) of less than 12 months [5, 6].
Currently, neoadjuvant chemotherapy, molecular targeted therapy and immunotherapy are the primary modes of treatment. Neoadjuvant chemotherapy involves administering drugs prior to surgery to reduce tumor size and facilitate surgical resection. Localized neoadjuvant chemotherapy has been shown to significantly reduce tumor stage, improve surgical success rates, and prolong patient survival [6]. Wilke previously demonstrated that cisplatin is highly effective in preoperative drug treatment for locally advanced GC, offering a chance for surgical intervention in patients with a poor prognosis [7]. However, a precise understanding of the timing of neoadjuvant chemotherapy is crucial, as it can impact subsequent surgical resection and potentially result in missed optimal surgical opportunities. This places significant demands on medical personnel. Furthermore, the systemic toxicity induced by chemotherapy poses a significant obstacle to treatment.
In comparison to neoadjuvant chemotherapy, molecular targeted therapy offers the advantage of targeted drug delivery at the site of disease, thereby minimizing damage to normal tissue [8]. The fundamental principle behind molecular targeted therapy is the utilization of drugs that specifically target some molecules to block signals that promote cancer cell growth, interfere with the regulation of cell cycle and induce cell death, with the aim of inhibiting or eliminating cancer cells [9]. Currently, the common molecular targeted therapies for GC treatment are: (1) Epidermal growth factor receptor (EGFR) blockade, which inhibits tumor proliferation, invasion, and distant metastasis; (2) Vascular endothelial growth factor (VEGF) blockade, which prevents tumor angiogenesis and consequently inhibits tumor growth [6]. Most targeted drugs presently only impact a single target, however, single-pathway targeted therapy may be insufficient for impeding tumor progression.
The immune system assumes a critical role in tumor development and metastasis [10, 11] by discerning tumor cells from normal cells based on distinctions in their biochemical composition, antigen structure, and biological behavior, consequently triggering recognition by the innate immune system. Antigen-presenting cells, such as dendritic cells, present antigens to activate CD4+ and CD8+ T cells within the adaptive immune system, facilitating their infiltration and subsequent elimination of tumor cells [12, 13]. Tumor cells release tumor-associated antigens, which are continuously recognized and captured, leading to the formation of a tumor immune cycle [13]. The current objective of immunotherapy for GC and other cancers is to restore a balance within the human immune system in order to eliminate tumor cells [14]. This is primarily achieved through the use of monoclonal antibodies [15, 16] and immune checkpoint inhibitors (ICIs). Currently, FDA-approved ICIs include CTLA-4, PD-1, and LAG-3 [14, 17]. The interaction between the immune system and tumor cells progresses through three stages: elimination, balance and escape. During the escape stage, tumor cells express immunosuppressive molecules on their surface to evade T cell recognition [12, 18]. Consequently, inhibiting tumor cell development becomes challenging under substantial physiological burden.
The prognosis for GC is especially grim in advanced stages, with the median OS of 10 ~ 12 months [19]. Researchers have constructed several prognostic prediction models for GC to facilitate targeted treatment for better outcomes. Yang et al. utilized cluster analysis and principal component analysis (PCA) to screen six lactic acid-related genes associated with GC development and created a model that predicts malignant progression and prognosis of GC [16]. Yu et al. constructed a prognostic model for GC by using multivariate Cox regression analysis. This model incorporates eight survival-associated genes (RNASE2, CGB5, INHBE, DUSP1, APOA1, CD36, PTGER3, CTLA4) to predict prognosis and identify patients who may benefit from ICIs [20]. Furthermore, ferroptosis, a non-apoptotic cellular death mechanism, is intricately linked to cancer development. Wei et al. developed a “4-ferroptosis-related lncRNA signature model” to accurately predict the prognosis of GC patients by analyzing lncRNA associated with ferroptosis and related to prognosis [21].
Myocyte enhancer factor 2c (MEF2C) is one of the transcriptional factors in the MADS-BOX family, having a critical function in the differentiation of cardiomyocytes [22, 23] and development of the nervous system [24, 25]. Human MEF2C gene is located on chromosome 5q14.3, encoding a protein encompassing five core domains (MADS, MEF2, TAD1, TAD2 and NLS domains) [26, 27]. Research findings indicate that MEF2C-mediated transcriptional regulation of interleukin 12 (IL-12) is imperative for macrophage phenotypic polarization and related immune pathologies. A previous study published in Cell Research demonstrated that transfection of a dominant negative form of MEF2C protects primed macrophage from cell death triggered by lipopolysaccharide [28]. Furthermore, M1 macrophages exert their pro-inflammatory effects by secreting cytokines such as IL-12 and IL-23, which contribute to the promotion of Th1 response (Th1 response refers to a specific type of immune response mediated by T-helper cells, particularly Th1 cells) [29–32] and cytotoxicity against tumor cells [33]. Moreover, MEF2C has been implicated in the pathogenesis of various cancers, including hepatocellular carcinoma [34], pancreatic ductal adenocarcinoma [35] and rhabdomyosarcoma [36]. Epithelial-mesenchymal transition (EMT) plays a crucial role in the invasion and metastasis of tumor cells [37]. Studies have demonstrated that MEF2C enhances invasiveness in hepatocellular carcinoma by upregulating transforming growth factor-beta 1 (TGF-β1) expression and promoting EMT [38]. Another research indicated that high expression of MEF2C led to poor event-free survival and OS in Indian T-acute lymphoblastic leukemia, and is a predictor of patient outcome [39]. And MEF2C overexpression was indicator for poor outcome in adult patients with acute myeloid leukemia by driving immune escape [40]. These findings suggest that MEF2C may also contribute to the development of other types of cancer. However, there is currently a dearth of research investigating the impact of MEF2C on the progression of GC and the prognosis of GC patients.
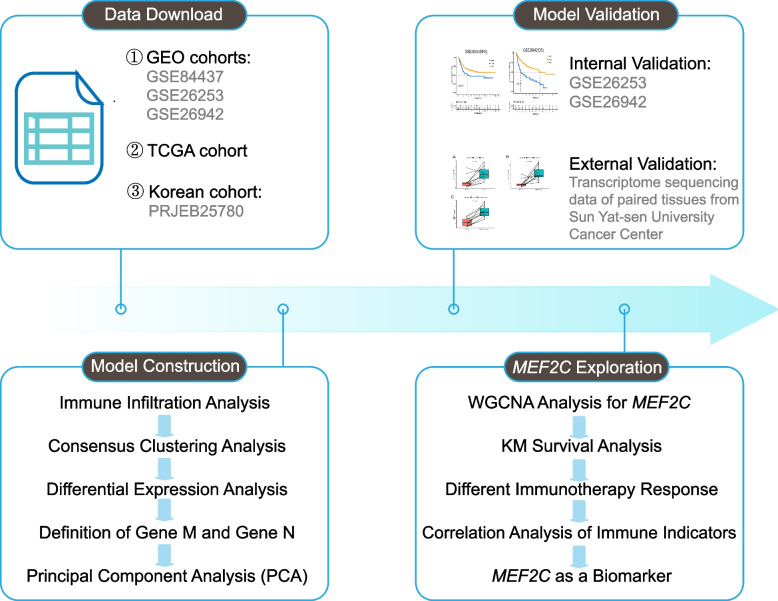
Therefore, our aim is to develop a prognostic model that predicts the prognosis of GC patients and initially explore the role of MEF2C in immunotherapy for GC, hoping to provide some insights for clinical treatment strategies. The study workflow is presented in Fig. 1.
Fig. 1.
The workflow of the whole study. (First, data download from public data sets; second, model construction using a series of bioinformatics methods; third, internal validation based on public data sets and external validation based on transcriptome sequencing data of paired tissues from Sun Yat-sen University Cancer Center; fourth, function and mechanism exploration of MEF2C using a series of bioinformatics methods.)
Materials and methods
Data download
In this study, we obtained transcriptome sequence data of GC from The Cancer Genome Atlas (TCGA) official website (https://portal.gdc.cancer.gov/), and subsequently converted the FPKM (Fragments Per Kilobase of transcript per Million mapped reads) data into the TPM (Transcripts Per Million) data. Additionally, we acquired the GSE84437, GSE26253, GSE26942 datasets of GC from the Gene Expression Omnibus (GEO) official website (https://www.ncbi.nlm.nih.gov/geo/). To eliminate batch effects, we employed SVA package [41] for data integration and merging of the two datasets. Furthermore, we downloaded transcriptome data from the PRJEB25780 cohort [42], which consists of GC patients treated with anti-PD-1 therapy. These transcriptome data were reprocessed and expression profiles were quantified for further analysis [43].
