ABSTRACT
Background
Acute lymphocytic leukemia (ALL), characterized by uncontrolled growth of abnormal lymphocytes, predominantly affects children. Genetic analysis focusing on genes and microRNAs reveals important information about the biology of ALL, enabling accurate diagnosis and treatment. This study examines gene and microRNA expression in B cell ALL to improve early diagnosis and personalized treatment.
Methods
Bone marrow samples were collected from patients both before and after the induction phase of chemotherapy. Comprehensive diagnostic techniques including flow cytometry, molecular assays, real‐time PCR for common translocations, karyotyping, and complete blood count (CBC) analysis were employed. These methods were utilized to determine the type and risk assessment of ALL, identify specific gene and microRNA expressions, and measure blood cell counts.
Results
The study comprised 12 patients, all under the age of 18. Post‐treatment RT‐PCR analysis revealed significant reductions in the expression of the ABCB1 gene, miR‐129‐5p, and miR‐9‐5p following the induction phase of chemotherapy. Karyotype analysis indicated that two patients were hypodiploid; unfortunately, both of these patients did not survive.
Conclusion
MicroRNAs and ABC genes serve as predictive and prognostic biomarkers in Acute Lymphoblastic Leukemia (ALL) and should be carefully reconsidered. It is more accurate to state that while microRNAs and ABC genes may potentially influence treatment response in ALL, further research is crucial to fully understand their roles in determining treatment outcomes.
Keywords: ABCB1, acute lymphoblastic leukemia, childhood, MicroRNA, relapse
MicroRNA and ABC gene serve as predictive and prognostic biomarkers influencing response to treatment in (ALL), and it is important to conduct further research to understand the importance of these factors in determining the outcome of treatment.
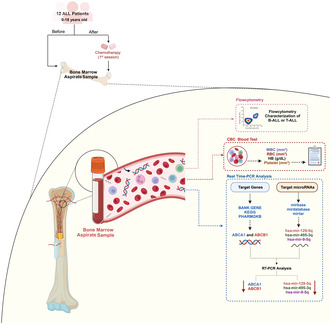
1. Introduction
Cancer remains the leading cause of death globally, and its incidence is steadily increasing, leading to increased mortality [1, 2, 3, 4]. Acute lymphocytic leukemia (ALL) is a type of cancer that involves the abnormal development of immature lymphocytes and their precursors, whether of B or T cell origin. This undeclared immunity leads to the conversion of normal bone marrow components and other lymphoid organs, creating the unique disease pattern characteristic of acute lymphocytic leukemia. Acute lymphocytic leukemia is slightly more common in men than in women and three times more common in whites than blacks [5]. It represents the most prevalent form of cancer in childhood, with the highest incidence occurring between the ages of 2 and 10. Acute Lymphocytic Leukemia is more frequently observed in children with Trisomy 21 (Down syndrome), neurofibromatosis Type 1, Bloom syndrome, and ataxia telangiectasia [6]. Patients often exhibit complications related to anemia, thrombocytopenia, and neutropenia due to the infiltration of the bone marrow by the tumor. These symptoms may manifest as fatigue, increased bruising/bleeding, and increased infections. B symptoms such as fever, night sweats, and loss of consciousness are common but may be mild. Hepatomegaly, splenomegaly, and lymphadenopathy may be observed in half of adults. Central nervous system (CNS) involvement is common and may be accompanied by heart disease or symptoms often associated with increased intracranial pressure [7, 8].
Due to the importance of childhood and the notable impact of (B‐ALL) on health and disease development at all stages of life, with the global spread of ALL, early research in screening and diagnosis is vital [9]. In this context, genetic analysis is the most powerful tool to identify genomic alterations for ALL diagnostics, risk assessment, and treatment selection in ALL, serving two purposes: discover new molecular targets to achieve a better understanding of ALL biological processes and therapeutic roles, aiming to discover effective changes in better disease management, is important [10]. Identifying the sequence of genes that play a role in childhood (ALL) can lead to the classification of their genetics profile, and at higher levels, investigating relevant epigenetic factors can provide more specificity for diagnosis and screening [11]. As the survival rate in children with ALL increases, the role of drug resistance and precision medicine is highly crucial [12]. Studies analyzing gene expression data have identified more than 20 genetic variants of B‐cell acute lymphocytic leukemia in pediatric patients [13]. One of the two main types in this group are (ABCB1) and (ABCA1).
ABC transporters are membrane‐spanning proteins that serve as carriers, transporting substances across various cellular compartments, including the cell interior and the cytoplasmic membrane. This type of transport occurs against the concentration gradient and relies on ATP hydrolysis. This large group has 49 members, and is divided into seven subfamilies labeled A through G. These carriers, are responsible for the transport of xenobiotic compounds, other toxic substances and natural anti‐cancer agents. Consequently, they contribute to resistance against chemotherapy in various cancers, including lymphoblastic and myeloid leukemias [14, 15, 16]. ABCB1 is related to the membrane transporter P‐glycoprotein (P‐gp), which is a product of the ABCB1 gene and serves dual roles as a functional barrier and an efflux transporter in various tissues, potentially affecting the pharmacokinetics of several anti‐cancer medications. Identification of these genes and potential polymorphisms may identify patients who may be more sensitive to various medications, including doxorubicin, vincristine, and prednisone [17, 18, 19]. The ABCA1 protein promotes the transfer of cellular cholesterol from the plasma membrane to apolipoprotein A‐I. Cancer cells often display high levels of intracellular cholesterol, and a deficiency in ABCA1 leads to an accumulation of mitochondrial cholesterol. This accumulation promotes cancer by stopping the release of cell death‐promoting molecules from mitochondria, and indicating that decreasing the ABCA1 gene plays a crucial role in the cancer phenotype [20]. MicroRNAs play a role in many processes such as apoptosis, tumorigenesis, cell growth and division, hematopoiesis, and drug resistance. The expression profile of microRNAs differs in the biological functions of different cells, leading to significant differences between normal and malignant hematopoietic diseases. This difference can be used to classify ALL subgroups. Additionally, microRNAs have the potential as diagnostic markers, prognostic indicators, and personalized therapeutic targets in childhood ALL [21]. Further studies will help unlock novel strategies for cancer management.
