Abstract
The family Daphniphyllaceae has a single genus, and no relevant comparative phylogenetic study has been reported on it. To explore the phylogenetic relationships and organelle evolution mechanisms of Daphniphyllaceae species, we sequenced and assembled the chloroplast and mitochondrial genomes of Daphniphyllum macropodum. We also conducted comparative analyses of organelles in Daphniphyllaceae species in terms of genome structure, phylogenetic relationships, divergence times, RNA editing events, and evolutionary rates, etc. Results indicated differences in the evolutionary patterns of the plastome and mitogenome in D. macropodum. The plastome had a more conserved structure but a faster nucleotide substitution rate, and the mitogenome showed a more complex structure while the mitotic genome shows a more complex structure but a slower nucleotide substitution rate. We identified several unidirectional protein-coding gene transfer events from the plastome to the mitogenome based on homology analysis, but no transfer events occurred from the mitogenome to the plastome. Multiple TE fragments existed in organelle genomes, and two organelles showed different preferences for nuclear TE insertion types. The estimation of divergence time indicated that the differentiation of Daphniphyllaceae and Altingiaceae at around 29.86 Mya might be due to the dramatic uplift of Tibetan Plateau during the Oligocene. About 75% of codon changes in organelles were found to be hydrophilic to hydrophobic amino acids. The RNA editing in protein-coding transcripts is the result of amino acid changes to increase their hydrophobicity and conservation in alleles, which may contribute to the formation of functional 3D structures in proteins. This study would enrich genomic resources and provide valuable insights into the structural dynamics and molecular biology of Daphniphyllaceae species.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-025-11213-9.
Keywords: Daphniphyllaceae, Organelle genomes, Structure dynamic, Phylogenetic relationships, Divergence time
Introduction
Higher plants possess organelle genomes, including plastome and mitogenome, which are controlled by the nuclear genome but have their own genetic systems; hence, these organelles are called semi-autonomous organelles. The mitochondrion and plastid serve as the sites of aerobic respiration and photosynthesis in the cell, respectively, and are essential for the proper conduct of cellular life activities. Although the plastome and mitogenome share many common characteristics, such as replication [1] and inheritance patterns [2], they also have notable differences. For example, the plastome typically has a single loop structure with a conserved quadripartite structure and is conserved in terms of gene content, gene order, and genome size. By contrast, the mitogenome displays greater variation in structure, gene number, and order. The mitogenome has a variety of structures, including cyclic, polycyclic, and linear ones [3, 4]. The size of the plastomes in higher plants ranges from about 100 to 200 kb [5], while the size of the mitogenome varies from 186 kb to 1 Mb [6]. The number of genes in plastomes generally hovers around 120, while the number of mitogenome genes ranges from 19 to 50 [7–9]. A precise and strict biological regulation mechanism exists between mitochondria and plastids. Organelle genomes have several properties such as structural simplicity and lack of recombination and uniparental inheritance. These genomes have been widely used in studies on phylogeny [10], phylogeography [11], and environmental adaptation [12] of different taxonomic orders. In recent years, the amount of organelle genomic data available, particularly for plastomes, has surged, making it possible to use organelle genomes as resources for studying adaptive evolution. Current studies are based on the hypothesis that sequence variation in plastomes is selectively neutral through genetic drift, that is, polymorphisms are selectively neutral [11]. However, an increasing number of studies suggested that positive selection may have played an important role in the adaptive evolution of plastomes [13]. In particular, many researchers have identified features of positive selection in the plastome sequences of different plant species through comparative genomic approaches; they suggested that plastomes may have undergone adaptive evolution in response to changes in their environments [12, 14]. Nevertheless, few studies have been conducted on extreme environmental adaptations at the mitogenome level, so this study should be extended to mitogenomes.
Daphniphyllum is the only genus of Daphniphyllaceae and is one of the oldest known genus. It is mainly distributed in East Asia, Southeast Asia, the Indian subcontinent, and Australasia [15]. This family is in a relatively isolated position in the modern classification of plants and is somewhat different from the other major plant families. Therefore, the study of this plant contributes to the understanding of the evolutionary history and taxonomic status of the Daphniphyllaceae family. The genus contains more than 34 species of dioecious evergreen shrubs and trees, which include D. oldhamii, D. macropodum, D. himalense, and D. calycinum [16]. All above-ground parts of D. macropodum contain large amounts of isoquinoline alkaloids. The first Daphniphyllum alkaloid was isolated by Yagi in 1909 as daphnimacrine from D. macropodum from Japan; since then, more than 320 Daphniphyllum alkaloids are known [16, 17]. Alkaloids, such as daphnicyclidin B and daphniglaucin C, show strong inhibitory activity against tumor cells. Thus, the study of this plant helps to understand the evolutionary history and taxonomic status of Daphniphyllum as well as the biosynthesis of medicinal constituents. Although data on Daphniphyllum alkaloids are well established, few phylogenetic and evolutionary studies on Daphniphyllaceae have been reported. Tang et al. [18] performed a multi-locus phylogenetic reconstruction of 55.6% (20/36) of the taxa in the genus by using chloroplast (psbA-trnH spacer region and trnL intron) and nuclear internal transcribed spacer (ITS: ITS1, 5.8S rDNA and ITS2) regions. The monophyly of any of the three groups proposed based on morphological classification schemes was overturned. Moreover, the ITS and chloroplast trees were inconsistent, and these topological conflicts may suggest hybridization events in the evolutionary history. Nevertheless, gene infiltration and gene exchange occur among species because of the similarity in the morphology and structure of Daphniphyllaceae species; as such, the phylogenetic relationships of Daphniphyllaceae are still ambiguous. Inclusive sampling and high-resolution phylogenies are needed for profile revision and species delimitation.
Analysis of the organelle genomes of D. macropodum may provide insights into the phylogenetic position of D. macropodum. Effective molecular markers can be developed for species identification and biogeographic studies using organelle genome data. In this study, we performed a systematic comparative genomic analysis of the plastome and mitogenome of D. macropodum by using third sequencing technology. We examined genome structure, gene number, transposable element type, intracellular gene transfer, and nucleic acid substitution rates in protein-coding genes (PCGs) and editing in organelle genomes. We also screened highly variable chloroplast regions by nucleic acid polymorphism analysis for subsequent molecular marker development. Our results are expected to provide valuable insights for future studies on the identification and phylogenetic evolution of Daphniphyllaceae species.
Materials and methods
Plant material
D. macropodum specimens were collected from Wuluo Town, Songtao Hmong Autonomous County, Guizhou Province, China (108.48°E, 28.0°N) and cultivated under natural conditions in the garden of Guizhou Education University. The samples were identified by Prof. Yuemei Zhao from Guizhou Education University, and the pressed voucher specimens were numbered (collection number: Dm_00977) and deposited at the herbarium of Guizhou Education University, China. DNA extraction and library construction were conducted using the methods of Zhang et al. [19]. 15 Kb fragment libraries constructed by the PacBio Sequel II platform were used to perform multiple rounds of sequencing on individual fragments to improve accuracy.
Sequencing, genome assembly, and annotation
New sequencing technologies and software updates have resulted in complete plant plastome and mitogenome assembly results. Here, we used the newly released PMAT v1.5.2 [4] to assemble PacBio HiFi reads with the default parameter to obtain the organelle genome of D. macropodum. The assembly results were polished using Pilon v1.23 [19]. Geneious Prime software [20] was used to annotate the plastome and mitogenome of D. macropodum. The mitogenome was annotated using Rhodiola crenulata (OP312067.1) [21] and Ribes nigrum (OR227936.1) [22] as references. The plastome was annotated using D. calycinum (NC_071199.1) as reference. tRNA annotation was performed using tRNAscan-SE software [23], and visualization was conducted using OGDRAW [24].
