Abstract
Respiratory syncytial virus (RSV) is a significant cause of acute lower respiratory tract infection (ALRTI) in children under five years of age. Between 2017 and 2021, 396 complete sequences of the RSV F gene were obtained from 500 RSV-positive throat swabs collected from ten hospitals across nine provinces in China. In addition, 151 sequences from China were sourced from GenBank and GISAID, making a total of 549 RSV F gene sequences subjected to analysis. Phylogenetic and genetic diversity analyses revealed that the RSV F genes circulating in China from 2017 to 2021 have remained relatively conserved, although some amino acids (AAs) have undergone changes. AA mutations with frequencies ≥ 10% were identified at six sites and the p27 region: V384I (site I), N276S (site II), R213S (site Ø), and K124N (p27) for RSV A; F45L (site I), M152I/L172Q/S173 L/I185V/K191R (site V), and R202Q/I206M/Q209R (site Ø) for RSV B. Comparing mutational frequencies in RSV-F before and after 2020 revealed minor changes for RSV A, while the K191R, I206M, and Q209R frequencies increased by over 10% in RSV B. Notably, the nirsevimab-resistant mutation, S211N in RSV B, increased in frequency from 0% to 1.15%. Both representative strains aligned with the predicted RSV-F structures of their respective prototypes exhibited similar conformations, with low root-mean-square deviation values. These results could provide foundational data from China for the development of RSV mAbs and vaccines.
Keywords: Human respiratory syncytial virus (RSV), Children, Fusion glycoprotein, Antigenic epitope, Variation
Highlights
-
•
This is a multi-center study that involves 10 hospitals of 9 provinces in China from 2017 to 2021.
-
•
F genes of RSV circulating in China from 2017 to 2021 are relatively conserved.
-
•
Mutational frequencies of I206M and Q209R in the binding site of nirsevimab are higher than global data.
-
•
The nirsevimab-resistant mutation, S211N, has emerged with a low frequency in China since 2020.
1. Introduction
Respiratory syncytial virus (RSV) is an enveloped virus with a genome of approximately 15.2 kb of single negative-strand RNA. The RSV genome consists of 10 genes, encoding 11 proteins, of which the attachment (G) and fusion (F) glycoproteins serve as the main surface antigens (Simoes, 1999; Efstathiou et al., 2020). According to discrepancies in antigenicity and the G gene sequence, RSV is classified into two subgroups (RSV A and RSV B). Differences in the G gene sequence between the two subgroups can reach up to 50%, whereas the F gene is relatively conserved (Fuentes et al., 2016; Pangesti et al., 2018). RSV F glycoprotein (RSV-F), which comprises 574 amino acids (AAs), mediates the fusion of viral envelopes with cell membranes. It facilitates the binding of RSV to host cell receptors, enabling viral entry into host cells and causing infected cells to fuse with neighboring cells to form syncytia (Hacking and Hull, 2002).
Clinically, RSV is one of the most common causes of acute lower respiratory tract infection (ALRTI) in children under the age of five years. Based on two global RSV epidemiological estimates, there are approximately 33 million RSV-related ALRTI cases and over 100,000 RSV-related deaths annually among children under five years old (Shi et al., 2017; Li et al., 2022). Furthermore, the disease burden of RSV in those over 60 years of age is comparable to that of influenza A virus (Ackerson et al., 2019; Korsten et al., 2021). The burden of RSV infection is particularly serious in low- and middle-income countries, where more than 95% of RSV-related ALRTI cases occur (Li et al., 2022). In China, RSV is also the most commonly detected virus in samples from patients with ALRTI and from children under 18 years old with community-acquired pneumonia (CAP) (Zhang et al., 2015; Zhu et al., 2021).