Patients and specimens
We enrolled 10 patients with advanced GC from Sun Yat-sen University Cancer Center, including paired tumor tissues, paracancerous tissues and peritoneal metastases. These tissues were ground in liquid nitrogen and then TRIZOL Reagent was added. After incubation at room temperature for 5min, add 200μL chloroform and vigorously shake for 15s. Subsequently, the mixture was incubated at room temperature for 10min and centrifuged at 12,000rpm at 4℃ for 15min. The aqueous phase containing RNA in the upper layer was transferred to a new tube and 500μL isopropanol was added for RNA precipitation. After incubation at room temperature for 10min and centrifugation at 12,000rpm at 4℃ for 15min, the supernatant was discarded. The RNA precipitation was washed with 1mL of 75% ethanol, then centrifuged at 7,500rpm at 4℃ for 5min. Dissolve the RNA in 20μL of DEPC water, and the concentration of RNA was determined using a spectrophotometer. Next, mRNA was enriched and purified using Oligo dT beads and fragmented. A poly-A tail was added to the 3’ ends, followed by ligation of sequencing junctions. Gel purification was performed to recover cDNA fragments ranging from 200 to 500bp. After PCR amplification, the sequencing library was prepared. The quality of the library was assessed, and sequencing was performed using Illumina HiSeq 2500 with the output in fastq format. Prior to participation in this study, all patients provided informed consent for the use of their information and specimens for research purposes.
Immune infiltration analysis
To evaluate the abundance of tumor infiltrating immune cells (TIICs) in each sample and elucidate the tumor immune microenvironment, we utilized the CIBERSORT algorithm. This immunological computational approach relies on a gene expression signature matrix comprising various annotated genes [44]. By employing linear support vector regression (SVR) as a machine learning method, it enables the deconvolution of gene expression profiles. Prior to CIBERSORT analysis, the original gene expression data from TCGA and GEO were subjected to normalization. Subsequently, we obtained a matrix of TIIC infiltration comprising 22 immune cell subtypes using the CIBERSORTx website (https://cibersortx.stanford.edu/). To enhance the accuracy of the deconvolution algorithm, we further considered the p-value and root mean square error provided by CIBERSORT, setting a filtering criterion of P < 0.05.
Immune microenvironment analysis
The ESTIMATE algorithm leverages the distinct transcriptional profiles of cancer samples to estimate the composition of tumor cells and infiltrating normal cells [45]. It specifically targets stromal cells and immune cells, as they represent the primary non-tumor constituents within tumor samples and exhibit characteristic signaling patterns associated with their infiltration into the tumor microenvironment. Ultimately, the algorithm predicts the abundance of infiltrating stromal and immune cells by calculating matrix and immune scores.
Consensus clustering analysis
Consensus clustering is a technique used to integrate the outputs of multiple clustering algorithms. The primary objective of clustering aggregation is to identify a single consensus cluster, which is deemed more appropriate than the existing clusters obtained from various input datasets. In this study, we employed the consensus clustering to conduct an initial analysis of the results generated by the CIBERSORT and ESTIMATE algorithms. CIBERSORT and ESTIMATE packages were applied to calculate immune cell content and tumor microenvironment scores. Then we used the “ConsensusClusterPlus” package for consensus “clustering (reps = 50, pItem = 0.8, pFeature = 1, clusterAlg = “km”). The cumulative distribution function and consensus heat map were used to evaluate the optimal number of clusters, and the optimal k value was 5. We utilized the limma package [46] to compare different subtypes of clustering and applied |logFC|> 1 and P < 0.05 as selection criteria for identifying differential expression genes. We extracted the expressions of differential genes and then performed clustering again using the same method described above. “ConsensusClusterPlus” package was applied again for consensus clustering (reps = 50, pItem = 0.8, pFeature = 1, clusterAlg = “km”). The cumulative distribution function and consensus heat map were used to evaluate the optimal number of clusters, and the optimal k value was 4. We then calculated the correlation between gene clustering and gene expression levels. Genes that display a positive correlation between their clustering results and gene expression levels are classified as gene M, while those that show an inverse correlation are classified as gene N.
Feature selection by boruta package
Boruta is a specialized feature selection method, specifically a permutation-based calculation approach within the framework of random forest [47]. Feature selection plays a pivotal role in predictive modeling, particularly when working with datasets containing multiple variables for model construction. The algorithmic concept of Boruta involves shuffling the original real features to construct shadow features. The real features and shadow features are concatenated together to form a feature matrix, which is then used for training. Ultimately, the feature importance score of the shadow features serves as the reference baseline for selecting a subset of real features that are truly correlated with the dependent variable. We employed the Boruta algorithm to further screen the differential expression genes obtained previously, which were subsequently utilized for subsequent PCA modeling.
Enrichment analysis
ClusterProfiler, a robust bioinformatics tool, facilitates the rapid exploration of functional characteristics in numerous species by leveraging current gene annotation data, encompassing both coding and non-coding genomic information [48]. This tool offers a versatile interface to acquire gene function annotation from diverse sources, enabling flexible utilization across various application scenarios. In this study, we utilized the clusterProfiler package to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis separately for gene M and gene N, which were previously identified through Boruta feature selection. We set the significance threshold at P < 0.05 for filtering purposes.
Gene set enrichment analysis (GSEA), also referred to as functional enrichment or pathway enrichment analysis, involves grouping genes together based on their participation in the same biological pathway or their proximity on the chromosome. The predefined sets of genes for this analysis can be accessed through the Molecular Signatures Database (MSigDB) (https://www.gsea-msigdb.org/gsea/msigdb). In this study, we downloaded the KEGG gene set and performed GSEA enrichment analysis to investigate the differences in enriched pathways based on high or low model scores.
PCA modeling
Principal component analysis (PCA) is a widely employed method for analyzing large datasets containing numerous dimensions or features per observation [49]. It aims to enhance data interpretability and preserve maximum information while enabling visualization of multi-dimensional data. PCA is a statistical technique that accomplishes dimensionality reduction by linearly transforming the data into a new coordinate system, wherein most variations can be described with fewer dimensions than the original data. In this study, we performed PCA dimensionality reduction on gene M and gene N, and then use the “Predict” function to predict the values of principal components, and selected a column of values that best represent the principal components of gene M and gene N, and took the difference to construct the ICI score model, as shown in the following formula:
Mutant-allele tumor heterogeneity (MATH) Scoring
The calculation of MATH score was done through the “infraHeterogeneity” function in the “maftoos” package. It was calculated based on individual samples. The lower the tumor heterogeneity, the lower the MATH score, and the lower the ICI score.
Construction and evaluation of nomogram
Nomogram is extensively utilized in cancer prognosis due to their ability to simplify statistical prediction models into a singular numerical estimation of event probability, such as death or recurrence, tailored to individual patient characteristics. Following univariate Cox regression analysis, multivariate Cox regression analysis was employed to identify independent prognostic factors for constructing a prognostic Nomogram map using the “rms” package in R [50]. The clinical factors incorporated in our nomogram included age, sex, T stage, N stage and model score. Subsequently, the calibration curve was generated using the “regplot” package in R [51]. This curve serves to compare the predicted probability of OS with the actual OS probability derived from the Nomogram, thereby visually assessing the discriminative capability of the line chart.
Drug sensitivity analysis
Genomics of Drug Sensitivity in Cancer (GDSC) dataset is a commonly used resource that includes drug sensitivity data IC50 (half maximal inhibitory concentration) for 1,000 cell lines, providing valuable information on drug sensitivity and resistance in GC cell lines [52]. The pRRophetic package in R was developed by Paul Geeleher et al. to predict drug response in cancer patients [53]. It utilizes the tissue gene expression profile to match with the IC50 values from cancer cell lines and applies the ridge regression algorithm to estimate drug response in a cohort of GC patients.
Tumor transcription factor database
Cistrome Cancer database (http://cistrome.org/CistromeCancer/) is a comprehensive resource for predicting transcription factor (TF) targets and enhancers in cancers. This database provides presumptive target genes for each cancer type, based on the active transcription of specific TFs. In this study, we retrieved tumor-related TFs from the Cistrome Cancer database and utilized our model to screen these TFs.
Weighted correlation network analysis (WGCNA)
WGCNA, an algorithm for high-throughput gene co-expression profile analysis, is extensively utilized to discover gene co-expression networks in diverse diseases. In comparison to conventional approaches, WGCNA excels in capturing gene association patterns and linking gene co-expression modules with clinical characteristics. The construction of co-expression modules using the WGCNA method involves the following primary steps: initially, a correlation coefficient matrix, known as the adjacency matrix, is constructed to represent gene–gene relationships; subsequently, the connection strength between each pair of nodes is calculated using the adjacency matrix (aij) through the following formula:
Vectors (bi and bj) represent gene expression values, while Pearson correlation coefficients of genes i and j (aij) indicate the strength of their connection in the co-expression network. To maintain a scale-free topology in the adjacency matrix, a suitable soft threshold power (β = 4) was chosen. Hierarchical clustering of the weighted coefficient matrix was applied to define modules, and functional modules containing pre-defined genes were identified in the co-expression network. Topology overlap measure (TOM) assessed the shared paracancerous genes concurrency using the following formula:
The weighted adjacency matrix (A) described earlier is utilized in the dissimilarity measurement using the TOM. The dissimilarity measurement based on TOM was applied to gene tree maps and average linkage hierarchical clustering. Similar expression profiles were grouped into the same gene modules using the dynamic tree-cutting package. To ensure module robustness, the minimum gene count was set to 100 per gene co-expression module, and the cutting height threshold for merging similar gene modules was established at 0.3. Subsequently, Pearson correlation analysis was conducted to verify the correlation between gene co-expression modules and clinical parameters. Therefore, the WGCNA algorithm facilitated identification of key gene modules that exhibited significant associations with clinical parameters, which were then used for subsequent analyses.