Comparison of complete organelles
The compositional skewness of each PCG in the organelle was calculated using the following formula: AT-skew = (A-T)/(G + C), GC-skew = (G-C)/(G + C). CodonW v1.4.4 [25] was employed to analyze relative synonymous codon usage (RSCU), and Geneious software [26] was used to determine GC content. RSCU value > 1.00 indicates that the codon is used more frequently than expected and vice versa. Effective number of codon (ENC) plots are commonly used to assess codon usage patterns in genes. The relationship between ENC and GC3s was visualized using R scripts (https://github.com/taotaoyuan/myscript). Predicted ENC values that lie on or above the expected curve can indicate that codon usage is primarily influenced by G + C mutations. If natural selection or other factors work, then the predicted ENC values will fall below the expected curve [27]. Shrinkage and expansion of IR boundaries were detected and visualized among the four major regions (LSC/IRb/SSC/IRa) of the plastome sequences of five Daphniphyllaceae species by using IRSCOPE (https://irscope.shinyapps.io/irapp/) [28]. The plastomes of five Daphniphyllaceae species were compared and annotated using the online website mVISTA [29], with reference to the plastome of D. macropodum selected. DnaSP v5 software [30] was used to calculate accounting polymorphisms among the chloroplast genomes of Daphniphyllaceae species.
Phylogenomic analysis
Considering the lack of systematic studies on Daphniphyllaceae species and mitogenome data, we obtained 67 plastome sequences from NCBI (https://pubmed.ncbi.nlm.nih.gov/) to reconstruct the phylogenetic relationships of Daphniphyllaceae, including 2 Cercidiphyllaceae species, 5 Daphniphyllaceae species, 27 Hamamelidaceae species, 3 Paeoniaae species, 5 Altingiaceae species, 5 Crassulaceae species, 1 Penthoraceae species, 7 Haloragidaceae species, 1 Iteaceae species, 3 Grossulariaceae species, 9 Saxifragaceae species, and 1 outgroup Vitaceae species. We extracted 79 PCGs by using PhyloSuite [31]. MAFFT v7.490 software [32] was used for CDS sequence comparison, and poorly matched regions were removed using the “automated1” parameter of BMGE software [33]. A supermatrix of all genes was constructed with FASconCAT-G [34] software and used to construct a phylogenetic tree. We determined the JC + I + G4 model by using ModelFinder [35] and inferred the Bayesian inference (BI) tree using MrBayes 3.2.7 [36]. We inferred the maximum likelihood (ML) tree by using the GTR + F + G4 model with 2000 ultrafast bootstraps and IQ-TREE [37].
Divergence times were estimated using the MCMCTree package from PAML v4.9j [38]. Fossil calibration points were mainly sourced from the Paleontological Database (https://paleobiodb.org/) and the Timetree5 website (http://www.timetree.org/). Three fossil calibration points were selected, namely, MRCA (110.2–120 Mya), Altingia excelsa–A. yunnanensis (23.4–37.7 Mya), and Cercidiphyllum japonicum–C. magnificum (6.487–6.52 Mya). The input tree file used the topology of the tree obtained in the previous step, and the input sequence used the trimmed nucleic acid sequence obtained in the previous step. A total of 1,000,000 generations were run, with sampling every 100 generations until the results converged. The convergence of the results was checked by Tracer v1.7.2 [39]. Finally, the results were visualized using the ChiPlot online website [40].
Identification of homologous sequences and transposable elements
We used TBtools software [41] to detect repetitive regions within the plastome and mitogenome of D. macropodum as well as the regions of covariance between the plastome and mitogenome. Given that the genera Ribes and Rhodiola are most closely related to Daphniphyllum, we chose Rhodiola wallichiana as a representative of polycyclic mitochondria and Ribes nigrum as a representative of monocyclic mitochondria for comparative analysis. Gepard software [42] was used to detect covariance between the plastomes of D. macropodum and R. crenlata and R. nigrum. MUMmer [43] and Generious software [20] were used to detect structural rearrangements and chromosome breaks between D. macropodum and R. crenlata and R. nigrum. The CENSOR web server [44] was used to detect transposable elements (TEs) in the D. macropodum organelle genome, with “Viridippantae” dataset as a reference.
Analysis of nucleotide substitution rate (NSR) and RNA editing
To assess NSR between the plastome and mitogenome of D. macropodum, we chose R. nigrum as a reference and used PCGs. Using PhyloSuite [31], we extracted 68 shared plastome PCGs and 24 mitogenome PCGs and computed synonymous (dS) and nonsynonymous (dN) substitution rates with KaKs_Calculator [45] and the yn00 model. The results were visualized using Chiplot web tools [40]. We initially filtered RNA data to identify RNA editing sites and employed Hisat-3N software [46] to map the filtered RNA data to the plastome and mitogenome. The results were visualized using Geneious prime [20], with exclusion criteria set at sequencing errors with depths < 5% and < 2.5% of RNA reads mapped to a specific fragment. Loci with RNA editing efficiency below 20% were excluded from the analysis.
Results
Characterization of organelle genomes
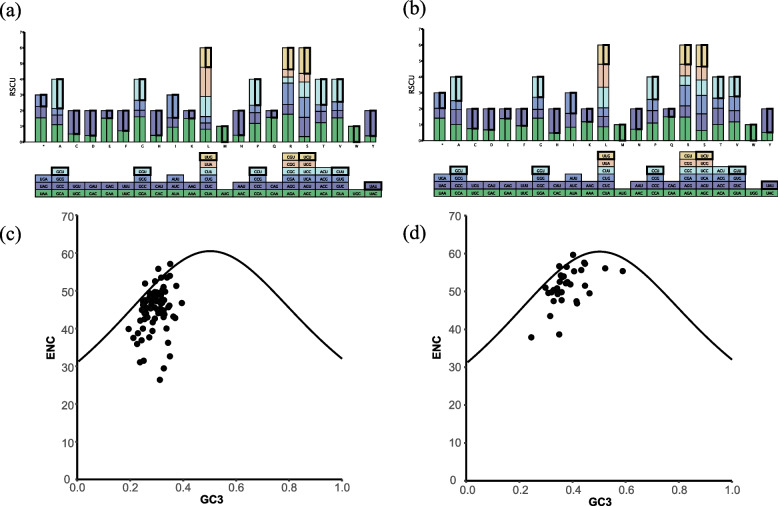
The plastome of D. macropodum exhibited typical quadripartite structure with a total length of 160,221 bp (Fig. 1a). It had 128 genes including 83 PCGs, 37 tRNA genes, and 8 rRNA genes. Among the PCGs, 47 genes were associated with photosynthesis, and 71 genes were related to self-replication (Table 1). The mitogenome of D. macropodum assembled into two circular chromosomes, with an atypical multi-loop conformation (Fig. 1b and c). The total length of the D. macropodum mitogenome was 804,571 bp, with the larger chromosome of 412,548 bp and the smaller one of 392,023 bp. The mitogenome included 35 PCGs, 19 tRNA genes, and 2 rRNA genes (Table 2). In the plastome, 30 codons had RSCU values greater than 1, 34 codons had RSCU values less than 1, and two codons had RSCU values equal to 1 (Fig. 2a). Among the 64 codons in the plastome, 16 ended with A, U, G, or C. In codons with RSCU values greater than 1, all codons ended in A/U. A similar pattern was observed in the mitogenome. Among codons with RSCU values greater than 1, except for two of the codons ending in G/C, all of the remaining codons ended in A/U (Fig. 2b; Table S1). This result is consistent with the report of Zhang et al. in the plastids of Aconitum species [19]. The ENc values of plastome PCGs ranged from 31 to 61 (Fig. 2c), while those of mitogenome PCGs ranged from 25.98 to 56.69 (Fig. 2d). Only four of plastome PCGs showed high codon bias (ENc < 35), while none of mitogenome PCGs had ENc values below 35. This finding indicates that these genes did not have a strong codon preference. The plastome and mitogenome of D. macropodum contained numerous TE fragments with a total length of 16,340 and 68,253 bp (Tables 3 and 4). The repeated sequence analysis indicated that all repeat fragments were shorter than 100 bp (28–89 bp) in the plastome, except for the two reverse repeat IR regions (Fig. S1, Table S2). Meanwhile, 259 repeat fragments with a full length of 15,271 bp were found in the mitogenome; of which the longest repeat sequence was 327 bp (Fig. S1, Table S2).