Despite the significant annual disease burden caused by RSV, effective antivirals are far from available. Although ribavirin was the only marketed antiviral against RSV, it is currently not recommended for routine clinical use (Efstathiou et al., 2020; Langedijk and Bont, 2023). Consequently, monoclonal antibodies (mAbs) and vaccines are the most promising preventive measures against RSV infection. To date, 33 drug candidates for preventing RSV infection are undergoing clinical evaluation (Mazur et al., 2023). The United States Food and Drug Administration (FDA) recently approved Arexvy, the first RSV vaccine, for use in seniors over 60 years old in the United States (Soni et al., 2023). As antibodies directed against the prefusion conformation of RSV-F provide the main neutralizing activity in serum, almost all mAbs and vaccines are designed to target this conformation (Rossey et al., 2018; Mazur et al., 2023). However, the development of viral escape, caused either by selective pressure or by natural polymorphisms of the virus, poses a potential risk to the efficacy of mAbs or vaccines. Therefore, surveillance on mutations in RSV F gene sequences is of great significance for the development of mAbs and vaccines.
Previously, we reported variations in RSV-F in China from 2014 to 2016 (Chen et al., 2018), and other studies have also documented the national or regional molecular prevalence of RSV-F in China (Qin et al., 2013; Zhang et al., 2013; Xia et al., 2014; Song et al., 2018; Sun et al., 2022). This present study analyzed mutations in RSV-F, compared mutation frequencies between the period before and after 2020, and investigated changes in RSV-F conformations, by sequencing RSV-positive samples collected from ten hospitals across nine provinces or municipalities in China from 2017 to 2021. This study aims to provide foundational data from China to aid in the development of RSV mAbs and vaccines.
2. Materials and methods
2.1. Samples collection
This multi-center study, conducted from February 2017 to February 2021, involved ten hospitals from Beijing, Chongqing, Guangdong, Guizhou, Hebei, Hunan, Ningxia, Shanghai, and Zhejiang provinces. Initially, throat swabs were collected by specially trained staff from children (under 18 years old, both boys and girls) hospitalized with ALRTI at the ten hospitals. All swabs were immediately transferred to tubes containing 3 mL of virus preservation solution (Yocon biotechnology Co., Ltd., China) and stored in a −80 °C freezer. Subsequently, these samples were routinely transferred to our laboratory using dry ice, followed by long-term storage in a −80 °C freezer. Finally, in our laboratory, these samples were centrally tested by a specialized group of staff.
2.2. Extraction of viral RNA
Viral RNA was extracted from the collected samples through nucleic acid extraction instrument (DaAnGene, China) and nucleic acid extraction kits (DaAnGene), followed by multiplex fluorescence real-time PCR and PCR amplification.
2.3. Determination of RSV-positive samples
A multiplex fluorescence real-time PCR kit (XABT, China) targeting 16 respiratory pathogens was performed to identify RSV-positive samples. A total of 500 RSV-positive throat swabs were selected for further testing based on factors such as the child's age, place of residence, time of onset, and RSV subgroups.
2.4. Determination of RSV subgroups and sequencing of RSV F genes
Nucleic acids from the RSV-positive samples were amplified using PrimeScript™ One Step RT-PCR Kit Ver. 2 (Takara, Dalian) with primers F2 and R2 (F2: 5′-ATGCCTATAACAAATGATCAGAAAAAGTT-3′; R2: 5′-GCAATGACCTCGAATTTCAAATT-3′) (Chen et al., 2018). Subsequently, products were sequenced by Sino Geno Max Co., Ltd. (China) using the same primers. Viral subgroups were determined based on BLAST search results on NCBI. Both amplification and sequencing of the full-length RSV F gene utilized the same amplification reagent and primer pairs A-F1/R1 (A-F1: 5′-ACACCAAAGGAAATCCARAACAMA-3′; A-R1: 5′-CCARCAAGGAGTATCWATTACACCATA-3′) and F2/R2 for RSV A-positive samples, or the primer pairs B–F1/R1 (B–F1: 5′-ATCCGAGCCCTCCACATCAAA-3′; B-R1: 5′- GTGCACAGAGGTGATGTGTGT-3′) and F2/R2 for RSV B-positive samples. The amplification program consisted of reverse transcription at 50 °C for 30 min, initial denaturation at 94 °C for 2 min, followed by 40 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min and 20 s, culminating in a final extension at 72 °C for 5 min. Full-length RSV F gene sequences were assembled using SeqMan 7.1.0 (DNAStar, Inc., USA) software.