Statistics
All statistical analyses in the field of bioinformatics were conducted using R software (version 4.2.2). Significance was determined based on a double-tailed test, with a threshold of P < 0.05. The Wilcoxon test was employed to compare differences between the low and high score groups. Spearman correlation analysis was utilized to assess the correlation between non-normally distributed quantitative variables. Survival curves were evaluated using the Kaplan–Meier (KM) method and Cox proportional hazard regression model, with differences analyzed through the log rank test. For survival and regression analysis, the “survival” [54] and “surminer” [55] packages in R were used.
Results
Classification of ICI subtypes and gene subtypes
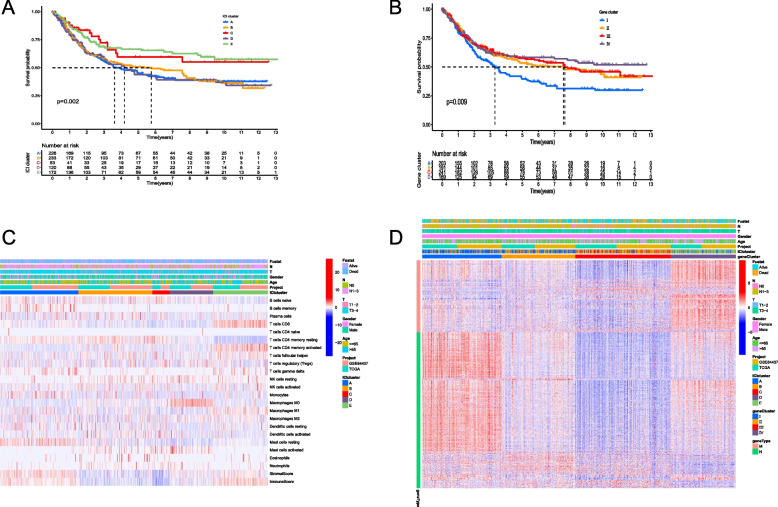
To investigate the tumor immune microenvironment status in GC patients, we utilized the ESTIMATE and CIBERSORT algorithms to analyze the immune microenvironment score and the number of immune-infiltrating cells, showing that memory B cells, CD8+ T cells, activated memory CD4+ T cells were significantly positively correlated with Immune Score, while regulatory T cells (Tregs), activated NK cells and macrophages M0 were significantly negatively correlated with Stroma Score and Immune Score (Supplementary Fig. 1A). Subsequently, we employed the consensus cluster package to classify GC patients into five subtypes (known as A, B, C, D and E; Supplementary Fig. 1B), with ICI subtypes C and E exhibiting higher OS based on the KM survival curve (P = 0.002, Fig. 2A). From the heat map of the immune microenvironment, it was observed that macrophages M0 clustered in ICI subtype D, while CD8+ T cells, resting memory CD4+ T cells and activated memory CD4+ T cells clustered in ICI subtype E. The latter cells play a crucial role in tumor cell killing and favorable prognosis, which aligns with the higher OS. In ICI subtype E, resting memory CD4+ T cells were negatively clustered, while activated memory CD4+ T cells were positively clustered. Additionally, the immune score in ICI subtype E was also higher (Fig. 2C). Furthermore, in order to identify molecular differences among different GC tumors, the limma package was used to perform pairwise differential analysis on the aforementioned five ICI subtypes, and all differential expression genes were extracted, as shown in Supplementary Table 1. Subsequent clustering based on gene expression levels using the consensus cluster package resulted in four gene subtypes (known as I, II, II and IV; Supplementary Fig. 1C), with significant differences in OS (P = 0.009, Fig. 2B). To identify the feature genes that determine the differences in GC, genes positively correlated with the gene subtypes (cor > 0, P < 0.05) were defined as gene M, while genes negatively correlated with the gene subtypes (cor < 0, P < 0.05) were defined as gene N (Fig. 2D).
Fig. 2.
Classification of the ICI subtypes and gene subtypes. A The difference of OS between the five ICI subtypes; B The difference of OS between the four gene subtypes; C Immune microenvironment analysis of the five ICI subtypes; D Consensus clustering analysis for classification of gene M and gene N
Construction and verification of a prognostic model of GC and cluster analysis for gene M and N
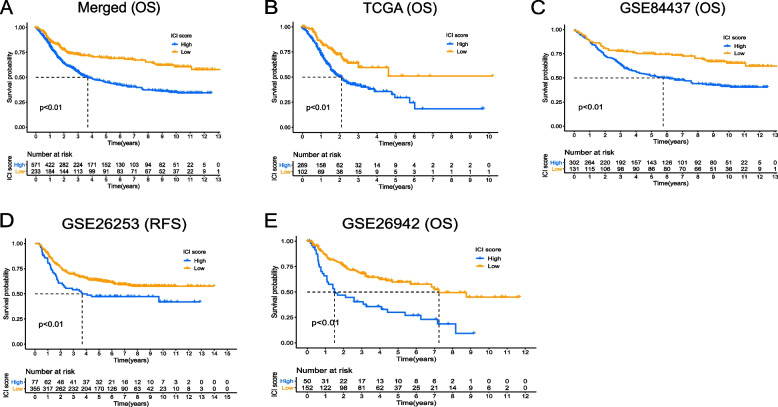
To differentiate the prognosis of GC patients, we developed an immune-related prognostic model. Initially, the Boruta algorithm was utilized for feature selection of gene M and gene N. By randomly disrupting each real feature in order, we evaluated the importance of each feature, and iteratively removed features with low correlation to find the best variable. Then, the PCA algorithm was applied to reduce the dimensionality of the selected transcriptome data, which was conducted using the ‘prcomp’ function with scaling and centering of variables. This approach focuses on the score of the set containing the most significantly related genes and involves scaling down the scores of genes not tracked to other members of the set. The difference of PCA1 scores between the two gene classes was defined as the ICI Score. For validation of the immune-related prognostic model for GC, separate models were constructed using transcriptome data from GSE84437, GSE26253, GSE26942, TCGA and their merged cohort, along with the generation of survival curves. Lower ICI Score was found to be correlated with improved prognosis in both the training and validation cohorts, providing preliminary validation of the model’s feasibility (all P < 0.01, Fig. 3A-E). Subsequently, GO and KEGG enrichment analyses on gene M and gene N revealed that gene M was enriched in chemokine signaling pathways and response to interferon-gamma. Interferon can induce abnormal DNA methylation or gene alterations in tumor cells, leading to tumor progression and recurrence (Supplementary Fig. 2A,B). Gene N exhibited enrichment in immune-related molecules such as cell adhesion molecules, which play a role in cell spreading, migration, and form the molecular basis for immune responses and tumor metastasis (Supplementary Fig. 2C,D). Finally, GSEA enrichment analysis was conducted on GC patients with high or low ICI Score separately, demonstrating that patients with higher ICI Score were primarily enriched in the KEGG-DNA-REPLICATION pathway (Supplementary Fig. 2E,F).
Fig. 3.
The differences of survival probability between the high and low ICI Score groups. A The difference of OS between the high and low ICI Score groups in the merged cohort; B The difference of OS between the high and low ICI Score groups in the TCGA cohort; C The difference of OS between the high and low ICI Score groups in the GSE84437 cohort; D The difference of relapse-free survival (RFS) between the high and low ICI Score groups in the GSE26253 cohort; E The difference of OS between the high and low ICI Score groups in the GSE26942 cohort
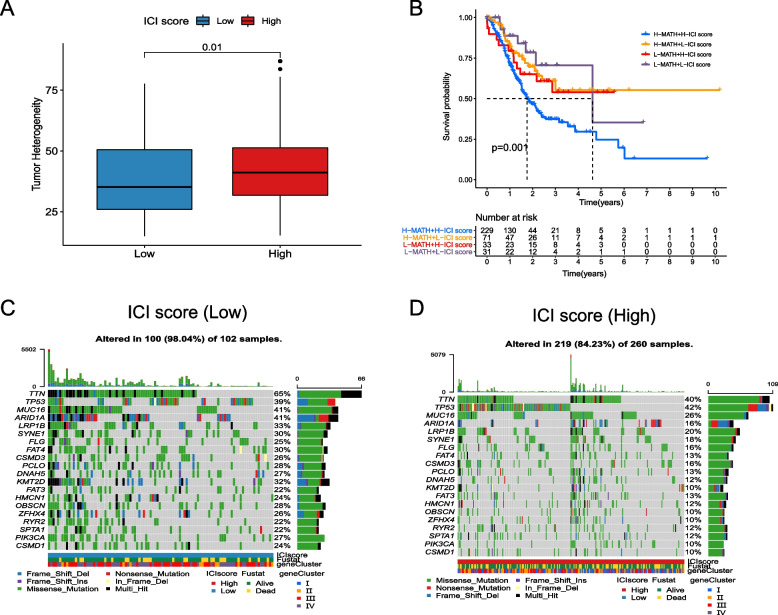
The correlation between ICI score and tumor mutation
As it is widely known, MATH is one of the characteristics of malignant tumors. The higher the MATH, the more likely the immune system’s anti-cancer ability is suppressed, leading to faster cancer progression [56, 57]. In our study, we found that lower MATH was associated with lower ICI Score (P = 0.01, Fig. 4A), which is consistent with the results of survival analysis. Furthermore, we conducted double-subgroup analysis (MATH and ICI Score) and found that patients with lower MATH and lower ICI Score had the best prognosis, while those with higher MATH and higher ICI Score had the worst prognosis (P = 0.001, Fig. 4B). Additionally, we observed significant differences in the mutation rate of tumor-related genes between the high and low ICI Score groups. For example, in the TCGA dataset, the mutation rate of the TTN gene in the high ICI Score group reached 65%, while it was only 40% in the low ICI Score group (Fig. 4C,D).