Fig. 1.
a Field photo of D. macropodum. b Gene map of the D. macropodum’s plastome. c and d Gene map of the D. macropodum’s mitogenome. Genes inside and outside the circle are transcribed clockwise and counterclockwise, respectively. Genes belonging to different functional groups are indicated by different colors
Table 1.
Gene composition in the plastome of D. macropodum
| Category for genes | Function | Gene list |
|---|---|---|
| Genes for photosynthesis | ATP synthase | atpA, atpB, atpD, atpE, atpF, atpH, atpI |
| ATP-dependent protease proteolytic subunit | clpP | |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK , psbL, psbM, psbN, psbT, psbZ | |
| Cytochrome b/f complex | petA, petB, petD, petG, petL, petN | |
| Rubisco large subunit | rbcL | |
| NADH dehydrogenase | ndhA, ndhB x2, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Self-replication | Ribosomal RNAs (rRNA) | rrn4.5 x2, rrn5 x2, rrn16 x2, rrn23 x2 |
| Transfer RNAs (tRNA) |
trnA-UGC x2, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-UCC x2, trnH-GUG, trnI-CAU x2, trnI-GAU x2, trnK-UUU, trnL-CAA, trnL-UAG, trnM-CAU x4, trnN-GUU x2, trnP-UGG, trnQ-UUG, trnR-ACG x2, trnR-UCU, trnS-GCU, trnS-GGA, trnT-GGU, trnT-UGU,trnV-GAC x2, trnV-UAC, trnW-CCA, trnY-GUA |
|
| Large subunit of ribosomal proteins (LSU) | rpl14, rpl16, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 | |
| Small subunit of ribosomal proteins (SSU) | rps2, rps3, rps4, rps7 x2, rps8, rps11, rps12, rps14, rps15, rps16, rps18, rps19 | |
| RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Translational initiation factor | infA | |
| Other genes | Maturase | matK |
| Envelope membrane protein | cemA | |
| Subunit of acetyl-CoA-carboxylase | accD | |
| Cytochrome complex assembly | ccsA | |
| Genes of unknown funcation | Hypothetical chloroplast reading frames | ycf1 x2,ycf2 x2, ycf3, ycf4 |
Table 2.
Gene composition in the mitogenome of D. macropodum
| Group of genes | Name of genes |
|---|---|
| Transport membrane protein | atp1, atp4, atp6, atp8, atp9 |
| Cytochrome c biogenesis | ccmB, ccmC, ccmFc, ccmFn |
| Ubichinol cytochrome c reductase | cob |
| Cytochrome c oxidase | cox1, cox2, cox3 |
| Maturases | matR |
| Transport membrane protein | mttB |
| NADH dehydrogenase | nad2, nad3, nad4, nad4L, nad6, nad7, nad9 |
| Large subunit of ribosome | rpl2, rpl5, rpl10, rpl16 |
| Small subunit of ribosome | rps1, rps3, rps4, rps12, rps13, rps14, rps19 |
| Ribosomal RNAs | rrn5, rrn18 |
| others | sdh4 |
| Transfer RNAs | trnC-GCA, trnD-GUC, trnE-TTC, trnF-GAA, trnG-GCC, trnH-GTG, trnK-UUU, trnM-CAT × 2, trnM-CAU, trnN-GUU, trnP-UGG × 3, trnQ-UUG, trnS-GCU, trnS-UGA, trnW-CCA, trnY-GUA |
Fig. 2.
a Codon usage in plastome genes. b Codon usage in mitogenome genes. c ENC map of plastome. d ENC map of mitogenome. The outer circle shows the gene location information, and the inner circle shows the repeat sequence information,the links on the innermost circles represent repeats identified by BLASTn
Table 3.
Comparation of TEs in R. juparensis organelle genomes
| Type | Plastome | Mitogenome | ||
|---|---|---|---|---|
| Fragments | Length | Fragments | Length | |
| DNA transposon | 97 | 10,603 | 162 | 10,968 |
| EnSpm/CACTA | 16 | 1646 | 37 | 2218 |
| Harbinger | 6 | 376 | 7 | 439 |
| Helitron | 28 | 3339 | 35 | 2734 |
| Mariner/Tc1 | \ | \ | 1 | 79 |
| MuDR | 30 | 3919 | 51 | 3581 |
| Novosib | \ | \ | 1 | 65 |
| hAT | 16 | 1214 | 27 | 1646 |
| LTR Retrotransposon | 53 | 5220 | 314 | 47,244 |
| Copia | 26 | 3403 | 144 | 17,783 |
| Gypsy | 27 | 1817 | 169 | 28,910 |
| Non-LTR Retrotransposon | 7 | 471 | 75 | 10,068 |
| L1 | 6 | 416 | 74 | 10,022 |
| SINE | 1 | 55 | 5 | 1307 |
| SINE2/tRNA | 1 | 55 | 5 | 280 |
| Total | 158 | 16,340 | 551 | 68,253 |
| Ratio | \ | 10.20% | \ | 8.48% |
The ratio was obtained by dividing the transposon sequence length by the genome length
Table 4.