This study aims to explore the expression of drug resistance‐related genes and associated microRNAs in B‐cell Acute Lymphoblastic Leukemia (B‐ALL). The findings may enhance diagnosis, identify treatments, and target drug resistance, contributing to more effective diagnostic tools and personalized therapies for improved patient outcomes.
2. Methods
We conducted fundamental and applied research involving 12 patients diagnosed with B‐cell acute lymphocytic leukemia through clinical and laboratory evaluations at referral centers at Rasoul‐e‐Akram Hospital, affiliated with Iran University of Medical Sciences in Tehran, Iran. The study was carried out from January 2021 to December 2023.
2.1. Patient Selection
Rasoul‐e‐Akram Hospital patients with B‐cell acute lymphocytic leukemia patients under 18 years of age, without any chronic diseases, diagnosed their cancer within the last 3 months, treatment just with the BFM protocol, with no history of previous cancer, and those who had not been using immunosuppressive drugs. Patients above 18 years old, with a history of prior cancer, individuals who had not received treatment with the BFM protocol, and patients with chronic diseases were not eligible to participate in this study (Figure 1).
FIGURE 1.
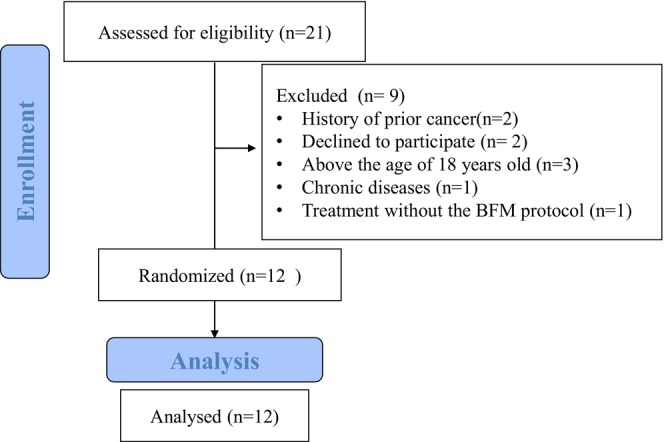
Flow diagram.
Bone marrow samples, each totaling 5 mL, were systematically collected from study participants both before and after the chemotherapy induction phase and stored at −80°C until use. A meticulously designed questionnaire was employed to compile comprehensive patient demographics information, such as gender and age. Moreover, flow cytometry and complete blood count (CBC) analysis were performed for each patient to assess their ALL type, counts of white blood cells (WBC), red blood cells (RBC), platelets, and the concentration of hemoglobin (HB). Karyotype analysis data for each patient were also obtained from their hospital records.
All the examinations were executed with the ethical code of (IR.IUMS.REC.1398.1198) certified by the ethical committee of the Iran University of Medical Sciences.
2.2. Flow Cytometry
To distinguish between acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) and further classify ALL into T‐cell ALL (T‐ALL) and B‐cell ALL (B‐ALL) along with specific B‐ALL subtypes, we used flow cytometry‐based analysis to measure the expression of cell surface and intracellular markers. Bone marrow aspirate samples were collected in Ethylenediaminetetraacetic Acid (EDTA) tubes and then stained with monoclonal antibodies against the following human proteins. In our study, we employed a comprehensive panel of antibodies to accurately differentiate between acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML). Additionally, we subclassified ALL into T‐cell ALL (T‐ALL) and B‐cell ALL (B‐ALL), further delineating specific B‐ALL subtypes, as detailed below:
ALL Antibody Panel: CD2, CD1a, CD3, CD5, CD7, CD4/8, HLA‐DR, CD10, CD19, CD20, CD22, CD13, CD33, CD34, TDT.
AML Antibody Panel: CD2, CD1a, CD3, CD5, CD7, CD4/8, HLA‐DR, CD10, CD19, CD20, CD22, CD13, CD33, CD34, CD117, CD14, CD64, CD15, MPO.
The immunophenotypic characterization of B‐ALL subtypes includes Early Pre‐B ALL (TdT+, CD19+, CD10−), Pre‐B ALL (CD10±, CD22, CD34, CD19, TdT, HLA‐DR positive), and Pro‐B ALL (CD19, CD34, CD22, TdT positive).
The intracellular markers included MPO, TDT, and cytoplasmic CD3, while the cell surface markers included CD34, CD10, CD1a, CD2, CD3, CD5, CD7, CD19, CD20, CD22, HLA‐DR, CD4, CD8, CD13, CD33, CD14, CD64, and CD15. All which were purchased from Biolegend, USA. For the cell surface staining, 100 μL samples were incubated with the appropriate concentration of antibodies for 30 min at 4°C in the dark. After incubation, we performed RBC lysis using 1× RBC Lysis Buffer (DACell, Iran) for 5 min at room temperature, followed by twice washes with PBS at 1500 rpm for 5 min at 4°C. Intracellular staining was performed using a permeabilization/fixation buffer. Initially, samples were subjected to 1x RBC Lysis Buffer and then washed twice with PBS. Intracellular markers were stained using the Intracell Solution Kit (Immunostep, Spain; Lot: 416686) according to the manufacturer's instructions. Finally, the samples were analyzed using Beckman Coulter FC500 equipped with CXP Software and Flowjo 7.6 software (Figure 2).