2.5. Obtaining of prototype and reference strain sequences
RSV F gene sequences for two prototype strains (accession number: JX198112.1 for Long and JX198143.1 for CH18537) and two reference strains (PP376579.1 and MZ516132.1) were downloaded from GenBank. The two reference strains were used exclusively to determine genotypes of strains clustered in A2. Additionally, 151 reference sequences collected in China from 2017 to 2021 (Supplementary Table S1) were downloaded from GenBank and GISAID database and included in this study.
2.6. Analysis of phylogenetic and genetic diversity
Multiple sequence alignments, pairwise distance, and phylogenetic analyses were performed using MEGA 11 (University Park, PA, USA). The neighborhood-joining and maximum likelihood methodologies were used to construct phylogenetic trees, utilizing a bootstrap value of 1000 to evaluate the accuracy of tree construction, with only values over 70% displayed. The phylogenetic trees embellishment was performed by iTOL version 6.8.1 (Letunic and Bork, 2021). AA sequences were deduced and translated using the standard genetic code within MEGA 11. Nucleotide and AA identity analyses of RSV-F were conducted with BioEdit version 7.2 (IBIS Biosciences, Inc., CA, USA).
2.7. Structural visualization and homology modeling of RSV-F
Structure diagrams of prefusion and postfusion RSV-F trimers were visualized using PyMOL version 2.5 (Schrödinger, LLC) with PDB files 5w23 and 3rrr, respectively (Sun et al., 2022). Homology models for prefusion and postfusion conformations of RSV-F protomers from both prototype strains (Long and CH18537) and isolate strains 20190024/BJ/CHN/2019 (abbreviated as 20190024), and CQ025/CQ/CHN/2021 (abbreviated as CQ025) were constructed by SWISS-MODEL (Waterhouse et al., 2018). The two isolate strains harbored all mutations with frequencies ≥ 10%. The templates for homology modeling included PDB files 5ude (for prefusion conformations of Long and strain 20190024), 6q0s (for prefusion conformations of CH18537 and strain CQ025), and 3rki (for all postfusion conformations). Next, the established RSV-F structure models were respectively aligned using PyMOL. Those structural outliers (certain peptides) were identified and excluded using programs. The root-mean-square deviation (RMSD) value represents the similarity between two protein structures, where a smaller value indicates higher similarity (Maiorov and Crippen, 1994).
3. Results
3.1. Sample information
In this study, a total of 396 (79.20%, 396/500) full-length RSV F genes from 500 RSV-positive throat swabs were successfully sequenced (GenBank accession numbers were listed on Supplementary Table S2). Of the 361 sequences for which case information was available, the male-to-female ratio was found to be 1.93:1 (238 boys and 123 girls). The age of these children ranged from 1 day to 14 years, with a median age of 0.58 years, and the majority were infants aged ≤ 1 year (69.25%, 250/361).
Combined with 151 reference strains (72 for RSV A and 79 for RSV B), a total of 547 RSV F gene sequences were analyzed in this study, including 242 (44.24%, 242/547) of subgroup A and 305 (55.76%, 305/547) of subgroup B. For RSV A, 136 sequences (56.20%, 136/242) were from Beijing, 38 (15.70%, 38/242) from Zhejiang, with the remaining 68 sequences (28.10%, 68/242) from other locations. The distribution of sequences collected between 2017 and 2021 was relatively balanced, peaking in 2019 at 33.47% (81/242) and reaching its nadir in 2020 at 9.92% (24/242) (Table 1). For RSV B, the majority of sequences were sourced from Hunan (24.59%, 75/305), Shanghai (21.97%, 67/305), Hubei (13.77%, 42/305), Beijing (13.11%, 40/305), and Chongqing (13.11%, 40/305); with a significant proportion of these sequences collected in 2021 (71.80%, 219/305) (Table 1).
Table 1.