Fig. 4.
The correlation between ICI Score and tumor mutation. A Tumor heterogeneity between the high and low ICI Score groups; B Double-subgroup KM analysis based on MATH and ICI Score; C The mutation rate of tumor-related genes in the low ICI Score group; D The mutation rate of tumor-related genes in the high ICI Score group
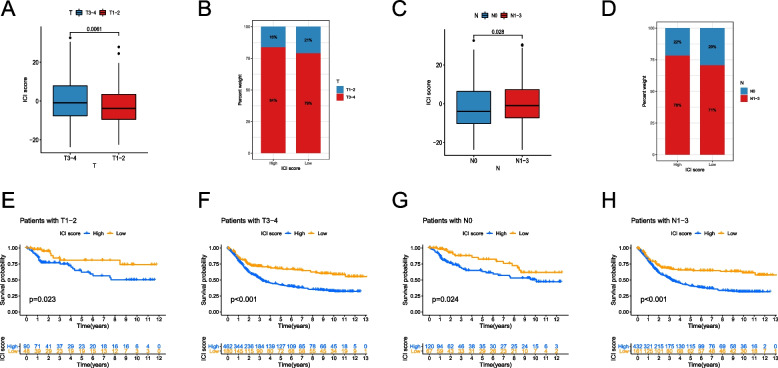
Distinguishing ability of ici score for subgroups of patients with different GC stages
To assess the prognostic discriminative ability of the ICI Score for patients with different GC stages, we conducted separate analyses for patients with T or N staging. We found that patients with stage T1-2 had lower ICI Scores compared to those with stage T3-4 (P = 0.0061, Fig. 5A). Additionally, we observed that the proportion of T3-4 stages was higher in both the high and low ICI Score groups (Fig. 5B). The corresponding KM survival curves indicated that regardless of the T staging, the low ICI Score group had higher OS compared to the high ICI Score group (both P < 0.05, Fig. 5E,F). Similarly, we found that patients with stage N0 had lower ICI Scores compared to those with stage N1-3 (P = 0.028, Fig. 5C). Furthermore, we observed that the proportion of N1-3 stages was higher in both the high and low ICI Score groups (Fig. 5D). Similar to the results of T staging, regardless of the N staging, the low ICI Score group had significantly higher OS compared to the high ICI Score group (both P < 0.05, Fig. 5G,H).
Fig. 5.
Distinguishing ability of ICI Score for subgroups of patients with different GC stages. A The difference of ICI Score between GC patients with stage T1-2 and those with stage T3-4; B The proportion of different T stages in the high or low ICI Score groups; C The difference of ICI Score between GC patients with stage N0 and those with stage N1-3; D The proportion of different N stages in the high or low ICI Score groups; E The difference of OS between the high and low ICI Score groups in GC patients with stage T1-2; F The difference of OS between the high and low ICI Score groups in GC patients with stage T3-4; G The difference of OS between the high and low ICI Score groups in GC patients with stage N0; H The difference of OS between the high and low ICI Score groups in GC patients with stage N1-3
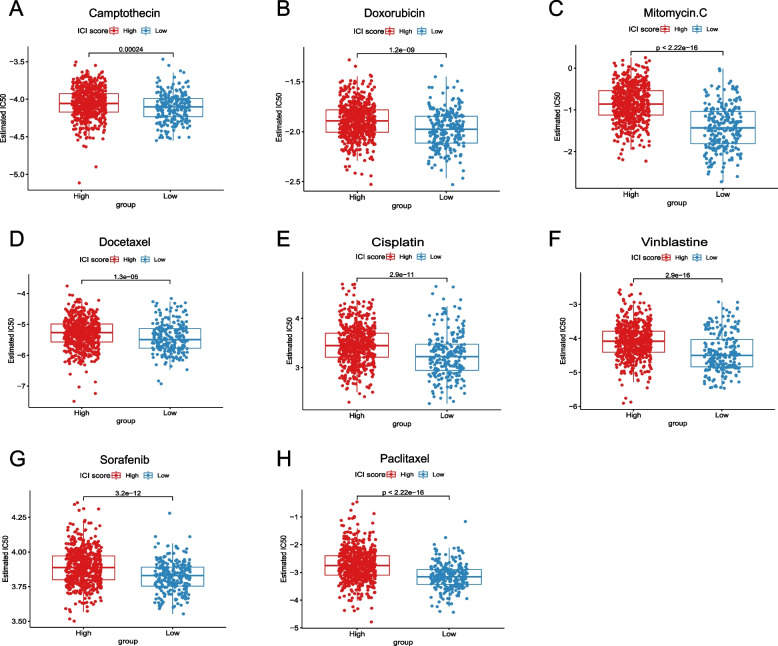
The correlation between ICI score and IC50 of different chemotherapeutic drugs
IC50 refers to the concentration of a certain drug that induces apoptosis in tumor cells by 50%. A lower IC50 value indicates a better drug efficacy and, in turn, demonstrates good treatment outcomes. In our study, we investigated the IC50 values of eight chemotherapy drugs: Camptothecin, Doxorubicin, Mitomycin, Docetaxel, Cisplatin, Vinblastine, Sorafenib and Paclitaxel. Consistently, for each drug, we found that the IC50 values were significantly lower in the low ICI Score group compared to the high ICI Score group (all P < 0.001, Fig. 6A-H). This further supports the notion that our constructed model can provide guidance for clinical drug selection.
Fig. 6.
The correlation between ICI Score and IC50 of different chemotherapeutic drugs. A Estimated IC50 of Camptothecin between the high and low ICI Score groups; B Estimated IC50 of Doxorubicin between the high and low ICI Score groups; C Estimated IC50 of Mitomycin between the high and low ICI Score groups; D Estimated IC50 of Docetaxel between the high and low ICI Score groups; E Estimated IC50 of Cisplatin between the high and low ICI Score groups; F Estimated IC50 of Vinblastine between the high and low ICI Score groups; G Estimated IC50 of Sorafenib between the high and low ICI Score groups; H Estimated IC50 of Paclitaxel between the high and low ICI Score groups
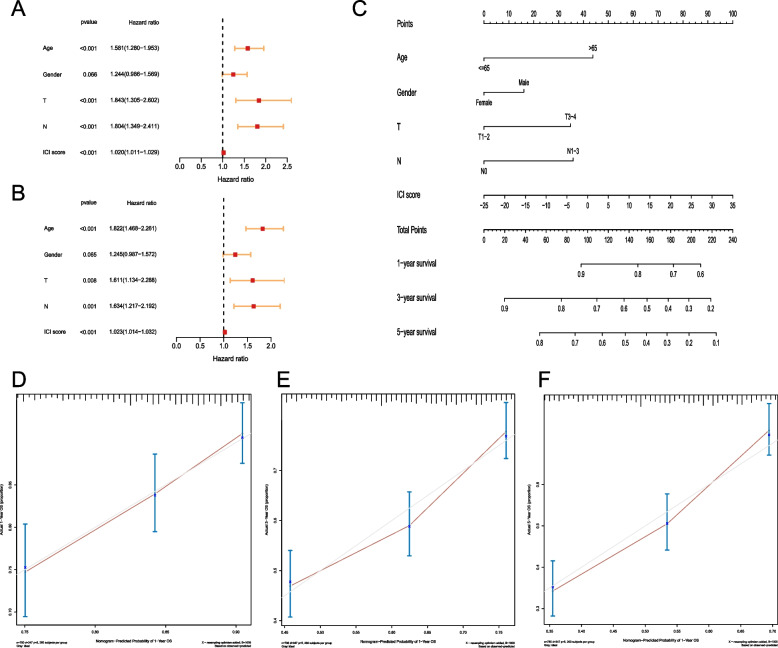
Evaluation for Prognosis of GC Patients by Nomogram
To explore the clinical significance of the model, we employed univariate and multivariate Cox regression to analyze the clinical characteristics (including age, gender, tumor size, metastasis) and ICI Score. The results showed that ICI Score was an independent prognostic factor for GC (both P < 0.001, Fig. 7A,B). Additionally, a Nomogram was developed to estimate OS at 1-, 3- and 5-year. The Nomogram indicated that, holding other factors constant, lower ICI Score corresponded to higher OS (Fig. 7C). To confirm the prediction accuracy, calibration curves were generated using the replog package, showing good agreement between predicted and actual probabilities (Fig. 7D-F).