FIGURE 2.
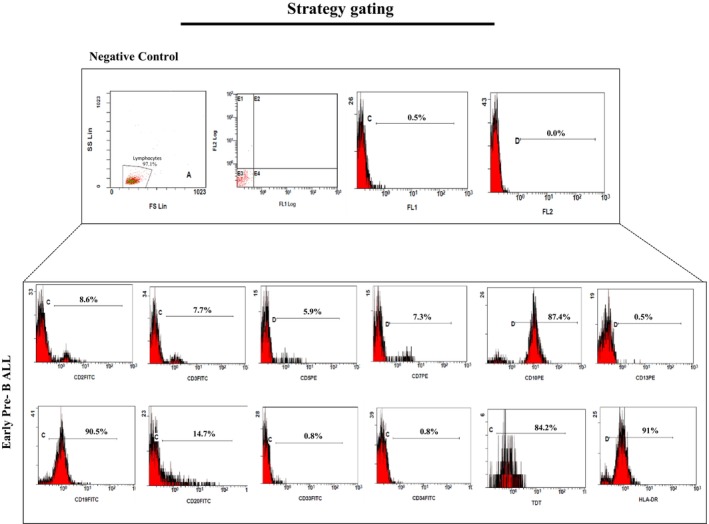
Strategy gating for ALL analysis by flow cytometry. Sequential gating strategy illustrating the identification and characterization of leukemic cell populations in early pre‐B‐ALL patient sample. Lymphocyte population selection based on FSC and SSC properties. Representative histograms of CD2, CD3, CD5, CD7, CD10, CD13, CD19, CD20, CD33, CD34, TDT and HLA‐DR expression on lymphocytes.
2.3. Identification of Target Genes and MicroRNA
The identification of target genes for the study was performed based on the patient's specific cancer type and the chemotherapy medications given. Genes identified through comprehensive literature searches on Google Scholar and thought to be responsive to various stages of treatment in clinical trials were selected as targets. To facilitate this, BANK GENE, KEGG, and PHARMGKB were used. Finally, ABCA1 and ABCB1 were selected as the study's target genes. For selecting the target microRNA, an exhaustive search was executed in articles and pertinent databases dedicated to microRNA, including mirbase, mirdatabase, and mirtar, which were specifically selected based on their high specificity percentage. Finally, hsa‐mir‐129‐5q, has‐mir‐495‐3q, and hsa‐mir‐9‐5q were selected as the target microRNAs of this study.
2.4. RNA Extraction and Real‐Time PCR
Total RNA was isolated from bone marrow samples using the TRIzol reagent (Tiangen, Cat. No. 15596‐026, Beijing, China). A total of 300 μL of chloroform was added to this solution, shaken for 30 s, incubated for 5 min at room temperature, and then centrifuged at 12,000 rpm at 4°C for 15 min. The resulting supernatant was transferred to a new RNase‐free microtube, combined with an equal volume of cold isopropanol, and stored at −80°C for 20 min. After centrifugation at 12,000 rpm at 4°C for 10 min and subsequent mixing with 70% ethanol, the sample was centrifuged again at 8000 rpm at 4°C for 10 min. The pellet was then combined with Tris‐EDTA and preserved at −80°C. The concentration of the extracted RNA was determined using a UV spectrophotometer (NanoDrop One Microvolume UV–Vis spectrophotometer, Thermofisher, USA). For cDNA synthesis, reverse transcription of RNA was performed using a kit (Anacell, LOT: MR0084) following the manufacturer's protocol. Two different protocols for the reverse transcription reaction were used including a cDNA synthesis MIR kit that works with steam loop primers, and for gene expression analysis random hexamer primer for cDNA synthesis was used. The reverse transcription reaction occurred at 37°C for 1 h, with subsequent inactivation of the reverse transcriptase at 70°C for 5 min. DNA polymerase activation was conducted at 95°C for 15 min, followed by 45 cycles of a three‐step PCR (95°C for 25 s, 58°C for 15 s, and 72°C for 40 s) then incubated at 35°C for 30 s [22, 23]. Dissociation curve analysis of hsa‐mir‐129‐5q, has‐mir‐495‐3q, and hsa‐mir‐9‐5q showed a single peak. The GADPH gene was used as a control in this study [24, 25, 26].
2.5. Statistical Analysis
Correlations between variables were analyzed using one‐sample and paired t‐tests. Continuous variables were presented as means with standard deviations (SD), while qualitative variables were expressed as frequency percentages. Statistical analysis was performed using SPSS 20 and GraphPad Prism 10.1.2 software. A significance level of p < 0.05 was adopted for determining statistical significance.
3. Results
3.1. Overall Analysis
The study population consisted of 12 patients, 7 males (58.3%) and 5 females (41.7%), aged between 3 and 16 years (mean age ± SD: 10.25 ± 4.15 years). The average at the time of diagnosis was 9.5 years. Flow cytometry analysis was used to determine the patients' subtype of acute lymphoblastic leukemia in patients. The results revealed four cases of early B‐ALL, five cases of pre B‐ALL, and three cases of pro B‐ALL. Karyotype analysis revealed that five patients were diploid, five patients were hyperdiploid, two patients were hypodiploid without any significant correlation and one patient was Philadelphia (Ph) chromosome positive. Participants were followed up for a period of up to 3 years, and the majority of them remained alive throughout the study period. Unfortunately, three patients succumbed to the disease as a result of relapse refractory disease (Figure 3).
FIGURE 3.