Spatiotemporal distribution of the number of RSV A/B fusion gene sequences included in this study.
| Province/Municipality | Year |
Total | ||||
|---|---|---|---|---|---|---|
| 2017 | 2018 | 2019 | 2020 | 2021 | ||
| Sequenced | ||||||
| Beijing | – | 31/4 a | 44/9 | 1/0 | – | 76/13 |
| Chongqing | – | – | – | – | 8/40 | 8/40 |
| Guizhou | 2/0 | 14/3 | 2/0 | – | – | 18/3 |
| Guangdong | 8/1 | 3/0 | 1/1 | – | 3/13 | 15/15 |
| Hebei | – | – | – | – | 1/1 | 1/1 |
| Hunan | – | – | – | – | 9/75 | 9/75 |
| Ningxia | 0/1 | 1/0 | 1/0 | – | – | 2/1 |
| Shanghai | – | – | – | – | 9/67 | 9/67 |
| Zhejiang | 32/11 | – | – | – | – | 32/11 |
| Downloaded | ||||||
| Beijing | – | 5/0 | 33/5 | 22/22 | – | 60/27 |
| Gansu b | 1/0 | – | – | – | – | 1/0 |
| Hubei b | – | 1/0 | – | 1/19 | 1/23 | 3/42 |
| Ningxia | 1/0 | – | – | – | – | 1/0 |
| Shandong b | – | 1/0 | – | – | – | 1/0 |
| Zhejiang | 2/4 | 4/5 | – | – | – | 6/9 |
| Unknown c | 0/1 | – | – | – | – | 0/1 |
| Total | 46/18 | 60/12 | 81/15 | 24/41 | 31/219 | 242/305 |
Numbers before and behind “/” indicated the number of RSV A and RSV B fusion glycoprotein gene sequences.
Gansu, Hubei, and Shandong Provinces were not included in the ten provinces.
Sequence downloaded from GenBank missing “Province/Municipality” information.
3.2. Phylogenetic analysis
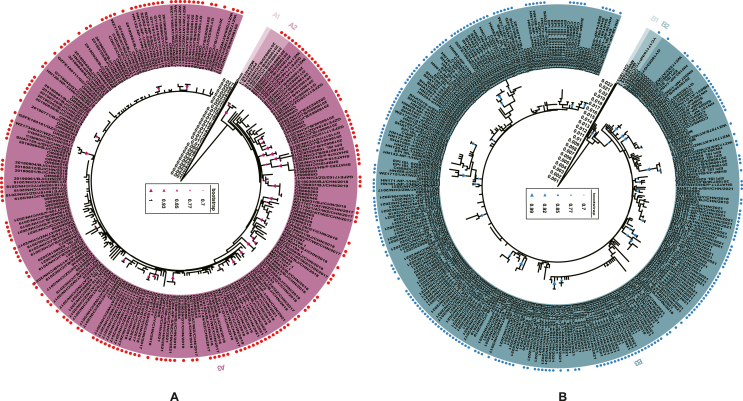
A total of 551 sequences of RSV F gene were analyzed, including 547 sequences, two from prototype strains, and two from reference strains utilized for the classification of cluster A2. Based on the mean p-distance between clusters A2 and A3 (0.010 ± 0.002) and B2 and B3 (0.009 ± 0.002), combined with the phylogenetic tree branching patterns, both RSV A and RSV B could be divided into 3 clusters. (Fig. 1A and 1B). Additionally, the maximum likelihood method verified these results (Supplementary Fig. S1). For RSV A, all sequences were clustered within A3, with the exception of Long, which was in cluster A1, and three isolates (20190825/BJ/CHN/2019, 20190845/BJ/CHN/2019, and 20190832/BJ/CHN/2019) clustered in A2 with two reference sequences. According to the RSV G gene of reference strains, clusters A2 and A3 both belonged to genotype ON1. For RSV B, CH18537 constituted cluster B1, one isolate (YC17110/NP/YC/CHN/2017) and three reference sequences (MW587044.1, MN163124.1, and OR666576) formed cluster B2, leaving the remaining sequences were in cluster B3. Similarly, according to the RSV G gene of reference strains, both clusters B2 and B3 belonged to genotype BA9.
Fig. 1.
Phylogenetic analysis of the fusion gene of RSV A (n = 245) and RSV B (n = 306). Phylogenetic trees of RSV A (A) and RSV B (B) were generated using the neighbor-joining method, with bootstrap values over 70% indicated. Strains marked with red (RSV A) and blue (RSV B) dots in front of their labels were obtained by sequencing, while those without these markings were acquired from GenBank and GISAID. Different clusters were distinctly colored and noted accordingly.