Fig. 7.
Evaluation for prognosis of GC patients. A Univariate Cox regression for analysis of the clinical characteristics and ICI Score; B Multivariate Cox regression for analysis of the clinical characteristics and ICI Score; C Nomogram for estimating OS at 1-, 3- and 5-year; D Calibration curves for predicted and actual probabilities of 1-year OS; E Calibration curves for predicted and actual probabilities of 3-year OS; F Calibration curves for predicted and actual probabilities of 5-year OS
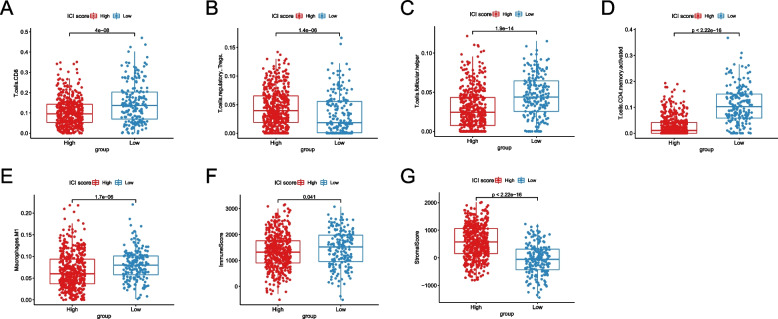
The difference of immune cell composition between high and low ICI score groups may affect the immunotherapy for GC patients
To assess the potential utility of our model in immunotherapy, we employed CIBERSORT deconvolution analysis on transcriptomic data to estimate immune cell composition. We found that Tregs were significantly enriched in the high ICI Score group (P < 0.001, Fig. 8B), which can suppress the activity of effector T cells and promote cancer progression, potentially limiting the efficacy of immunotherapy. Conversely, the low ICI Score group exhibited elevated proportions of tumor-suppressive macrophages M1, CD8+ T cells, activated memory CD4+ T cells, and follicular helper T cells (all P < 0.001, Fig. 8A,C,D,E). Analysis of the tumor immune microenvironment revealed a higher Immune Score in the low ICI Score group (P = 0.041, Fig. 8F) and a higher Stromal Score in the high ICI Score group (P < 0.001, Fig. 8G), implying that the low ICI Score group could potentially benefit more from immunotherapy.
Fig. 8.
The differences of immune cell composition between high and low ICI Score groups. A The difference of CD8+ T cells content between high and low ICI Score groups; B The difference of Tregs content between high and low ICI Score groups; C The difference of follicular helper T cells content between high and low ICI Score groups; D The difference of activated memory CD4+ T cells content between high and low ICI Score groups; E The difference of macrophages M1 content between high and low ICI Score groups; F The difference of Immune Score between high and low ICI Score groups; G The difference of Stromal Score between high and low ICI Score groups
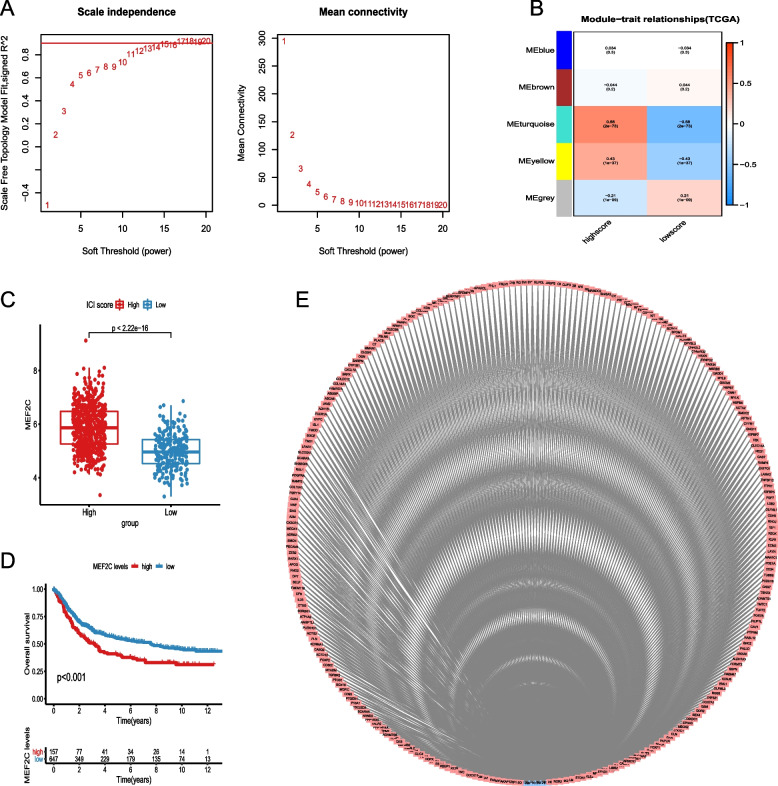
MEF2C screened out by WGCNA analysis
To identify transcription factors strongly associated with immunotherapy in GC, we performed WGCNA with a soft threshold set at 15 (Fig. 9A). Genes were grouped into five modules based on their correlation and assigned different colors. We also examined the correlation between these modules and the ICI Score, which revealed that the yellow and turquoise modules exhibited the strongest correlation with the ICI Score (Fig. 9B). Subsequently, tumor-related transcription factors were obtained from the Cistrome Cancer database, and MEF2C was selected for further investigation based on the correlation analysis (Fig. 9E). We assessed the MEF2C expression level in relation to the ICI Score and observed that the low ICI Score group had significantly lower MEF2C expression compared to the high ICI Score group (P < 0.001, Fig. 9C). Additionally, the MEF2C low expression group demonstrated a significantly better OS compared to the high expression group (P < 0.001, Fig. 9D).
Fig. 9.
MEF2C screened out by WGCNA analysis. A Soft threshold set of WGCNA analysis; B The correlation between the five modules and ICI Score; C MEF2C expressions between the high and low ICI Score groups; D The difference of OS between the MEF2C high and low expression groups; E The correlation between tumor transcriptional factors and related gene expressions
MEF2C as a marker to predict the efficacy of immunotherapy
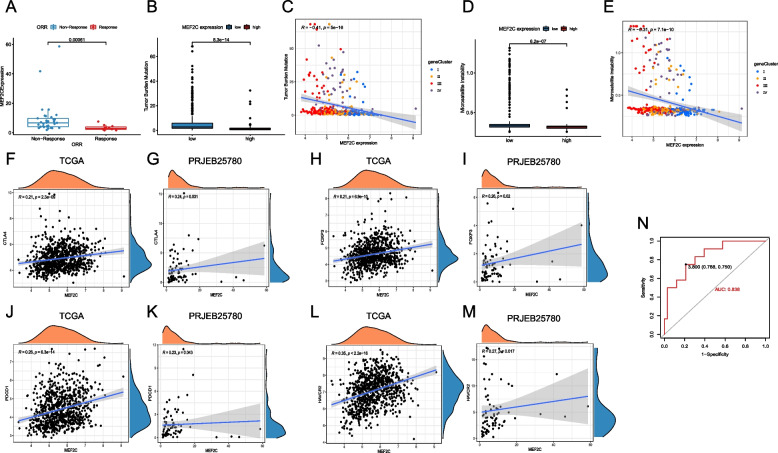
To assess the predictive role of MEF2C in the efficacy of immunotherapy for GC, we conducted an analysis on a cohort of patients undergoing immunotherapy. Our findings revealed a significant downregulation of MEF2C expression in patients who achieved complete response (CR) or partial response (PR) compared to those patients with no response (P < 0.001, Fig. 10A). Furthermore, our investigation into the correlation between MEF2C and tumor mutation burden (TMB), a widely utilized indicator for evaluating immunotherapy, indicated that the MEF2C high expression group exhibited lower TMB than the low expression group (P < 0.001, Fig. 10B), with a negative correlation confirmed through regression analysis (P < 0.001, Fig. 10C). Likewise, our study on the correlation between MEF2C and microsatellite instability (MSI) demonstrated that the MEF2C high expression group also displayed lower MSI (P < 0.001, Fig. 10D), with a negative correlation observed (P < 0.001, Fig. 10E). Additionally, we explored the correlation between MEF2C and immune-related molecules, identifying a positive correlation between MEF2C and immunosuppressive molecules such as CTLA4, FOXP3, PDCD1 and HAVCR2 (all P < 0.05, Fig. 10F-M). Building upon these results, we proposed the hypothesis that MEF2C may exert its influence on tumor development by upregulating immunosuppressive molecules, thereby suppressing the immune process. Utilizing receiver operator characteristic (ROC) curve analysis, we found that MEF2C expression can effectively predict the efficacy of immunotherapy for GC (Fig. 10N), suggesting its potential as a predictive marker for the efficacy of immunotherapy.