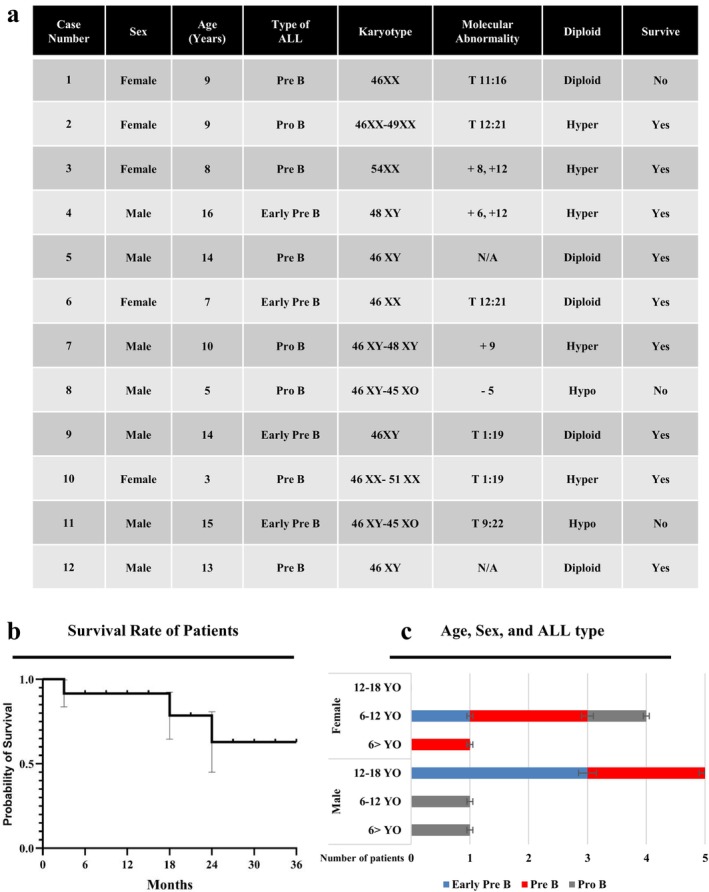
(a), (c) The demographic and clinical information of patients including age, sex, type of ALL, karyotype, and molecular abnormality were demonstrated. YO means years old. T means translocation. (+) indicates the presence of an extra chromosome, while (−) indicates the absence of a chromosome. N/A means without abnormality. (b) Kaplan–Meier survival curves of each patient were demonstrated for 36‐month follow‐up. Three patients did not survive during the study.
3.2. Complete Blood Count Analysis
Blood tests revealed abnormal patient characteristics; In eight individuals, the white blood cell (WBC) count was above the normal range that was reported in various studies (4.5 to 11.0 × 109/L) [27]. A total of 10 patients had a red blood cell (RBC) count below normal (4.5 to 5 × 109/L) [28] and 11 patients had hemoglobin (Hb) below normal (12 g/dL) [29]. Additionally, the platelet count was lower than normal (150–450 × 109/L) [30] in nine patients. The mean ± SD of WBC, RBC, HB, and platelet of patients were evaluated (58.21 ± 128.8), (3.69 ± 0.64), (8.85 ± 2.14), (102.45 ± 131.87), respectively (Figure 4).
FIGURE 4.
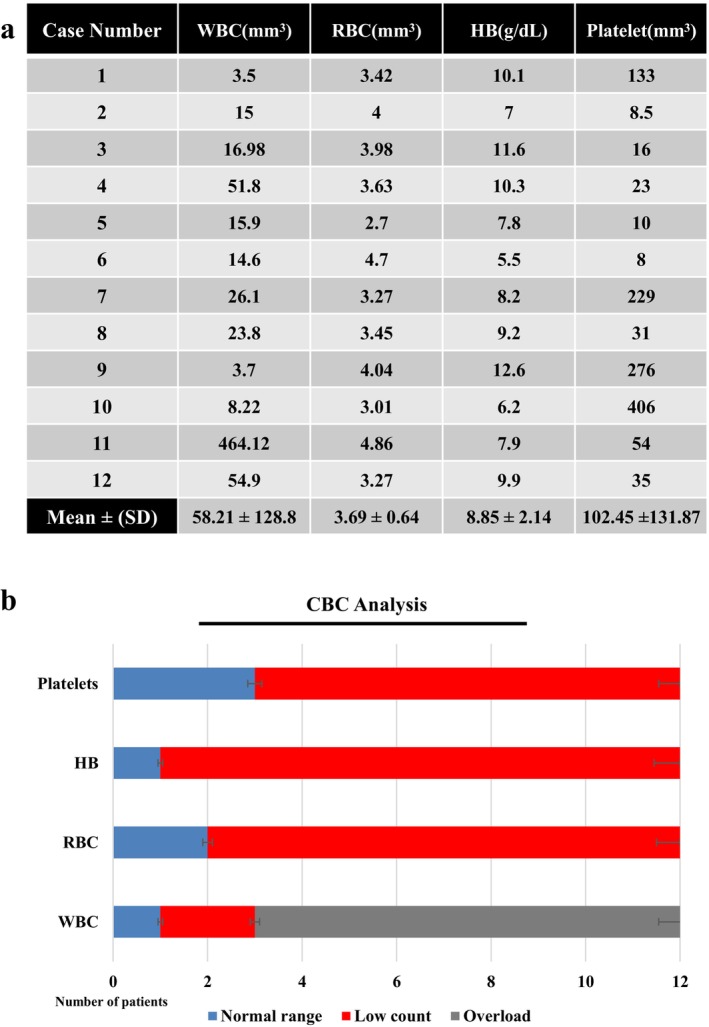
(a) Table displayed CBC information of patients with a mean of each factor. (b) The number of patients based on the normal range of each factor in CBC analysis.