Compared to Long and CH18537, the mean p-distances were 0.0488 ± 5.146E-03 for RSV A and 0.0377 ± 4.359E-03 for RSV B, respectively; the nucleotide (amino acid) sequence identities were 94.7%–95.3% (97.5%–98.2%) for RSV A and 95.7%–96.5% (97.3%–98.4%) for RSV B, respectively. Excluding the prototype strains, nucleotide (amino acid) sequence identities were 98.4%–100% (98.7%–100%) among the 242 RSV A sequences and 98.4%–100% (98.6%–100%) among the 305 RSV B sequences, with mean distances calculated within groups of 0.0051 ± 5.948E-04 for RSV A and 0.0048 ± 5.466E-04 for RSV B.
3.3. Analysis of variations in sites of RSV-F
All mutations in the RSV F gene involved nucleotide substitutions compared to their respective prototype strains; no deletions or insertions were observed. For RSV A and RSV B, 60 and 58 AA changes were identified, respectively (Fig. 2). As the number of monoclonal antibodies targeting different sites of RSV-F increases, classifying RSV-F antigenic sites has become increasingly complex. Rossey et al. identified six sites based on the secondary structure of RSV-F: sites Ø and I–V (Table 2) (Rossey et al., 2018). Additionally, although p27 is cleaved during RSV-F maturation, it is of great significance in the process of RSV infection and warrants further analysis (Krzyzaniak et al., 2013). In this study, 24 AA alterations were identified at six sites and within the p27 region for RSV A, and 32 changes for RSV B, with most occurring at frequencies below 10% (Table 2). For RSV A, main AA changes included: sites Ø (R213S, 100%), I (V384I, 100%), II (N276S, 90.5%), and p27 (K124N, 100%). For RSV B, these changes included: sites Ø (R202Q, 100%; I206M, 98.03%; Q209R, 97.7%), I (F45L, 100%), and V (M152I, 100%; L172Q, 100%; S173L, 100%; I185V, 100%; K191R, 97.7%) (Figs. 2 and 3).
Fig. 2.
Amino acid mutations of the full-length (574 amino acids) fusion glycoprotein of RSV A (n = 242) and RSV B (n = 305) compared respectively with Long and CH18537. Amino acid changes detected at frequencies of ≥ 10% were displayed as colored columns (red for RSV A and blue for RSV B). The form of amino acid changes is shown as “A position B”.
Table 2.
Amino acid mutations at six sites and in the p27 region of the fusion glycoprotein of RSV A and RSV B based on deduced amino acid sequences.
| Sites on F protein a | AA location of sites | RSV A (n = 242) |
RSV B (n = 305) |
||
|---|---|---|---|---|---|
| V b | F (%) | V | F (%) | ||
| Ⅰ | 26–45; 313–319; 379–390 |
T29I | 0.41 | F45L | 100 |
| E31K | 0.41 | ||||
| V40I | 0.42 | ||||
| V384I | 100 | ||||
| K390R | 1.24 | ||||
| Ⅱ | 254–277 | N276S | 90.5 | M264I | 0.33 |
| Ⅲ | 46–54; 301–312; 345–352; 367–378 |
T50P | 0.41 | null | null |
| V349I | 1.65 | ||||
| Ⅵ | 422–471 | S451T | 0.41 | C439F | 0.33 |
| N466S | 2.3 | ||||
| L467F | 0.33 | ||||
| Ⅴ | 55–62; 146–194; 287–300 |
S55R | 0.41 | I59V | 0.33 |
| M152I | 100 | ||||
| L172Q | 100 | ||||
| S173L | 100 | ||||
| V178A | 0.33 | ||||
| I291T | 0.83 | I185V | 100 | ||
| K191R | 97.7 | ||||
| E295G | 0.33 | ||||
| Ø | 63–96; 195–227 |
N67S | 0.41 | E66D | 0.66 |
| N67T | 0.83 | D73E | 0.33 | ||
| L93W | 0.83 | N197D | 1.97 | ||
| R202Q | 100 | ||||
| K201R | 0.41 | I206M | 98.03 | ||
| Q209R | 97.7 | ||||
| R213S | 100 | S211N | 0.98 | ||
| p27 | 110–136 | L111I | 1.65 | Y114H | 0.33 |
| R113S | 1.24 | M115I | 0.66 | ||
| M115V | 0.33 | ||||
| T118A | 0.83 | N120Y | 0.33 | ||
| N120D | 0.41 | T121A | 0.33 | ||
| K123N | 1.31 | ||||
| T122A | 4.13 | N124S | 0.33 | ||
| K123E | 2.07 | N124D | 0.66 | ||
| N124Y | 0.33 | ||||
| K124N | 100 | L125P | 2.95 | ||
| L129F | 0.41 | V127I | 0.33 | ||
| I129L | 0.66 | ||||
Abbreviations: V, variation; F, frequency.