Fig. 10.
MEF2C as a marker to predict the efficacy of immunotherapy. A MEF2C expression in patients who achieved CR/PR compared to those with no response; B The difference of TMB between the MEF2C high and low expression groups; C The correlation between MEF2C expression and TMB; D The difference of MSI between the MEF2C high and low expression groups; E The correlation between MEF2C expression and MSI; F The correlation between MEF2C expression and CTLA4 in the TCGA cohort; G The correlation between MEF2C expression and CTLA4 in the PRJEB25780 cohort; H The correlation between MEF2C expression and FOXP3 in the TCGA cohort; I The correlation between MEF2C expression and FOXP3 in the PRJEB25780 cohort; J The correlation between MEF2C expression and PDCD1 in the TCGA cohort; K The correlation between MEF2C expression and PDCD1 in the PRJEB25780 cohort; L The correlation between MEF2C expression and HAVCR2 in the TCGA cohort; M The correlation between MEF2C expression and HAVCR2 in the PRJEB25780 cohort; N ROC curve of predicting the efficacy of immunotherapy for GC based on MEF2C expression
Validation based on external dataset
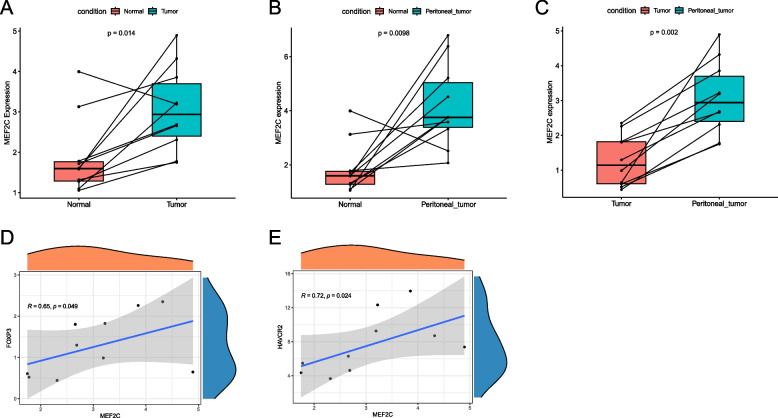
We conducted transcriptome sequencing on 10 paired tumor tissues, paracancerous tissues and peritoneal metastases from 10 GC patients to verify the role of MEF2C in tumor development. Our analysis revealed that the MEF2C expression was significantly lower in paracancerous tissues compared to tumor tissues (P = 0.014, Fig. 11A) and peritoneal metastases (P = 0.0098, Fig. 11B), respectively. Furthermore, the MEF2C expression was also lower in tumor tissues compared to peritoneal metastases (P = 0.002, Fig. 11C), indicating a potential positive association between MEF2C expression and tumor invasiveness. We then examined the correlation between MEF2C expression in tumors and immunosuppressive molecules, finding that MEF2C is positively correlated with FOXP3 and HAVCR2 (both P < 0.05, Fig. 11D,E). It indicates that MEF2C may promote the development of tumors by upregulating immunosuppressive molecules and ultimately inhibiting the efficacy of immunotherapy.
Fig. 11.
Validation based on external dataset. A The difference of MEF2C expressions between paracancerous tissues and tumor tissues; B The difference of MEF2C expressions between paracancerous tissues and peritoneal metastases; C The difference of MEF2C expressions between tumor tissues and peritoneal metastases; D The correlation between MEF2C expression and FOXP3; E The correlation between MEF2C expression and HAVCR2
Discussion
Early-stage GC is often asymptomatic, resulting in a low diagnostic rate and progression to advanced stages in the majority of patients (> 70%) [6]. Advanced GC is associated with poor OS and high metastatic potential, leading to a bleak prognosis [5, 6]. Obtaining preoperative information on GC patients’ physical condition, recovery ability, and OS can facilitate targeted treatments, potentially improving prognosis and extending OS. In this study, we collected a large dataset of GC and utilized the CIBERSORT and ESTIMATE algorithms to analyze the immune microenvironment. Differential analysis was performed using the limma package to identify critical genes that contribute to the variations in GC. Through multiple iterations of consensus clustering, we defined two gene categories (M and N) and quantified the PCA difference between them as the ICI Score. The feasibility of the ICI Score was validated by KM survival curves obtained from GC data downloaded from the TCGA and GSO databases. Our findings showed that lower ICI Score was significantly associated with better prognosis in GC patients. Consequently, based on the ICI Score, we can effectively distinguish GC patients with varying prognosis, even for patients in different stages, enabling targeted and individualized treatment. And the ICI Score was an independent prognostic factor for GC according to univariate and multivariate Cox regression, with good agreement between predicted and actual probabilities. Furthermore, we identified MEF2C as a transcription factor strongly correlated with immunotherapy for GC, and it can serve as a potential biomarker for assessing GC patients’ tumor immune status and eventually predicting the efficacy of immunotherapy.
Genetic variations, environmental disparities and cellular characteristics contribute to the phenotypic and functional heterogeneity within a same type of tumor [58]. Furthermore, inter-patient heterogeneity among tumors arises from differences in the tumor microenvironment and unique cell mutations [59]. Tumor heterogeneity presents a significant challenge in the management of cancer, as it leads to complexity and diversity in tumor characteristics, treatment approaches and associated difficulties. Through an examination of the correlation between our constructed model and tumor heterogeneity, we found that lower ICI Score is associated with reduced tumor heterogeneity. Lower levels of tumor heterogeneity have favorable implications for patient treatment and prognosis, providing some validation for the accuracy and correctness of our model. Additionally, we observed a lower mutation rate of tumor-associated genes in the low ICI Score group, indicating increased drug sensitivity in tumor cells. To evaluate the discriminative capacity of our model for GC patients, we conducted statistical analyses on GC patients at different stages. Irrespective of the stage (early or advanced), patients with higher OS demonstrated lower ICI Score. In essence, the ICI Score enables effective differentiation of GC patients with distinct prognoses, facilitating the implementation of targeted and individualized treatment strategies to optimize therapeutic outcomes. Meanwhile, in our study, we investigated the IC50 values of eight chemotherapy drugs (Camptothecin, Doxorubicin, Mitomycin, Docetaxel, Cisplatin, Vinblastine, Sorafenib and Paclitaxel). We found that the IC50 values were significantly lower in the low ICI Score group compared to the high ICI Score group, which may guide the choice of drugs to a certain extent.
During the screening for factors strongly associated with immunotherapy for GC, we discovered the transcription factor MEF2C. The low ICI Score group, which demonstrated better OS, had a significantly lower MEF2C expression compared to the high ICI Score group. To investigate the potential role of MEF2C in tumor progression, we assessed its expression levels in paired tumor tissues, paracancerous tissues and peritoneal metastases. MEF2C expression was found to be lowest in paracancerous tissues and highest in peritoneal metastases. Some previous research has shown that MEF2C can enhance invasiveness in other types of cancer. Tian et al. found that MEF2C was significantly upregulated in prostate cancer and breast cancer when bone metastases occurred [60]. Ostrander et al. were the first to identify the MEF2C expression in breast cancer cells and concluded that it promotes cell proliferation and tumor metastasis [61]. Zhang et al. discovered that MEF2C weakens the inhibition of tumor metastasis by affecting the expression of Yin Yang-1 [35]. While there have been various findings in different tumor fields regarding the role of MEF2C in promoting tumor invasion and metastasis, there is still a lack of relevant research in GC. Cheng et al. found that MEF2C can promote invasion and metastasis in diffuse GC through the MAPK signaling pathway [62], which is consistent with our result.