3.3. Real Time PCR Analysis
RT‐PCR analysis demonstrated a decreasing trend in all expressions of gene and microRNA of the study between before and after treatments. A delta cycle threshold (ΔCT) average of each gene and microRNA were demonstrated in Figure 5 before and after chemotherapy induction. Moreover, significant correlations were investigated in reducing the expressions of the ABCB1 gene (p value < 0.0001), mir‐129‐5q (p value < 0.001), and mir‐9‐5q (p value < 0.05), between before and after induction chemotherapy. Seven patients (58%) had not any ABCB1 expressions after the induction phase of chemotherapy without any significant correlation with survival rate (Figure 5). It was notable that all deceased patients had lower level of mir‐129‐5q and mir‐9‐5q expressions after the chemotherapy induction. This points to a potential prognostic value of these microRNAs in predicting patient outcomes following treatment.
FIGURE 5.
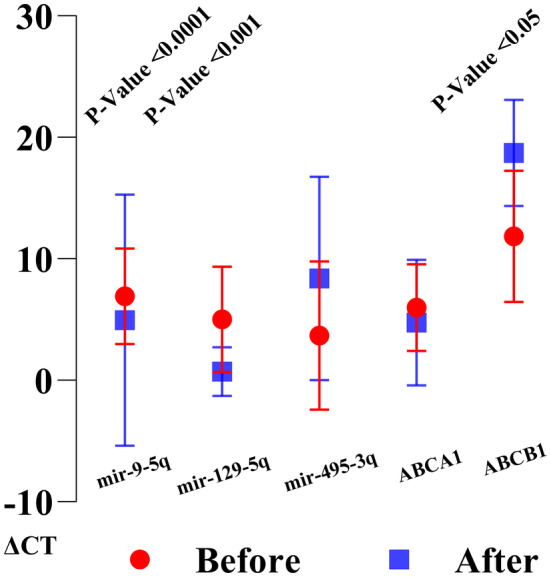
RT‐PCR analysis of patients was depicted, showcasing the CT values. Following treatment, a paired t‐test revealed a significant decrease in the expression of the ABCB1 gene, mir‐129‐5q, and mir‐129‐5q.
4. Discussion
Leukemia is the most common malignancy affecting children, with acute lymphoblastic leukemia emerging as the predominant form among childhood leukemia cases. A study by Moussavi focused on CBC screening in the diagnosis of childhood ALL, reported that a significant number of patients exhibited leukocytosis. The results of this study showed that the white blood cell (WBC) count increased in 66.6% of the patients [31]. study conducted in 2018, anemia was defined as the initial symptom in ALL patients, and it was found that the hemoglobin (HB) level of patients diagnosed with B‐cell ALL was 7.9 at the time of diagnosis [32]. Additionally, a 2011 study reported that pediatric ALL patients often exhibit low red blood cell (RBC) counts, and the use of iron chelators may exacerbate this symptom [33]. These findings were consistent with our study in which 83.3% of patients showed low red blood cell count. Additionally, in our study, the HB level was 8.8% and 91% of the participants had hemoglobin deficiency. Straub's research shows that low platelet counts were common in acquired thrombocytopenia such as acute lymphocytic leukemia. A common hypothesis was that during leukemic progression, immature blasts suppressed megakaryocytic progenitor cells in the bone marrow, leading to decreased peripheral platelet counts [34]. Also, this investigation demonstrated that platelet count was low in 75% of participants.
ATP‐binding cassette (ABC) transporters play multiple roles in cancer biology and drug resistance. They are responsible for transporting various inflammatory mediators and lipids that directly contribute to tumor progression [35, 36]. Hedditch's study reported that in tumors with an aggressive phenotype, ABCA1 was increasingly expressed more prominently in the tumor than in the stromal cells [37]. In addition, ABCB1, the most extensively studied ATP‐binding cassette transporter, encodes the multi‐drug efflux pump P‐glycoprotein (P‐gp) and is implicated in the transport of a broad spectrum of anti‐cancer drugs [38]. A 2013 study reported that increased ABCB1 expression makes cancer cells more resistant to taxanes. The investigation also suggested that heightened ABCB1 expression in primary tumors may impact the time to relapse, particularly in the subgroup of patients with significant residual tumor post‐surgery [39]. In this study, the expression of ABCA1 and ABCB1 expressions were both downregulated after chemotherapy with a significant correlation in ABCB1 expression that 58% of patients did not have any ABCB1 expression after treatment. The reduction in ABCB1 expression post‐chemotherapy is significant as this gene is often linked to drug resistance. Its downregulation may indicate a reduced capability of the cancer cells to expel chemotherapeutic agents, potentially enhancing treatment efficacy.
MicroRNAs (miRNAs) constitute a family of 21‐ to 25‐nucleotide, noncoding small RNAs primarily serving as gene regulators. The revelation that these diminutive molecules, with their remarkable diversity and biological significance, remained undiscovered for so many years is surprising. Their discoveries ushered in a new era in cancer research. In recent years, studies of miRNAs in oncology have increased rapidly using new molecular techniques. miRNA abnormalities have become an important topic in cancer research. Researchers are actively investigating the value of miRNAs in cancer classification and prognosis, and new treatment strategies targeting miRNAs have been developed [40]. One of the most important microRNAs in cancer is MiR‐129‐5p. Various studies have reported that MiR‐129‐5p promoted cell proliferation, facilitated the formation of colonies, promoted invasion, induced epithelial‐mesenchymal transition (EMT) marker expression, and improved the tumorigenicity of cancer cells [41, 42, 43]. The modulation of DNA damage by miR‐9‐5p, particularly concerning altered TOP2b/180 protein levels, contributes to acquired drug resistance, notably to etoposide, in high‐risk lymphoblastic leukemia cells [44]. Moreover, our results show a significant decrease in the level of these biomarkers during treatment.