Mitogenome characteristics of 10 Saxifragales species
| Species | Genome size (bp) | Number of genes | Protein genes | tRNA genes | rRNA genes |
|---|---|---|---|---|---|
| P. lactiflora | 181,688 | 48 | 28 | 17 | 3 |
| P. suffruticosa | 203,077 | 51 | 29 | 17 | 5 |
| D. macropodum chr1 | 412,548 | 31 | 18 | 11 | 2 |
| D. macropodum chr2 | 392,023 | 25 | 17 | 8 | 0 |
| R. crenulata | 194,106 | 33 | 22 | 8 | 3 |
| R. wallichiana chr1 | 118,787 | 24 | 14 | 7 | 3 |
| R. wallichiana chr2 | 82,073 | 19 | 13 | 6 | 0 |
| R. sacra chr1 | 128,593 | 19 | 12 | 4 | 3 |
| R. sacra chr2 | 80,477 | 18 | 15 | 3 | 0 |
| R. nigrum | 450,223 | 62 | 39 | 20 | 3 |
| R. alpinum | 375,267 | 60 | 38 | 19 | 3 |
| T. polyphylla chr1 | 430,435 | 42 | 24 | 16 | 0 |
| T. polyphylla chr2 | 126,946 | 14 | 10 | 1 | 3 |
| T. polyphylla chr3 | 55,296 | 5 | 4 | 1 | 0 |
| H. parviflora | 542,954 | 84 | 42 | 28 | 7 |
Comparative plastome analysis
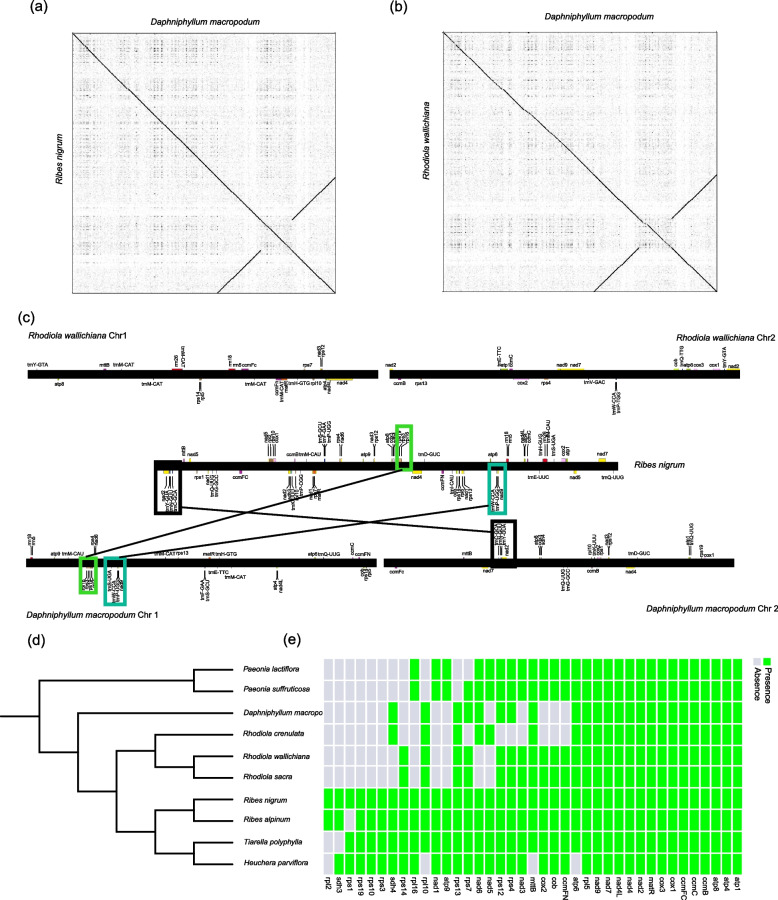
To investigate the variability among plastome sequences within the family Daphniphyllaceae, we compared the plastome sequences of five Daphniphyllaceae species by using mVISTA software with the plastome of D. macropodum as a reference (Fig. 3a). The results revealed intra-generic sequence variation in the plastome, with highly divergent regions mainly located in intergenic regions (psbE-petL, ndhF-rpl32, and trnT-GUU-trnL-UAA) and a variant region of the ndhF gene in the coding region (Fig. S1a). Overall, the similarity in the structure and gene order of the five plastomes suggested genome-wide evolutionary conservation of the plastome. Furthermore, we comprehensively compared the IR-SSC and IR-LSC boundaries of the plastome of the five Daphniphyllaceae species (Fig. S1b). The rps19, ycf1, ycf1, and rpl2 genes are located at the junctions of the LSC/IRb, IRb/SSC, SSC/IRa, and IRa/LSC boundaries, respectively. The rps19 gene located at the LSC/IRb junction transferred to the IRb region in all five species, while the ycf1 gene in D. macropodum and D. oldhamii shifted to the SSC region. In addition, we identified three hotspots (trnT-GUU-trnL-UAA, psbE-petL, and ndhF-rpl32) in the plastomes of the Daphniphyllaceae by PI analysis (Fig. S1c, Table S3). These identified hotspot regions hold the potential to serve as valuable molecular markers and barcodes for Daphniphyllaceae. This result lays the foundation for future phylogenetic analyses and species identification.
Fig. 3.
Syntenic analyses of organelle genomes. a Syntenic regions between D. macropodum and R. nigrum plastomes. b Syntenic regions between D. macropodum and R. wallichiana plastomes. c Syntenic regions of mitogenomes among D. macropodum, R. nigrum, and R. wallichiana. d plastome-based phylogeny, mitogenome genes gain and loss. On the left is a phylogenetic tree constructed using genes coding for plastome, and on the right is the loss of mitochondrial genes in 10 species
Synteny and structural rearrangement analysis
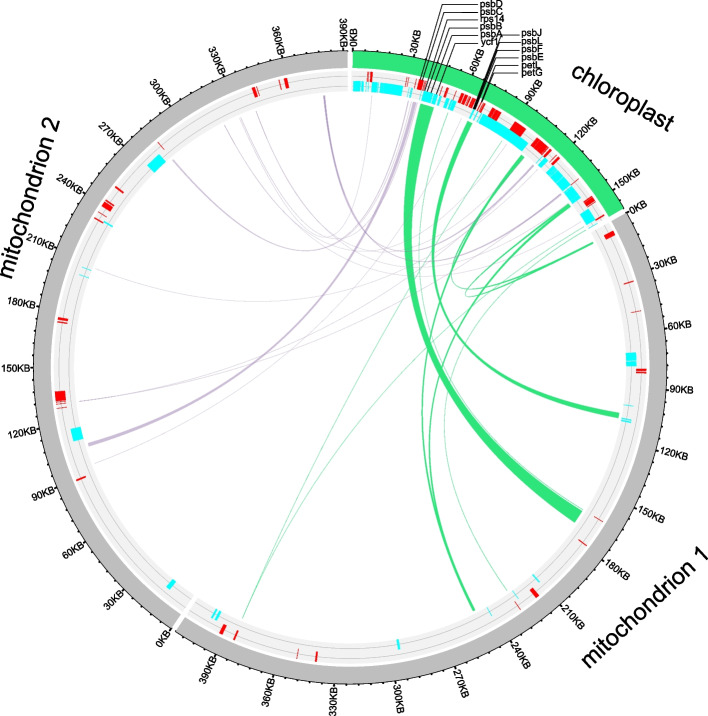
Collinearity block region analysis is commonly used to determine the evolutionary relationships among closely related species at the genome level. We performed collinearity block analysis to explore structural differences in the mitogenomes of Saxifragales species. The results based on “Phylogenetic analysis and divergence time estimation” section indicated that D. macropodum was closely related to Crassulaceae and Grossulariaceae, so we detected the homologous regions among the three organelle genomes. The plastomes of D. macropodum, R. wallichiana, and R. nigrum showed a high degree of collinearity (Fig. 3a and b). Among these mitogenomes, 106 collinearity blocks were identified between D. macropodum and R. wallichiana and 343 collinearity blocks were found between D. macropodum and R. nigrum (Table S4). The mitogenomes of the three species exhibited a complex collinearity structure (Fig. 3c). The presence of a chromosomal fission in the D. macropodum mitogenome formed two subgenomes, resulting in reduced collinearity with R. wallichiana. These collinearity blocks were dispersed throughout the genomes, suggesting the occurrence of structural rearrangements in the mitogenomes of Crassulaceae and Daphniphyllaceae species. We also compared the positions of the orthologous genes of the three Saxifragales species and found that structural rearrangements disrupted gene clusters in the mitogenome (Fig. 3c). However, some gene clusters were preserved between D. macropodum and R. nigrum, including nad2-trnY-GUA-trnN-GUU-trnC-GCA, rps19-rps3-rpl16, and nad9-trnP-UGG-trnW-GCA gene clusters. However, we did not find conserved gene clusters among R. wallichiana and D. macropodum or R. wallichiana and R. nigrum. We found that the PCGs of mitogenomes were significantly absent in species that underwent structural rearrangements, including atp9, nad1, rpl2, rpl12, rps1, rps3, rps10, rps19, and sdh3 (Fig. 3d). Previous studies reported the presence of plastome gene residues in the mitogenome, implying significant sequence transfer between the two organelles [19, 21, 47]. To identify potential gene transfer fragments between the plastome and mitogenome, we searched using blastN and obtained 27 fragments (Fig. S2, Table S5). The longest fragment was 7,763 bp, and the total length of these sequences was 19,663 bp. After annotation, the mitogenome contained intact plastome-derived PCGs, including eight psb type genes, rps14, petL, petG, and ycf1, which are referred to as mitochondrial plastid DNAs (MTPTs) (Fig. 4). However, we did not find any intact mitogenome-derived PCGs in the plastome. Additionally, we observed that several tRNA genes (trnw-CCA and trnP-UGG) were highly similar in sequence between the plastome and mitogenome.