We found that Tregs were significantly enriched in the high ICI Score group, which can suppress the activity of effector T cells and promote cancer progression, potentially limiting the efficacy of immunotherapy. It implies that the low ICI Score group could potentially benefit more from immunotherapy. Tregs play a crucial role in regulating the immune response and preventing attacks on self-tissues by immune cells such as T cells and B cells. Tregs achieve this through three characteristic functions: (1) inhibiting the activation of non-Tregs [63], (2) impairing T cell receptor (TCR) signaling transmission [64], and (3) inducing an “anergic” state [65]. These functions are dependent on the protein CTLA4. The extracellular domain of CTLA4 enables Tregs to inhibit the activation and proliferation of non-Tregs, while damage to the intracellular domain of CTLA4 impairs TCR signal transmission, leading to T cell “anergy” [65]. However, the immune blockade mediated by CTLA4-expressing Tregs can contribute to tumor cell escape and progression. In contrast, FOXP3, another regulatory protein, exhibits a complex role in tumors [66]. FOXP3 can inhibit the expression of the oncogene SKP2 [67], but it can also be induced by HBZ-activated TGF-β/Smad signaling, promoting the proliferation of pathogenic Treg-like cells and tumor cell escape [68]. Co-expression of LAG3 and PDCD1 significantly reduces the proliferation and secretion function of T cells [69]. Likewise, HAVCR2 can inhibit T cell immune responses by inducing BAT3 release through binding to galectin [70, 71]. Additionally, HAVCR2 can disrupt immune synapses by binding to receptor phosphatases CD45 and CD148 [72]. In this study, we investigated the correlation between MEF2C and immunosuppressive molecules, including CTLA4, FOXP3, LAG3, PDCD1 and HAVCR2. Our correlation analysis revealed that higher MEF2C expression was associated with increased presence of these immunosuppressive molecules. Based on these findings, we hypothesize that MEF2C may upregulate the expression of immunosuppressive molecules in tumors, weakening the inhibitory effect of the immune system on tumor cells and promoting tumor development and immune escape.
However, there are some limitations in this study. First, this is a retrospective study, and bioinformatics research for this work is conducted partly on publicly available data sets. We need to ensure that the results of this survey are accurate by using clinical trial participants in a prospective study design. Second, how MEF2C influences the prognosis of GC through the immune system is not yet fully understood. Further cytological and animal experimental studies in vivo and in vitro are needed.
Conclusion
In this study, we innovatively utilized the difference of PAC results to construct a model based on ICI Score for predicting the prognosis of GC patients. The model identified MEF2C as a transcription factor associated with immunotherapy in gastric cancer. The ICI Score exhibited significant correlations with tumor heterogeneity, mutation rate of tumor-related genes, different stages of GC, TMB and immune cell infiltration. Notably, the identified transcription factor MEF2C3 can serve as a biomarker for assessing the efficacy of immunotherapy for GC. The prognostic model effectively predicted patient outcomes and facilitated patient stratification, offering valuable insights for clinical decision-making. It is important to note that the predictive accuracy of our model requires validation through future prospective clinical trials, as well as further cytological and animal experiments to elucidate the deeper mechanisms.
Supplementary Information
Supplementary Material 1: Supplementary Fig. 1. Classification of ICI subtypes and gene subtypes. (A) Immune microenvironment score and the number of immune-infiltrating cells; (B) Five ICI subtypes of GC patients by consensus cluster package; (C) Four gene subtypes of GC patients by consensus cluster package.
Supplementary Material 2: Supplementary Fig. 2. Enrichment analyses. (A) GO enrichment analysis on gene M; (B) KEGG enrichment analysis on gene M; (C) GO enrichment analysis on gene N; (D) KEGG enrichment analysis on gene N; (E) GSEA enrichment analysis in the high ICI Score group; (F) GSEA enrichment analysis in the low ICI Score group.
Acknowledgements
Not applicable.
Abbreviations
- GC
Gastric cancer
- OS
Overall survival
- RFS
Relapse-free survival
- EGFR
Epidermal growth factor receptor
- VEGF
Vascular endothelial growth factor
- ICIs
Immune checkpoint inhibitors
- PCA
Principal component analysis
- MEF2C
Myocyte enhancer factor 2c
- IL
Interleukin
- EMT
Epithelial-mesenchymal transition
- TGF
Transforming growth factor
- TCGA
The Cancer Genome Atlas
- FPKM
Fragments Per Kilobase of transcript per Million mapped reads
- TPM
Transcripts Per Million
- GEO
Gene Expression Omnibus
- TIIC
Tumor infiltrating immune cell
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GSEA
Gene set enrichment analysis
- GDSC
Genomics of Drug Sensitivity in Cancer
- IC50
Half maximal inhibitory concentration
- TF
Transcription factor
- WGCNA
Weighted correlation network analysis
- TOM
Topology overlap measure
- KM
Kaplan-Meier
- MATH
Mutant-allele tumor heterogeneity
- Tregs
Regulatory T cells
- CR
Complete response
- PR
Partial response
- TMB
Tumor mutation burden
- MSI
Microsatellite instability
- ROC
Receiver operator characteristic
- TCR
T cell receptor
Authors’ contributions
Si-yu Wang, Yu-xin Wang, Lu-shun Guan: Investigation, Project design, Methodology, Software, Experiment implement, Writing—original draft, Writing—review & editing. Ao Shen, Run-jie Huang, Shu-qiang Yuan, Yu-long Xiao: Methodology, Resources. Li-shuai Wang, Dan Lei, Yin Zhao, Chuan Lin, Chang-ping Wang: Data curation, Resources. Zhi-ping Yuan (corresponding author): Conceptualization, Resources, Supervision.
Funding
Not applicable.
Data availability
The datasets generated during and/or analyzed during the current study are available in the official websites, showing as follows: TCGA [https://portal.gdc.cancer.gov/]; GEO [https://www.ncbi.nlm.nih.gov/geo/]; CIBERSORTx [https://cibersortx.stanford.edu/]; MSigDB [https://www.gsea-msigdb.org/gsea/msigdb]; Cistrome Cancer database [http://cistrome.org/CistromeCancer/].
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Prior to participation in this study, all patients provided informed consent for the use of their information and specimens for research purposes.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Si-yu Wang, Yu-xin Wang and Lu-shun Guan contributed equally to this work and share first authorship.
References
- 1.Lauren P. The Two Histological Main Types of Gastric Carcinoma: Diffuse and So-Called Intestinal-Type Carcinoma An Attempt at a Histo-Clinical Classification. Acta Pathol Microbiol Scand. 1965;64:31–49. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 3.Wong MCS, Huang J, Chan PSF, et al. Global Incidence and Mortality of Gastric Cancer, 1980–2018. JAMA Netw Open. 2021;4(7):e2118457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swan R, Miner TJ. Current role of surgical therapy in gastric cancer. World J Gastroenterol. 2006;12(3):372–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Machlowska J, Baj J, Sitarz M, et al. gastric cancer: epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci. 2020;21(11):4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song Z, Wu Y, Yang J, et al. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39(7):1010428317714626. [DOI] [PubMed] [Google Scholar]
- 7.Wilke H, Preusser P, Fink U, et al. Preoperative chemotherapy in locally advanced and nonresectable gastric cancer: a phase II study with etoposide, doxorubicin, and cisplatin. J Clin Oncol. 1989;7(9):1318–26. [DOI] [PubMed] [Google Scholar]
- 8.Padma VV. An overview of targeted cancer therapy. Biomedicine (Taipei). 2015;5(4):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YT, Tan YJ, Oon CE. Molecular targeted therapy: Treating cancer with specificity. Eur J Pharmacol. 2018;834:188–96. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Wang C, Xu M, et al. Preoperative NLR for predicting survival rate after radical resection combined with adjuvant immunotherapy with CIK and postoperative chemotherapy in gastric cancer. J Cancer Res Clin Oncol. 2017;143(5):861–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, Dubois RN. Immunosuppression associated with chronic inflammation in the tumor microenvironment. Carcinogenesis. 2015;36(10):1085–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbott M, Ustoyev Y. Cancer and the immune system: the history and background of immunotherapy. Semin Oncol Nurs. 2019;35(5):150923. [DOI] [PubMed] [Google Scholar]
- 13.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39(1):1–10. [DOI] [PubMed] [Google Scholar]
- 14.Ibrahim R, Saleh K, Chahine C, et al. LAG-3 inhibitors: novel immune checkpoint inhibitors changing the landscape of immunotherapy. Biomedicines. 2023;11(7). [DOI] [PMC free article] [PubMed]
- 15.De Guillebon E, Roussille P, Frouin E, et al. Anti program death-1/anti program death-ligand 1 in digestive cancers. World J Gastrointest Oncol. 2015;7(8):95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang H, Zou X, Yang S, et al. Identification of lactylation related model to predict prognostic, tumor infiltrating immunocytes and response of immunotherapy in gastric cancer. Front Immunol. 2023;14:1149989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abril-Rodriguez G, Ribas A. SnapShot: Immune Checkpoint Inhibitors. Cancer Cell. 2017;31(6):848-848 e1. [DOI] [PubMed] [Google Scholar]