Upregulation of miR‐495‐3p in BCR‐ABL‐positive patients resulted in reduced leukemic cell proliferation and improved patient response to therapeutic agents, especially tyrosine kinase inhibitors [45]. Within the leukemia context, miR‐495‐3p has been identified as an important factor in controlling cell proliferation and response to medications. Research indicates that elevated levels of miR‐495‐3p can hinder the growth of leukemia cells, especially in instances involving the presence of the BCR‐ABL fusion gene, a condition frequently linked to unfavorable prognoses and treatment resistance [46]. However, our study did not show a correlation between the miR‐495‐3p expression and overall survival or treatment. The significant decreases in mir‐129‐5q and mir‐9‐5q, along with their association with patient survival, suggest that these microRNAs could serve as valuable biomarkers for assessing the effectiveness of chemotherapy and predicting patient outcomes.
Although the mechanisms driving the results of this study are not fully understood, miRNAs may contribute to pathways involved in cell growth. However, the possibility of using miRNAs as prognostic biomarkers deserves deeper investigation in larger studies.
5. Conclusion
Understanding the roles of microRNAs and ABC transporter genes in B‐ALL is vital for advancing diagnosis, prognosis, and treatment strategies. Chemoresistance poses a significant challenge to effective treatment outcomes in pediatric leukemia. Their combined analysis not only offers insights into the molecular underpinnings of the disease but also paves the way for the development of personalized therapies aimed at overcoming drug resistance and improving patient outcomes. Further research is essential to fully elucidate their complex interactions and to translate these findings into clinical practice.
Author Contributions
Reza Nekouian: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing – original draft, writing – review and editing. Pooya Faranoush: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing – original draft, writing – review and editing. Fatemeh Khesali: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, writing – review and editing. All authors provided critical review and editing to this manuscript. Parisa Shams: writing – original draft, writing – review and editing, formal analysis, methodology, software. Mohammad Reza Foroughi‐Gilvaee: data curation, funding acquisition, investigation, methodology, resources, software. Negin Sadighnia: data curation, funding acquisition, investigation, methodology, resources, software, writing – original draft, writing – review and editing. Dorsa Fallah Azad: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, investigation. MohammadAli Ehsani: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology. Mohammad Faranoush: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing – original draft, writing – review and editing. All authors provided critical review and editing to this manuscript. All authors provided critical review and editing to this manuscript. In addition, all authors have read and approved the final version of this manuscript.
Ethics Statement
All the examinations were executed with the ethical code of (IR.IUMS.REC.1398.1198) certified by the ethical committee of the Iran University of Medical Sciences.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding: The authors received no specific funding for this work.
Pooya Faranoush and Fatemeh Khesali have contributed equally to this work and share first authorship.
Contributor Information
Reza Nekouian, Email: nekouian.r@iums.ac.ir.
Mohammad Faranoush, Email: faranoush47@gmail.com.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Khalaji A., Yancheshmeh F. B., Farham F., et al., “Don't Eat Me/Eat Me Signals as a Novel Strategy in Cancer Immunotherapy,” Heliyon 9, no. 10 (2023): e20507, 10.1016/j.heliyon.2023.e20507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tahavvori A., Alizadeh F., Mousavian A., et al., “Achievements, Challenges, and Limitations of Car‐T Cell Therapy in Cancer Treatment, A Comprehensive Review,” Journal of Pharmaceutical Negative Results 13 (2022): 11145–11152. [Google Scholar]
- 3. Mokhtari Dowlatabad H., Mamdouh A., Yousefpour N., et al., “High‐Frequency (30 MHz–6 GHz) Breast Tissue Characterization Stabilized by Suction Force for Intraoperative Tumor Margin Assessment,” Diagnostics 13, no. 2 (2023): 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arian Y., Faranoush P., Ehsanipour F., Sadighnia N., Elahinia A., and Faranoush M., “Clinical Outcomes of Pediatric Cancer Patients with COVID‐19: A Cross‐Sectional Study,” International Journal of Hematology‐Oncology and Stem Cell Research 18, no. 4 (2024), 10.18502/ijhoscr.v18i4.16756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Puckett Y. and Chan O., “Acute Lymphocytic Leukemia,” in StatPearls (Treasure Island (FL): StatPearls Publishing, 2024). [Google Scholar]