Determination of these sites was referred to reference (Rossey et al., 2018).
The form of amino acid changes was shown as “A position B”.
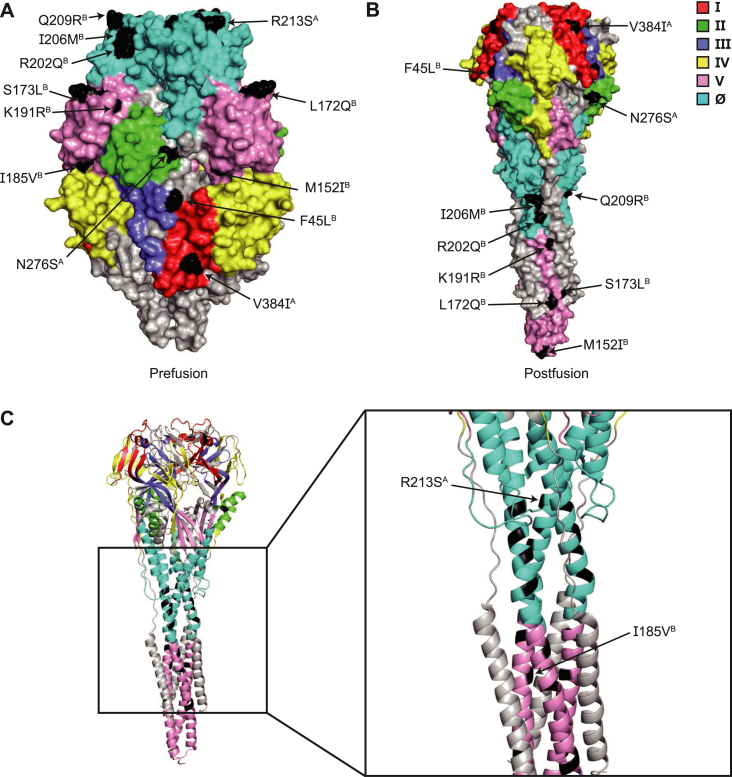
Fig. 3.
Visualization of amino acid variations at six sites of the fusion glycoprotein in different conformations. A and B Structure models of the prefusion and postfusion conformations of the fusion glycoprotein indicated the location of sites I (red), II (green), III (purple), IV (yellow), V (pink), and Ø (cyan), and mutated amino acids (black). C The left image presented a cartoon structure of the postfusion conformation, while the right image displayed a locally magnified structure showing the positions of the changed amino acids. Changed amino acids detected at ≥ 10% frequencies are displayed. The form of amino acid changes is shown as “A position B”.