- 18.Stanley JO, Mohamed SA. Cancer immunotherapy: a brief review of the history, possibilities, and challenges ahead. J Can Metastasis Treat. 2017;3:250–61. [Google Scholar]
- 19.Digklia A, Wagner AD. Advanced gastric cancer: Current treatment landscape and future perspectives. World J Gastroenterol. 2016;22(8):2403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu M, Zhang Y, Mao R, et al. A Risk Model of Eight Immune-Related Genes Predicting Prognostic Response to Immune Therapies for Gastric Cancer. Genes (Basel). 2022;13(5):720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei J, Zeng Y, Gao X, et al. A novel ferroptosis-related lncRNA signature for prognosis prediction in gastric cancer. BMC Cancer. 2021;21(1):1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madugula K, Mulherkar R, Khan ZK, et al. MEF-2 isoforms’ (A-D) roles in development and tumorigenesis. Oncotarget. 2019;10(28):2755–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134(23):4131–40. [DOI] [PubMed] [Google Scholar]
- 24.Assali A, Harrington AJ, Cowan CW. Emerging roles for MEF2 in brain development and mental disorders. Curr Opin Neurobiol. 2019;59:49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Radford JC, Ragusa MJ, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci U S A. 2008;105(27):9397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Zhao Y. Progress on the roles of MEF2C in neuropsychiatric diseases. Mol Brain. 2022;15(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong C, Yang XZ, Zhang CY, et al. Myocyte enhancer factor 2C and its directly-interacting proteins: A review. Prog Biophys Mol Biol. 2017;126:22–30. [DOI] [PubMed] [Google Scholar]
- 28.Fu W, Wei J, Gu J. MEF2C mediates the activation induced cell death (AICD) of macrophages. Cell Res. 2006;16(6):559–65. [DOI] [PubMed] [Google Scholar]
- 29.Boutilier AJ, Elsawa SF. Macrophage Polarization States in the Tumor Microenvironment. Int J Mol Sci. 2021;22(13):6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verreck FA, De Boer T, Langenberg DM, et al. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc Natl Acad Sci U S A. 2004;101(13):4560–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1–m2 polarization balance. Front Immunol. 2014;5:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao X, Di Q, Liu H, et al. MEF2C promotes M1 macrophage polarization and Th1 responses. Cell Mol Immunol. 2022;19(4):540–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai X, Wu L, Liang T, et al. Overexpression of myocyte enhancer factor 2 and histone hyperacetylation in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2008;134(1):83–91. [DOI] [PubMed] [Google Scholar]
- 35.Zhang JJ, Zhu Y, Xie KL, et al. Yin Yang-1 suppresses invasion and metastasis of pancreatic ductal adenocarcinoma by downregulating MMP10 in a MUC4/ErbB2/p38/MEF2C-dependent mechanism. Mol Cancer. 2014;13:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M, Zhu B, Davie J. Alternative splicing of MEF2C pre-mRNA controls its activity in normal myogenesis and promotes tumorigenicity in rhabdomyosarcoma cells. J Biol Chem. 2015;290(1):310–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19(11):1438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu W, Huang C, Wang Q, et al. MEF2 transcription factors promotes EMT and invasiveness of hepatocellular carcinoma through TGF-beta1 autoregulation circuitry. Tumour Biol. 2014;35(11):10943–51. [DOI] [PubMed] [Google Scholar]
- 39.Singh J, Kumar R, Verma D, et al. MEF2C expression, but not absence of bi-allelic deletion of TCR gamma chains (ABD), is a predictor of patient outcome in Indian T-acute lymphoblastic leukemia. Am J Blood Res. 2020;10(5):294–304. [PMC free article] [PubMed] [Google Scholar]
- 40.Menshawy NE, El-Ghonemy MS, Ebrahim MA, et al. Aberrant ecotropic viral integration site-1 (EVI-1) and myocyte enhancer factor 2 C gene (MEF2C) in adult acute myeloid leukemia are associated with adverse t (9:22) & 11q23 rearrangements. Ann Hematol. 2024;103(7):2355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noh MG, Yoon Y, Kim G, et al. Practical prediction model of the clinical response to programmed death-ligand 1 inhibitors in advanced gastric cancer. Exp Mol Med. 2021;53(2):223–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24(9):1449–58. [DOI] [PubMed] [Google Scholar]
- 44.Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kursa MB, Rudnicki WR. Feature Selection with the Boruta Package. J Stat Softw. 2010;36(11):1–13. [Google Scholar]
- 48.Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenacre M, Groenen PJF, Hastie T, et al. Principal component analysis. Nature Reviews Methods Primers. 2022;2(1):100. [Google Scholar]
- 50.Harrell Jr FE. _rms: Regression Modeling Strategies_. R package version 6.7–1. 2023. https://CRAN.R-project.org/package=rms.
- 51.Marshall R. _regplot: Enhanced Regression Nomogram Plot_. R package version 1.1. 2020. https://CRAN.R-project.org/package=regplot.
- 52.Iorio F, Knijnenburg TA, Vis DJ, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166(3):740–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE. 2014;9(9):e107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Therneau T. _A Package for Survival Analysis in R_. R package version 3.5–7. 2023. https://CRAN.R-project.org/package=survival.
- 55.Kassambara A, Kosinski M, Biecek P. _survminer: Drawing Survival Curves using 'ggplot2'_. R package version 0.4.9. 2021. https://CRAN.R-project.org/package=survminer.
- 56.Mroz EA, Tward AD, Hammon RJ, et al. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015;12(2):e1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ran X, Xiao J, Zhang Y, et al. Low intratumor heterogeneity correlates with increased response to PD-1 blockade in renal cell carcinoma. Ther Adv Med Oncol. 2020;12:1758835920977117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes[J]. Science. 2013;339(6127):1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tian Q, Lu Y, Yan B, et al. Integrative Bioinformatics Analysis Reveals That miR-524-5p/MEF2C Regulates Bone Metastasis in Prostate Cancer and Breast Cancer. Comput Math Methods Med. 2022;2022:5211329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ostrander JH, Daniel AR, Lofgren K, et al. Breast tumor kinase (protein tyrosine kinase 6) regulates heregulin-induced activation of ERK5 and p38 MAP kinases in breast cancer cells. Cancer Res. 2007;67(9):4199–209. [DOI] [PubMed] [Google Scholar]
- 62.Chen S, Xu J, Yin S, et al. Identification of a Two-Gene Signature and Establishment of a Prognostic Nomogram Predicting Overall Survival in Diffuse-Type Gastric Cancer. Curr Oncol. 2022;30(1):171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hosseini A, Gharibi T, Marofi F, et al. CTLA-4: From mechanism to autoimmune therapy. Int Immunopharmacol. 2020;80:106221. [DOI] [PubMed] [Google Scholar]
- 64.Gough SC, Walker LS, Sansom DM. CTLA4 gene polymorphism and autoimmunity. Immunol Rev. 2005;204:102–15. [DOI] [PubMed] [Google Scholar]
- 65.Tai X, Van Laethem F, Pobezinsky L, et al. Basis of CTLA-4 function in regulatory and conventional CD4(+) T cells. Blood. 2012;119(22):5155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J, Gong R, Zhao C, et al. Human FOXP3 and tumour microenvironment. Immunology. 2023;168(2):248–55. [DOI] [PubMed] [Google Scholar]
- 67.Kim HK, Won KY, Han SA. The antioncogenic effect of Beclin-1 and FOXP3 is associated with SKP2 expression in gastric adenocarcinoma. Medicine (Baltimore). 2021;100(33):e26951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Higuchi Y, Yasunaga JI, Mitagami Y, et al. HTLV-1 induces T cell malignancy and inflammation by viral antisense factor-mediated modulation of the cytokine signaling. Proc Natl Acad Sci U S A. 2020;117(24):13740–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi AP, Tang XY, Xiong YL, et al. Immune Checkpoint LAG3 and Its Ligand FGL1 in Cancer. Front Immunol. 2021;12:785091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rangachari M, Zhu C, Sakuishi K, et al. Bat3 promotes T cell responses and autoimmunity by repressing Tim-3-mediated cell death and exhaustion. Nat Med. 2012;18(9):1394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wolf Y, Anderson AC, Kuchroo VK. TIM3 comes of age as an inhibitory receptor. Nat Rev Immunol. 2020;20(3):173–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clayton KL, Haaland MS, Douglas-Vail MB, et al. T cell Ig and mucin domain-containing protein 3 is recruited to the immune synapse, disrupts stable synapse formation, and associates with receptor phosphatases. J Immunol. 2014;192(2):782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Supplementary Fig. 1. Classification of ICI subtypes and gene subtypes. (A) Immune microenvironment score and the number of immune-infiltrating cells; (B) Five ICI subtypes of GC patients by consensus cluster package; (C) Four gene subtypes of GC patients by consensus cluster package.
Supplementary Material 2: Supplementary Fig. 2. Enrichment analyses. (A) GO enrichment analysis on gene M; (B) KEGG enrichment analysis on gene M; (C) GO enrichment analysis on gene N; (D) KEGG enrichment analysis on gene N; (E) GSEA enrichment analysis in the high ICI Score group; (F) GSEA enrichment analysis in the low ICI Score group.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in the official websites, showing as follows: TCGA [https://portal.gdc.cancer.gov/]; GEO [https://www.ncbi.nlm.nih.gov/geo/]; CIBERSORTx [https://cibersortx.stanford.edu/]; MSigDB [https://www.gsea-msigdb.org/gsea/msigdb]; Cistrome Cancer database [http://cistrome.org/CistromeCancer/].
All data generated or analyzed during this study are included in this published article and its supplementary information files.