Fig. 4.
Homology sequences between plastome and mitogenome. On the circle plot, the red bars represent genes in the counter clockwise direction and the cyan bars represent genes in the clockwise direction. The shaded links represent identified homologous sequences. In homologous sequences, intact PCGs are highlighted with a broken line
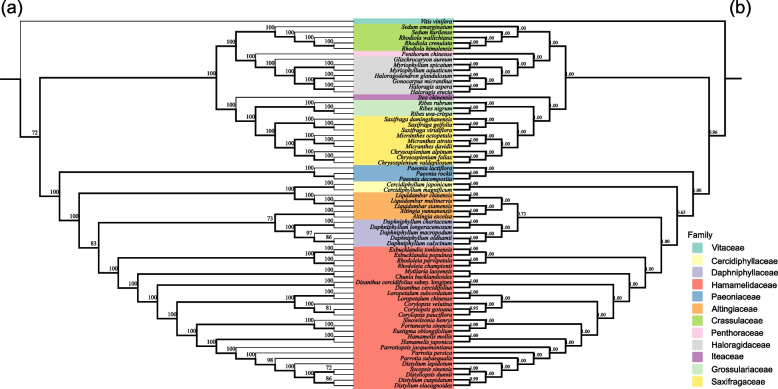
Phylogenetic analysis and divergence time estimation
To determine the position of D. macropodum on the phylogenetic tree, we selected chloroplast genomic data and reconstructed the phylogenetic relationships of Daphniphyllaceae. Seventy-nine PCGs from 68 species were used to construct ML and BI trees (Fig. 5). The phylogenetic trees constructed by the two methods had consistent topological relationships and high node bootstrap values. Overall, all species of Saxifragales were divided into two clades; the family of Crassulaceae, Penthoraceae, Haloragidaceae, Iteaceae, Grossulariaceae, and Saxifragaceae formed a monophyletic clade. Although the results obtained by the two methods are basically the same in terms of topology, the results of the BI tree support a monophyletic clade with a high degree of support for Daphniphyllaceae. The ml tree supports that Daphniphyllaceae and Altingiaceae form a non-monophyletic clade, and the branch node of Daphniphyllaceae and Altingiaceae have a support rate of only 73. Therefore, we should recognize Daphniphyllaceae as a monophyletic clade and has the closest relationship to Altingiaceae. Within Daphniphyllaceae, species of the genus Daphniphyllum are divided into two clades, with D. chartaceum clustered into a single clade with D. longeracemosum and D. oldhamii clustered into a single clade with D. calycinum and D. macropodum. V. vinifera was used as an outgroup to predict the time of differentiation. Daphniphyllaceae and Altingiaceae underwent rapid radial evolution at about 29.86 Mya (Fig. 6), whereas species differentiation within Daphniphyllaceae occurred at about 2.01 Mya. In the phylogenetic tree, D. macropodum was more closely related to D. oldhamii and D. calycinum was more closely related and diverged at about 1.78 Mya (Fig. 6).
Fig. 5.
ML tree and BI tree constructed based on plastome. The ML tree is on the left and the BI tree is on the right. Numbers below the lines represent ML bootstrap proportions BI posterior probabilities
Fig. 6.
Chronogram showing divergence times among 68 plants with node age and 95% confidence intervals (green bars). The black numbers above the branches represent Divergence times
Analysis of NSR and positive selection in Daphniphyllaceae species
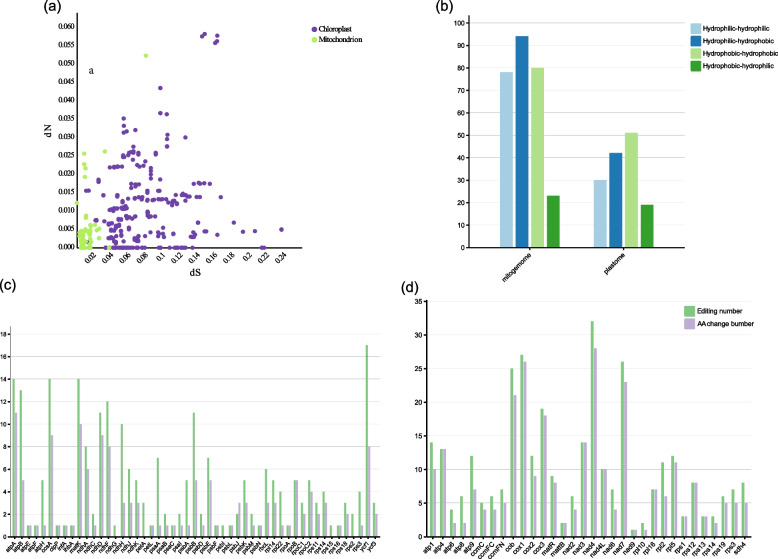
Mitochondria and chloroplasts perform different functions in plants and are exposed to different types of environmental selection pressures. We selected R. nigrum as a reference and used the yn00 module [36] in PAML v4.9j software to calculate pairwise NSRs for plastome and mitogenome genes. The genes in the plastome of D. macropodum had a higher dS value than those in the mitogenome. The relatively high dS suggested that the D. macropodum plastome is in a stable state and undergo evolution at a slow pace. Furthermore, D. macropodum organelles exhibited dN/dS values below 1, which provided evidence for purifying selection acting on PCGs within D. macropodum organelles. In summary, plastome genes in D. macropodum had a faster mutation rate than mitogenome genes (Fig. 7a, Table S6). We further performed a Mann–Whitney U test for the dN and dS values of plastome PCGs and mitogenome PSGs. The values were significantly higher for plastome PCGs than for mitogenome PCGs (p-value of dS = 5.157e-07; p-value of dN = 0.00623). These results suggest that plastome genes tend to mutate at faster rates than mitogenome genes, despite some genes in the two organelle genomes mutating at similar rates.
Fig. 7.
Variation in sequence divergence across species and organelles. a Comparison of dN and dS values across organelles. b Comparison of edit sites that lead to a change in hydrophobicity/hydrophilicity of the resulting amino acid via non-synonymous RNA editing. c Comparison of edit sites that lead to AA change in the PCGs of plastome. d Comparison of edit sites that lead to AA change in the PCGs of mitogenome
RNA editing and damage repair
After manual inspection, we identified 241 RNA editing sites in the plastome (Fig. 7c; Table S7) and 327 RNA editing sites in the mitogenome (Fig. 7d; Table S7), suggested that the mitogenome has more editing sites. Among the plastome PCGs, 51 genes had editing sites, with 94.6% of the edits occurring at codon 1 and 2 positions. Furthermore, 139 (57.8%) edits resulted in nonsynonymous amino acid changes (Fig. 7a). By contrast, 32 mitogenome PCGs had editing sites, with 97.2% of the edits occurring at codon 1 and 2 positions. Furthermore, 327 (82.9%) editing sites resulted in nonsynonymous amino acid changes (Fig. 7b, Table S8). The ycf1 gene of the plastome had the highest number of editing sites (17 sites), followed by matK, ccsA, and atpA, with 14 editing sites, respectively. Among the mitogenome PCGs, nad4 had the highest number of editing sites with 32, followed by cox1 and nad7, both with more than 25 editing sites. We also counted the effects of editing on the hydrophobicity of coding amino acids. The vast majority of nonsynonymous editing sites in the plastome (57.8%) and mitogenome (82.9%) converted codons of hydrophilic amino acids to hydrophobic codons (Table S8).