3.4. The difference in mutational frequencies of RSV-F between the time before and after 2020
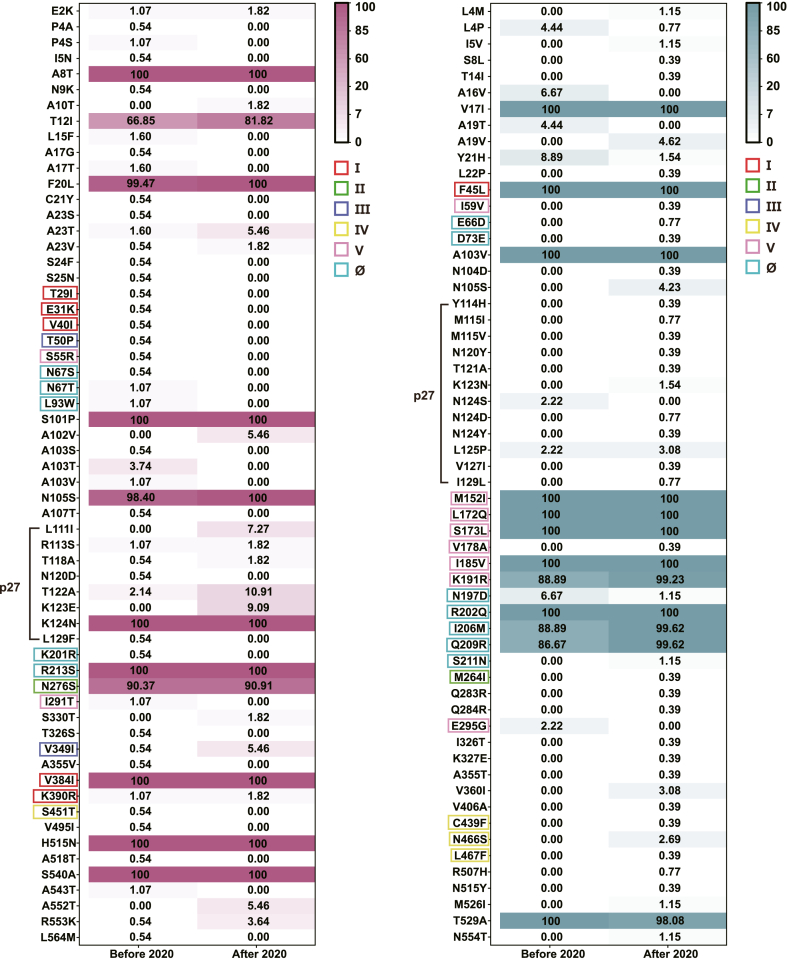
It has been reported that mutational frequencies at nirsevimab-binding site of RSV-B changed before and after 2020 in the United States (Rios-Guzman et al., 2024). To investigate whether similar changes occurred in China, we analyzed 547 RSV-F sequences collected during two periods: 2017–2019 (before 2020) and 2020–2021 (after 2020). Mutational frequencies at all AA positions were assessed. However, most frequencies exhibited slight changes, with absolute differences < 5% (Fig. 4). Specifically, for RSV A, frequencies increased by more than 5% at positions T12I, A102V, L111I, T122A, K123E, and A552T. For RSV B, frequencies increased by more than 10% at positions K191R, I206M, and Q209R, while frequencies decreased by more than 5% at positions A16V, Y21H, and N197D.
Fig. 4.
Differences in mutational frequencies of the fusion glycoprotein between the time before and after 2020. Heat maps displayed differences in the frequency of all mutations in the fusion glycoprotein of both RSV A [n = 242, n (before) = 187 and n (after) = 55] and RSV B [n = 305, n (before) = 45 and n (after) = 260] between the time before (2017–2019) and after (2020–2021) 2020. Each cell in the heat maps represented the frequency (%) of a mutation (shown on the left of the heat maps) in a specific time group (shown at the bottom of the heat maps). The color intensity of each cell corresponded to the frequency value, with darker colors indicating higher frequencies. Mutations located at sites I (red), II (green), III (purple), IV (yellow), V (pink) and Ø (cyan) were outlined in corresponding colors, while those located in the p27 region were marked with black lines. The form of amino acid changes is shown as “A position B”.
Across six sites of RSV-F, mutational frequencies changed slightly for RSV A. However, the mutation V349I at site Ш increased from 0.54% to 5.46%, suggesting a potential evolutionary trend. Notably, for RSV B, mutational frequencies obviously increased for I206M (from 88.89% to 99.62%) and Q209R (from 86.67% to 99.62%) in site Ø, while N197D decreased (from 6.67% to 1.15%). In site V, K191R showed a notable increase (from 88.89% to 99.23%).
In the p27 region, mutational frequencies obviously increased for RSV A at L111I (from 0% to 7.27%), T122A (from 2.14% to 10.91%), and K123E (from 0% to 9.09%). In contrast, all mutational frequencies in p27 changed slightly for RSV B.