- 6. Roberts K. G., “Genetics and Prognosis of ALL in Children vs Adults,” Hematology. American Society of Hematology. Education Program 2018, no. 1 (2018): 137–145, 10.1182/asheducation-2018.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jain T. and Litzow M. R., “No Free Rides: Management of Toxicities of Novel Immunotherapies in ALL, Including Financial,” Hematology. American Society of Hematology. Education Program 2018, no. 1 (2018): 25–34, 10.1182/asheducation-2018.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dinner S. and Liedtke M., “Antibody‐Based Therapies in Patients With Acute Lymphoblastic Leukemia,” Hematology. American Society of Hematology. Education Program 2018, no. 1 (2018): 9–15, 10.1182/asheducation-2018.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johnston W. T., Lightfoot T. J., Simpson J., and Roman E., “Childhood Cancer Survival: A Report From the United Kingdom Childhood Cancer Study,” Cancer Epidemiology 34, no. 6 (2010): 659–666, 10.1016/j.canep.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 10. Tasian S. K. and Hunger S. P., “Genomic Characterization of Paediatric Acute Lymphoblastic Leukaemia: An Opportunity for Precision Medicine Therapeutics,” British Journal of Haematology 176, no. 6 (2017): 867–882, 10.1111/bjh.14474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pui C. H., Yang J. J., Hunger S. P., et al., “Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration,” Journal of Clinical Oncology 33, no. 27 (2015): 2938–2948, 10.1200/jco.2014.59.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aberuyi N., Rahgozar S., Ghodousi E. S., and Ghaedi K., “Drug Resistance Biomarkers and Their Clinical Applications in Childhood Acute Lymphoblastic Leukemia,” Frontiers in Oncology 9 (2020): 1496, 10.3389/fonc.2019.01496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. den Boer M. L., Cario G., Moorman A. V., et al., “Outcomes of Paediatric Patients With B‐Cell Acute Lymphocytic Leukaemia With ABL‐Class Fusion in the Pre‐Tyrosine‐Kinase Inhibitor Era: A Multicentre, Retrospective, Cohort Study,” Lancet Haematology 8, no. 1 (2021): e55–e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aberuyi N., Rahgozar S., and Moafi A., “The Role of ATP‐Binding Cassette Transporter A2 in Childhood Acute Lymphoblastic Leukemia Multidrug Resistance,” Iranian Journal of Pediatric Hematology and Oncology 4, no. 3 (2014): 118–126. [PMC free article] [PubMed] [Google Scholar]
- 15. Fukuda Y. and Schuetz J. D., “ABC Transporters and Their Role in Nucleoside and Nucleotide Drug Resistance,” Biochemical Pharmacology 83, no. 8 (2012): 1073–1083, 10.1016/j.bcp.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steinbach D., Gillet J. P., Sauerbrey A., et al., “ABCA3 as a Possible Cause of Drug Resistance in Childhood Acute Myeloid Leukemia,” Clinical Cancer Research 12, no. 14 Pt 1 (2006): 4357–4363, 10.1158/1078-0432.Ccr-05-2587. [DOI] [PubMed] [Google Scholar]
- 17. Arrigoni E., Galimberti S., Petrini M., Danesi R., and di Paolo A., “ATP‐Binding Cassette Transmembrane Transporters and Their Epigenetic Control in Cancer: An Overview,” Expert Opinion on Drug Metabolism & Toxicology 12, no. 12 (2016): 1419–1432, 10.1080/17425255.2016.1215423. [DOI] [PubMed] [Google Scholar]
- 18. Pan S. T., Li Z. L., He Z. X., Qiu J. X., and Zhou S. F., “Molecular Mechanisms for Tumour Resistance to Chemotherapy,” Clinical and Experimental Pharmacology & Physiology 43, no. 8 (2016): 723–737, 10.1111/1440-1681.12581. [DOI] [PubMed] [Google Scholar]
- 19. Gregers J., Gréen H., Christensen I. J., et al., “Polymorphisms in the ABCB1 Gene and Effect on Outcome and Toxicity in Childhood Acute Lymphoblastic Leukemia,” Pharmacogenomics Journal 15, no. 4 (2015): 372–379, 10.1038/tpj.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith B. and Land H., “Anticancer Activity of the Cholesterol Exporter ABCA1 Gene,” Cell Reports 2, no. 3 (2012): 580–590, 10.1016/j.celrep.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ultimo S., Martelli A. M., Zauli G., Vitale M., Calin G. A., and Neri L. M., “Roles and Clinical Implications of microRNAs in Acute Lymphoblastic Leukemia,” Journal of Cellular Physiology 233, no. 8 (2018): 5642–5654, 10.1002/jcp.26290. [DOI] [PubMed] [Google Scholar]
- 22. Faranoush P., Jahandideh A., Nekouian R., and Mortazavi P., “Evaluation of the In Vitro and In Vivo Effect of Liposomal Doxorubicin Along With Oncolytic Newcastle Disease Virus on 4T1 Cell Line: Animal Preclinical Research,” Veterinary Medicine and Science 9, no. 3 (2023): 1426–1437, 10.1002/vms3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sadighnia N., Arfaee F., Tavakoli A., and Jahandideh A., “Dextranase Enzyme and Enterococcus faecium Probiotic Have Anti‐Biofilm Effects by Reducing the Count of Bacteria in Dental Plaque in the Oral Cavity of Dogs,” Journal of the American Veterinary Medical Association 261, no. 10 (2023): 1525–1530, 10.2460/javma.23.03.0162. [DOI] [PubMed] [Google Scholar]
- 24. Naeini F. B., Hassanpour S., and Asghari A., “Resveratrol Exerts Anxiolytic‐Like Effects Through Anti‐Inflammatory and Antioxidant Activities in Rats Exposed to Chronic Social Isolation,” Behavioural Brain Research 438 (2023): 114201. [DOI] [PubMed] [Google Scholar]
- 25. Zarebavani M., Baghaei Naeini F., Farahvash A., Moradi F., and Dashti N., “Resveratrol Attenuates Chronic Social Isolation Stress‐Induced Affective Disorders: Involvement of NF‐κB/NLRP3 Axis,” Journal of Biochemical and Molecular Toxicology 37, no. 5 (2023): e23311. [DOI] [PubMed] [Google Scholar]
- 26. Moradi F., Dashti N., Farahvash A., Naeini F. B., and Zarebavani M., “Curcumin Ameliorates Chronic Toxoplasma Gondii Infection‐Induced Affective Disorders Through Modulation of Proinflammatory Cytokines and Oxidative Stress,” Iranian Journal of Basic Medical Sciences 26, no. 4 (2023): 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matter F., Hematology (Seventh Edition), eds. Hoffman R., Benz E. J., Silberstein L. E., et al. (Elsevier, 2018), i–ii. [Google Scholar]