Discussion
Genome features of organelles
The plastome of D. macropodum is consistent with previous studies on Rhodiola in terms of size, structure, and gene order, indicating that the plastomes of Daphniphyllaceae species were conserved [22]. However, comparative genomic analysis revealed greater sequence differences in non-coding regions compared with those in plastome PCGs, such as in the trnT-GUU-trnL-UAA, psbE-petL, and ndhF-rpl32 regions. Additionally, the ndhF gene in the coding region was a high mutation hotspot region. These newly discovered high mutation hotspot genes and intergenic regions could provide useful molecular markers for phylogeographic and population genetics studies within Daphniphyllaceae. Tang et al. [18] used two chloroplast regions (psbA-trnH spacer and trnL intron regions) for phylogenetic analysis of Daphniphyllaceae, which resulted in the presence of multiple lineages. Our results indicate that the psbA-trnH spacer and trnL intron regions are very conserved (Fig. S1), which may account for the presence of concatenated lineages. Therefore, we can utilize trnT-GUU-trnL-UAA, psbE-petL, and ndhF-rpl32 regions as molecular markers to improve the resolution among Daphniphyllaceae species. The mitogenomes showed significant variations in the structure, number, and order of genes arranged. For instance, the mitogenome of D. macropodum had two loops and had experienced gene loss of up to 15 genes (Fig. 3). Interestingly, we found that gene loss was prevalent in species in the families Paeoniaceae [48], Daphniphyllaceae, and Crassulaceae [20]; however, these lost genes were generally retained in the families Grossulariaceae and Saxifragaceae (Fig. 5e). Based on the phylogenetic tree of the species and the results of differentiation time, this loss event occurred independently after the differentiation of the families (Figs. 3d and 5). Although the cause of these loss events remain unknown, we speculate that the corresponding mitochondrial genes may have been transferred to the nuclear genome to adapt to the environment. These conjectures will be confirmed with the subsequent publication of the nuclear genome data of the corresponding species. Furthermore, most PCGs in the plastome and mitogenome of D. macropodum were found to be below the expected ENc curve, indicating that natural selection was the main factor in shaping codon usage preferences [49]. This finding is particularly true for photosynthesis-related and respiration-related genes, which are subject to strong environmental selection pressures (i.e., intense UV radiation and intense light). However, not all photosynthesis-related and respiration-related genes showed ENc values on the expected curve, suggesting that mutations played a minor role in shaping codon preferences. Hence, natural selection is the primary factor that determined codon usage preferences in D. macropodum, particularly for important genes, such as those involved in photosynthesis and respiration [50]. We also noted that the plastome had a faster NSR than the mitogenome. The different genetic features between the two organelles might result from the fact that they possess different genomic repair mechanisms [51]. Overall, this study would contribute to the subsequent understanding of organelle evolution in Daphniphyllaceae species and could serve as a basis for further investigations into the genetic mechanisms that shaped the genome structure and function of the plastome and mitogenome in plants.
Gene loss, intracellular gene transfer, and structural rearrangement
Unlike animals, which typically have a nearly constant number of mitogenome genes, the loss of mitogenome genes are common in many land plant lineages [21]. The number of mitogenome genes varies widely, from 19 in Viscum scurruloideum to over 50 in Marchantia polymorpha [7–9]. The loss of mitogenome genes, especially ribosomal protein genes, is a relatively frequent and persistent phenomenon in angiosperms [52, 53]. Most of the gene losses probably occurred after the functional transfer of the genes to the nucleus, although gene loss did not necessarily imply a functional transfer to the nucleus [54, 55]. For instance, almost all of Zostera’s ribosomal genes were lost, but only a small fragment of the genes were found in the nucleus [52]. The mitogenome genes are probably replaced by homologous genes derived from the plastome or nuclear DNA [55].
In our study, a large number of gene loss events were found in D. macropodum. In the mitogenomes of Daphniphyllaceae and Crassulaceae, a spectrum-specific loss of genes appears to be correlated with breaks in the mitogenome. However, no research has suggested a necessary link between the occurrence of the two events. As far as the results of the present study are concerned, gene loss is often accompanied by mitogenome breaks. However, this finding needs to be verified by subsequent large amounts of mitogenome data in plants. Further analysis of homologous sequences revealed the presence of several intact plastome-derived PCGs in the mitogenome, but no intact mitogenome-derived PCGs were found in the plastome. These results are consistent with previous studies that suggested prevalent DNA transfer events from the plastome to the mitosome in species [21]. These transfers might play a role in the loss of certain mitogenome genes in Saxifragales. The direction of gene transfer was found to be unidirectional, supporting the idea that plastomes are more receptive to DNA transfer than mitogenomes [56, 57]. Gene transfer between organelles is an important process in plant evolution, and transfer between organelles in plant cells is essential for growth and development. Cheng et al. showed that the transfer of the Arabidopsis plastome AtRNH1C gene into the mitogenome contributed to the maintenance of mitochondrial R-loop homeostasis and genomic stability during embryonic development [58]. This finding highlights the importance of gene transfer between organelles in maintaining individual plant development and environmental adaptation.
In addition to frequent gene loss and gene transfer, structural rearrangement is another characteristic of plant mitogenomes, and it reflects rapid differentiation at the structural level. Environmental stress, such as intense light and UV radiation, contribute to mitogenome rearrangement, as well as nuclear gene variation [58–61]. For instance, Xu et al. found that chromosomal rearrangements in MSH1 mutants could alter plastid properties, leading to the protection of plants from intense light damage [60]. Therefore, chromosomal rearrangements present in Daphniphyllaceae and Crassulaceae species might contribute to their adaptation to environment conditions. This finding needs to be verified by subsequent experiments.
Phylogenetic analysis and divergence time estimation
The phylogenetic analyses revealed that the ML and BI trees displayed a consistent topology, supporting Daphniphyllaceae species as a monophyletic branch. At the order level, all Saxifragales species were divided into two clades. Although the results obtained by the two methods are basically the same in terms of topology, the results of the BI tree support a monophyletic clade with a high degree of support for Daphniphyllaceae. Our results support the phylogenetic results of Tang et al. [18] based on two chloroplast regions, namely, D. macropodum clustered with D. oldhamii and D. calycinum. Given the chloroplast data used by Tang et al. did not include D. chartaceum, we cannot compare the phylogenetic relationships between D. chartaceum and D. longeracemosum. Considering the higher node support of the BI tree, this study is more supportive of Daphniphyllaceae as a monophyletic taxon. The BI tree was also used for the subsequent analysis of divergence times. In the present study, Daphniphyllaceae species diverged from Altingiaceae at about 29.86 Mya. At about 2.01 Mya, differentiation within Daphniphyllaceae began to occur. At 30–40 Mya, the Central Valley and the upper crust, where the Lhasa lithosphere is located, were uplifted to a height of about 4500 m by the coupling of multiple address movements. At about 25–15 Mya, the subducted Indian continental lithosphere below the Himalayas and the subducted Eurasian continental lithosphere below the Cocoanutian–Kunlun Mountains in northern Tibet successively underwent dismantling and sinking because of the continuous subduction of the Indian continent; the Himalayas and the Kunlun Mountains were successively uplifted to the modern height, forming the current Tibetan Plateau [62]. According to the distribution of species, Daphniphyllaceae species are widely distributed in southeastern China and Southeast Asia; among which, D. himalense is widely distributed in Tibet and Yunnan [62]. Altingia excelsa, A. yunnanensis, and other species under the family Altingiaceae are widely distributed in southeastern and southwestern Yunnan and Mutuo in southeastern Tibet [63]. Liquidambar chinensis, L. siamensis, and L. multinervis under the family Altingiaceae are widely distributed in China, such as Guangdong, Guangxi, Guizhou, Yunnan, Hainan, and other places in China [64]. Therefore, we hypothesize that the divergence of Daphniphyllaceae species from Altingiaceae at ca. 29.86 Mya may have resulted from the uplift of the Tibetan Plateau.
RNA editing and damage repair
RNA editing is essential for regulating gene expression, RNA splicing, and plant growth and development. Mitochondrial RNA editing evolves in a variety of eukaryotic organisms, such as slime molds [64], land plants, and dinoflagellates [65]. Plastid RNA editing has only been reported in land plants, peridinin, and fucoxanthin dinoflagellates [66, 67], but the main type of RNA editing in mitochondria and plastid is C-to-U transformation. In the present study, 80 and 346 RNA editing sites were identified from the plastome and mitogenome, respectively, suggesting that the mitochondrial DNA (mtDNA) mutational spectrum was broader and more variable than that of chloroplast DNA (cpDNA) [68]. Notably, 94.6% of RNA editing in the plastome occurred at the first two codon positions, while 97.2% of editing in the mitogenome occurred at the first two codon positions. This finding is consistent with the findings of Grimes et al. [66, 69] and Zheng et al. [70], which suggested that most RNA editing sites resulted in amino acid changes in the first two codon positions [49, 71]. We also observed that RNA editing primarily converted amino acids to hydrophobic acids (65.5% in plastids and 63.3% in the mitogenome). Hydrophobicity is one of the main drivers of correct protein folding [47, 72]. Therefore, extensive RNA editing led to an increase in the number of hydrophobic amino acids that might contribute to the translation of mRNA into polypeptides with the correct folding structure; this finding is the structural basis for proteins to exercise specific functions [73].
Conclusion
Comparative genomic analysis revealed that the plastome was more conserved in structure, gene number, and gene order but evolved at a faster rate than the mitogenome. By contrast, the mitogenome had more gene loss and chromosome rearrangements and evolved at a slower rate. Furthermore, gene transfer between organelles appeared to be unidirectional from the plastome to the mitogenome, with no transfer events found from the mitogenome to the plastome. Additionally, both organelle genomes harbored multiple nuclear TEs. The plastome preferred DNA transposons, and the mitogenome preferred retrotransposons. Phylogenetic analysis supported Daphniphyllaceae as a monophyletic clade and confirmed its close relationship with Altingiaceae. The estimation of divergence time indicated that the differentiation of Daphniphyllaceae and Altingiaceae at around 29.86 Mya might be due to the dramatic uplift of Tibetan Plateau during the Oligocene. Furthermore, RNA editing analysis revealed more editing sites in the mitogenome, and extensive RNA editing led to an increase in hydrophobic amino acids. This phenomenon might help translate mRNA into a correctly folded structure at the appropriate position polypeptide, ultimately leading to the formation of a 3D structure with a specific function. Overall, this study provides valuable insights into the phylogeny and genome evolution of Daphniphyllaceae and the first mitochondrial genomic data for Daphniphyllaceae species.
Supplementary Information
Acknowledgements
Thanks to Ms. Jiao Chen for her help in life.
Authors’ contributions
Formal analysis, T. Y., RX. Z. and Y. L.; Investigation, YM. Z. and NY. X.; Sample collection, SW. L.; Experiment, XM. G.; Revision, Y. L.; Writing-original draft, T. Y.; Writing-review & editing, Y. L., SW. L. and T. Y. All Authors have read and agreed to published version of the manuscript.
Funding
This work was funded by the Science and Technology Support Plan of Guizhou Province (Qiankehe Support [2021] General 253).
Data availability
The plastome and mitogenome sequences generated in this study were deposited in GenBank database under accession numbers PP986955, PP986956, and PP986957.
Declarations
Ethics approval and consent to partticpate
Not Applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this article was revised: there was a typographical error in Ying Liu's name.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rongxiang Zhang and Yin Liu contributed equally to this work.
Change history
1/29/2025
A Correction to this paper has been published: 10.1186/s12864-025-11289-3
References
- 1.Marechal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010;186(2):299–317. [DOI] [PubMed] [Google Scholar]
- 2.Birky CW Jr. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc Natl Acad Sci. 1995;92(25):11331–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen J, Zhang Y, Havey MJ, et al. Copy numbers of mitochondrial genes change during melon leaf development and are lower than the numbers of mitochondria. Horticulture Research. 2019;6:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi C, Qu Y, Hou J, et al. Deciphering the multi-chromosomal mitochondrial genome of Populus simonii. Front Plant Sci. 2022;13: 914635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daniell H, Lin CS, Yu M, et al. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oda K, Yamato K, Nakamura Y, et al. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA: a primitive form of plant mitochondrial genome. Mol Biol. 1992;223:1–7. [DOI] [PubMed] [Google Scholar]
- 7.Skippington E, Barkman TJ, Rice DW, et al. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc Natl Acad Sci. 2015;112(27):E3515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston IG, Williams BP. Evolutionary inference across eukaryotes identifies specific pressures favoring mitochondrial gene retention. Cell Syst. 2016;2(2):101–11. [DOI] [PubMed] [Google Scholar]
- 9.Mower JP. The PREP suite: Predictive rna editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009;37:W253–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X, Jiang W, Tan W, et al. Deciphering the organelle genomes and transcriptomes of a common ornamental plant Ligustrum quihoui reveals multiple fragments of transposable elements in the mitogenome. Int J Biol Macromol. 2020;165(Pt B):1988–99. [DOI] [PubMed] [Google Scholar]
- 11.Shaw J, Shafer HL, Leonard OR, et al. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. Am J Bot. 2014;101:1987–2004. [DOI] [PubMed] [Google Scholar]
- 12.Jiang P, Shi FX, Li MR, et al. Positive selection driving cytoplasmic genome evolution of the medicinally important Ginseng Plant Genus Panax. Front Plant Sci. 2018;9:359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H, Li DZ, Ma PF. Unprecedented variation pattern of plastid genomes and the potential role in adaptive evolution in Poales. BMC Biol. 2024;22:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao DN, Ren Y, Zhang JQ. Conservation and innovation: Plastome evolution during rapid radiation of Rhodiola on the Qinghai-Tibetan Plateau. Mol Phylogenet Evol. 2020;144: 106713. [DOI] [PubMed] [Google Scholar]
- 15.Tang MS, Yang YP, Tsai CC, et al. The diversity of pistillate flowers and its taxonomic value to the classification of Daphniphyllum (Daphniphyllaceae). Bot Stud. 2012;53:509–24. [Google Scholar]
- 16.Zhong J, Wang H, Zhang Q, et al. The chemistry of Daphniphyllum alkaloids. Alkaloids Chemistry Biology. 2021;85:113–76. [DOI] [PubMed] [Google Scholar]
- 17.Yagi S. Daphniphyllum Alkaloid Kyoto Igaku Zasshi. 1909;6:208–22. [Google Scholar]
- 18.Tang MS, Tsai CC, Yang YP, et al. A Multilocus Phylogeny of Daphniphyllum (Daphniphyllaceae). Ann Mo Bot Gard. 2022;107:137–52. [Google Scholar]
- 19.Zhang R, Xiang N, Qian C, et al. Comparative analysis of the organelle genomes of Aconitum carmichaelii revealed structural and sequence differences and phylogenetic relationships. BMC Genomics. 2024;25:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kearse M, Moir R, Wilson A, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu XL, Wei P, Chen ZYF, et al. Comparative analysis of the organelle genomes of three Rhodiola species provide insights into their structural dynamics and sequence divergences. BMC Plant Biologyogy. 2023;23:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pikunova A, Goryunova S, Golyaeva O, et al. Plastome Data of Red Currant and Gooseberry Reveal Potential Taxonomical Issues within the Ribes Genus (Grossulariaceae). Horticulturae. 2023;9(9):972. [Google Scholar]
- 23.Chan PP, Lowe TM. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol Biol. 2019;1962:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47(W1):W59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peden JF. Analysis of codon usage. PhD Thesis, University of Nottingham, UK. 1999.