- 28. Pagana K. D., Mosby Diagnostic and Laboratory Test Reference, 7th ed. (St. Louis, Mo: Elsevier, Mosby, 2005). [Google Scholar]
- 29. Pluncevic Gligoroska J., Gontarev S., Dejanova B., Todorovska L., Shukova Stojmanova D., and Manchevska S., “Red Blood Cell Variables in Children and Adolescents Regarding the Age and Sex,” Iranian Journal of Public Health 48, no. 4 (2019): 704–712. [PMC free article] [PubMed] [Google Scholar]
- 30. Zulkafli Z., Janaveloo T., Wan Ab Rahman W. S., Hassan M. N., and Abdullah W. Z., “Extreme Thrombocytosis in a Child: Laboratory Approaches and Diagnostic Challenges,” Oman Medical Journal 34, no. 4 (2019): 336–340, 10.5001/omj.2019.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moussavi F., Hosseini S., Saket S., and Derakhshanfar H., “The First CBC in Diagnosis of Childhood Acute Lymphoblastic Leukemia,” International Journal of Medical Investigation 3, no. 1 (2014): 310–319. [Google Scholar]
- 32. Jaime‐Pérez J. C., García‐Arellano G., Herrera‐Garza J. L., Marfil‐Rivera L. J., and Gómez‐Almaguer D., “Revisiting the Complete Blood Count and Clinical Findings at Diagnosis of Childhood Acute Lymphoblastic Leukemia: 10‐Year Experience at a Single Center,” Hematology, Transfusion and Cell Therapy 41, no. 1 (2019): 57–61, 10.1016/j.htct.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eng J. and Fish J. D., “Insidious Iron Burden in Pediatric Patients With Acute Lymphoblastic Leukemia,” Pediatric Blood & Cancer 56, no. 3 (2011): 368–371, 10.1002/pbc.22851. [DOI] [PubMed] [Google Scholar]
- 34. Strauß G., Vollert C., von Stackelberg A., Weimann A., Gaedicke G., and Schulze H., “Immature Platelet Count: A Simple Parameter for Distinguishing Thrombocytopenia in Pediatric Acute Lymphocytic Leukemia From Immune Thrombocytopenia,” Pediatric Blood & Cancer 57, no. 4 (2011): 641–647, 10.1002/pbc.22907. [DOI] [PubMed] [Google Scholar]
- 35. Nieman K. M., Kenny H. A., Penicka C. V., et al., “Adipocytes Promote Ovarian Cancer Metastasis and Provide Energy for Rapid Tumor Growth,” Nature Medicine 17, no. 11 (2011): 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pyragius C. E., Fuller M., Ricciardelli C., and Oehler M. K., “Aberrant Lipid Metabolism: An Emerging Diagnostic and Therapeutic Target in Ovarian Cancer,” International Journal of Molecular Sciences 14, no. 4 (2013): 7742–7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hedditch E. L., Gao B., Russell A. J., et al., “ABCA Transporter Gene Expression and Poor Outcome in Epithelial Ovarian Cancer,” JNCI Journal of the National Cancer Institute 106, no. 7 (2014): 149–161, 10.1093/jnci/dju149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Orina J. N., Calcagno A. M., Wu C.‐P., et al., “Evaluation of Current Methods Used to Analyze the Expression Profiles of ATP‐Binding Cassette Transporters Yields an Improved Drug‐Discovery Database,” Molecular Cancer Therapeutics 8, no. 7 (2009): 2057–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnatty S. E., Beesley J., Gao B., et al., “ABCB1 (MDR1) Polymorphisms and Ovarian Cancer Progression and Survival: A Comprehensive Analysis From the Ovarian Cancer Association Consortium and the Cancer Genome Atlas,” Gynecologic Oncology 131, no. 1 (2013): 8–14, 10.1016/j.ygyno.2013.07.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang W., Dahlberg J. E., and Tam W., “MicroRNAs in Tumorigenesis: A Primer,” American Journal of Pathology 171, no. 3 (2007): 728–738, 10.2353/ajpath.2007.070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zuo Y., Li Y., Zhou Z., Ma M., and Fu K., “Long Non‐coding RNA MALAT1 Promotes Proliferation and Invasion via Targeting miR‐129‐5p in Triple‐Negative Breast Cancer,” Biomedicine & Pharmacotherapy 95 (2017): 922–928, 10.1016/j.biopha.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 42. Gao J., Yuan Y., Zhang L., et al., “Inhibition of ZEB1‐AS1 Confers Cisplatin Sensitivity in Breast Cancer by Promoting microRNA‐129‐5p‐Dependent ZEB1 Downregulation,” Cancer Cell International 20 (2020): 90, 10.1186/s12935-020-1164-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43. Hwang‐Verslues W. W., Chang P. H., Wei P. C., et al., “miR‐495 Is Upregulated by E12/E47 in Breast Cancer Stem Cells, and Promotes Oncogenesis and Hypoxia Resistance via Downregulation of E‐Cadherin and REDD1,” Oncogene 30, no. 21 (2011): 2463–2474, 10.1038/onc.2010.618. [DOI] [PubMed] [Google Scholar]
- 44. Carvajal‐Moreno J., Hernandez V. A., Wang X., Li J., Yalowich J. C., and Elton T. S., “Effects of Hsa‐miR‐9‐3p and Hsa‐miR‐9‐5p on Topoisomerase IIβ Expression in Human Leukemia K562 Cells With Acquired Resistance to Etoposide,” Journal of Pharmacology and Experimental Therapeutics 384, no. 2 (2023): 265–276, 10.1124/jpet.122.001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rittavee Y., Artus J., and Desterke C., “miR‐495‐3p Sensitizes BCR::ABL1 Expressing Leukemic Cells to Tyrosine Kinase Inhibitors by Targeting Multidrug Resistance 1 Gene Including in T315I Mutated Cells,” bioRxiv 118 (2022): 40–45, 10.1101/2022.10.17.512501. [DOI] [PubMed] [Google Scholar]
- 46. Rittavee Y., Artus J., Desterke C., et al., “miR‐495‐3p Sensitizes BCR‐ABL1‐Expressing Leukemic Cells to Tyrosine Kinase Inhibitors by Targeting Multidrug Resistance 1 Gene in T315I Mutated Cells,” Experimental Hematology 118 (2023): 40–52, 10.1016/j.exphem.2022.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