- 26.Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9(11): e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87:23–9. [DOI] [PubMed] [Google Scholar]
- 28.Ali A, Jaakko H, Peter P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34(17):3030–1. [DOI] [PubMed]
- 29.Frazer KA, Pachter L, Poliakov A, et al. mVISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;1((32)(Web Server issue)):W273-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–2. [DOI] [PubMed] [Google Scholar]
- 31.Zhang D, Gao F, Jakovlić I, et al. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecule Ecology Resource. 2020;20(1):348–55. [DOI] [PubMed] [Google Scholar]
- 32.Katoh K, Kuma K, Toh H, et al. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evolution Biology. 2010;10:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuck P, Longo GC. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool. 2014;11(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronquist F, Teslenko M, Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen LT, Schmidt HA, von Haeseler A, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology Evolution. 2015;32(1):268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology Evolution. 2007;24(8):1586–91. [DOI] [PubMed] [Google Scholar]
- 39.Rambaut A, Drummond AJ, Xie D, et al. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst Biol. 2018;67(5):901–4. [DOI] [PMC free article] [PubMed]
- 40.Xie JM, Chen YR, Cai GJ, et al. Tree Visualization By One Table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023;51(W1):W587–W592. [DOI] [PMC free article] [PubMed]
- 41.Chen C, Chen H, Zhang Y, et al. TBtools: an integrative Toolkit developed for interactive analyses of big Biological Data. Mol Plant. 2020;13(8):1194–202. [DOI] [PubMed] [Google Scholar]
- 42.Krumsiek J, Arnold R, Rattei T. Gepard: A rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23(8):1026–8. [DOI] [PubMed] [Google Scholar]
- 43.Kurtz S, Phillippy A, Delcher AL, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohany O, Gentles AJ, Hankus L, et al. Annotation, submission and screening of repetitive elements in repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, Li J, Zhao XQ, et al. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4(4):259–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Park C, Bennett C, et al. Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res. 2021;31:1290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li ZC, Liu ZQ, Zhong WQ, et al. Large-scale identification of human protein function using topological features of interaction network. Sci Rep. 2016;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang P, Ni Y, Li J, Lu Q, Liu C, Guo J. The Complete Mitochondrial Genome of Paeonia lactiflora Pall. (Saxifragales: Paeoniaceae): Evidence of Gene Transfer from Chloroplast to Mitochondrial Genome. Genes (Basel). 2024;15(2):239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Shen ZN, Meng XR, et al. Codon usage patterns across seven Rosales species. BMC Plant Biol. 2020;22(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia X, Liu SY, Zhao H, et al. Non-uniqueness of factors constraint on the codon usage in Bombyx mori. BMC Genomics. 2015;16(1):356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith DR, Keeling PJ. Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci. 2015;112(33):10177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petersen G, Cuenca A, Moller IM, et al. Massive gene loss in mistletoe (Viscum, Viscaceae) mitochondria. Sci Rep. 2015;5(1):17588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zervas A, Petersen G, Seberg O. Mitochondrial genome evolution in parasitic plants. BMC Ecology and Evolution. 2019;19(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams KL, Ong HC, Palmer JD. Mitochondrial gene transfer in pieces: fission of the ribosomal protein gene rpl2 and partial or complete gene transfer to the nucleus. Mol Biol Evol. 2001;18(12):2289–97. [DOI] [PubMed] [Google Scholar]
- 55.Adams KL, Qiu YL, Stoutemyer M, et al. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci. 2002;99(15):9905–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao C, Ren X, Mason AS, et al. Horizontal gene transfer in plants. Funct Integr Genomics. 2014;14(1):23–9. [DOI] [PubMed] [Google Scholar]
- 57.Smith DR, Asmail SR. Next-generation sequencing data suggest that certain nonphotosynthetic green plants have lost their plastid genomes. New Phytologist. 2014;204(1):7–11. [DOI] [PubMed] [Google Scholar]
- 58.Cheng L, Wang W, Yao Y, et al. Mitochondrial RNase H1 activity regulates R-loop homeostasis to maintain genome integrity and enable early embryogenesis in Arabidopsis. PLoS Biol. 2021;19(8): e3001357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shedge V, Davila J, Arrieta-Montiel MP, et al. Extensive rearrangement of the Arabidopsis mitochondrial genome elicits cellular conditions for thermotolerance. Plant Physiol. 2010;152(4):1960–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu YZ, Arrieta-Montiel MP, Virdi KS, et al. MutS HOMOLOG1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. Plant Cell. 2011;23(9):3428–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Virdi KS, Wamboldt Y, Kundariya H, et al. MSH1 is a plant organellar DNA binding and thylakoid protein under precise spatial regulation to alter development. Mol Plant. 2016;9(2):245–60. [DOI] [PubMed] [Google Scholar]
- 62.Yoshida K, Yokochi Y, Hisabori T. New light on chloroplast redox regulation: molecular mechanism of protein thiol oxidation. Front Plant Sci. 2019;10:1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ickert-Bond SM, Wen J. A taxonomic synopsis of Altingiaceae with nine new combinations. PhytoKeys. 2013;31:21–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Byrne EM, Gott JM. Unexpectedly complex editing patterns at dinucleotide insertion sites in Physarum mitochondria. Mol Biol Cell. 2004;24(18):7821–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waller RF, Jackson CJ. Dinoflagellate mitochondrial genomes: Stretching the rules of molecular biology. BioEssays. 2009;31(2):237–45. [DOI] [PubMed] [Google Scholar]
- 66.Lin S, Zhang H, Spencer DF, et al. Widespread and extensive editing of mitochondrial mRNAS in dinoflagellates. J Mol Biol. 2002;320(4):727–39. [DOI] [PubMed] [Google Scholar]
- 67.Howe CJ, Nisbet RER, Barbrook AC. The remarkable chloroplast genome of dinoflagellates. J Exp Bot. 2008;59(5):1035–45. [DOI] [PubMed] [Google Scholar]
- 68.Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311(5768):1727–30. [DOI] [PubMed] [Google Scholar]
- 69.Grimes BT, Sisay AK, Carroll HD, et al. Deep sequencing of the tobacco mitochondrial transcriptome reveals expressed ORFs and numerous editing sites outside coding regions. BMC Genomics. 2014;15:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng P, Wang D, Huang Y, Chen, et al. Detection and analysis of C-to-U RNA editing in rice mitochondria-encoded ORFs. Plan Theory. 2020;9:1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang A, Jiang X, Zhang F, et al. Dynamic response of RNA editing to temperature in grape by RNA deep sequencing. Funct Integr Genomics. 2020;20:421–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moelbert S, Emberly E, Tang C. Correlation between sequence hydrophobicity and surface-exposure pattern of database proteins. Protein Sci. 2004;13:752–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yura K, Go M. Correlation between amino acid residues converted by RNA editing and functional residues in protein three-dimensional structures in plant organelles. BMC Plant Biologyogy. 2008;8:79–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The plastome and mitogenome sequences generated in this study were deposited in GenBank database under accession numbers PP986955, PP986956, and PP986957.