3.5. Analysis of RSV-F conformations
According to chemical structures, no changes in the AA polarity were observed in the primary mutations of RSV A. In contrast, RSV B strains exhibited altered AA polarities at sites V (M152I, L172Q, and S173L) and Ø (I206M).
Furthermore, to investigate the influence of the main mutations on protein conformation, structure predictions of RSV-F were performed with homology modeling. The results of structure alignments revealed that both isolate strains, 20190024 (RSV A) and CQ025 (RSV B), closely overlapped with their respective prototype strains in both the prefusion and postfusion conformations. The RSMD values for RSV A were 0.107 (prefusion) and 0.061 (postfusion), and for RSV B were 0.070 (prefusion) and 0.068 (postfusion), indicating a high structural similarity between the isolate and prototype strains (Fig. 5A). However, some secondary structure changes were observed in the isolate strains. In isolate 20190024, α-helix to random coil transitions were detected at positions AA 35–36 and AA 97–98 in the prefusion conformation, with no changes observed in the postfusion conformation. However, no changes were found in the postfusion conformation. Conversely, in isolate CQ025, all transitions involved random coil to α-helix changes, specifically at positions AA 369–371 in the prefusion conformation and AA 355–357 and AA 384–385 in the postfusion conformation (Fig. 5B).
Fig. 5.
Analysis of changes in the fusion glycoprotein conformation. A Cartoon structures displayed the alignment results of Long with isolate strain 20190024/BJ/CHN/2019 (RSV A, left) and CH18537 with isolate strain CQ025/CQ/CHN/2021 (RSV B, right). Images (c) and (d) were obtained by rotating Images (a) and (b) 180° along the X-axis, respectively. B Local magnifications of the alignment result revealed changes in the secondary structure of the fusion glycoprotein. Modified regions were marked with black lines and annotations. Structural outliers of isolate strains were colored with red (RSV A) or blue (RSV B). Root-mean-square deviation values were indicated.
However, all findings presented herein are preliminary predictions generated by software, relying on deduced AA sequences. Assessing the actual impact of these mutations on protein conformations is complex and necessitates further investigation through high-resolution structural analyses and comparative biological function studies.
4. Discussion
RSV is a primary pathogen responsible for ALRTI in children, posing significant threats to their health and lives. Currently, effective antiviral treatments for RSV are lacking. RSV-F serves as a critical target protein for vaccine and monoclonal antibody (mAb) development. Therefore, monitoring mutations in RSV-F is crucial for the advancement and evaluation of vaccines and mAbs.
Phylogenetic analysis revealed that both RSV A and RSV B formed three clusters based on the RSV F gene. Clusters A2 and A3 were genotype ON1, while clusters B2 and B3 were genotype BA9. RSV B strains exhibited greater variability in AA polymorphisms compared to RSV A strains, consistent with prior studies (Hause et al., 2017). Across six sites and in the p27 region, four mutations were detected at ≥ 10% frequencies in RSV A strains, and nine in RSV B strains. Among these, R213S (site Ø, RSV A), M152I, and I185V (site V, RSV B) are reported for the first time in this study, while other mutations have been previously documented (Chen et al., 2018; Song et al., 2018; Sun et al., 2022). Importantly, these three mutations are not novel variants.
Between 2014 and 2016 in China, we observed that the mutational frequencies of K124N (RSV A) and R202Q (RSV B) consistently remained at 100%. Conversely, the mutational frequencies of F45L, L172Q, S173L (RSV B), and V384I (RSV A) increased from 94.6%, 91.3%, 73.9%, and 99.3%–100%, respectively (Chen et al., 2018). It might be due to natural evolution. This also reflects that mutations at these sites might be conducive to the spread of the virus. Notably, mutations L172Q and S173L in RSV B have been associated with resistance to suptavumab (Simões et al., 2021). L172Q and S173L are natural polymorphisms of RSV B, with historically reported frequencies of less than 1%, rather than mutations developed under selective pressure with suptavumab (Simões et al., 2021; Langedijk and Bont, 2023). However, the frequency of both mutations found simultaneously in historical isolates was unavailable until 2015. The concurrent appearance of the two mutations was first reported in China in 2015, and they globally predominated over 3 RSV seasons (Chen et al., 2018; Simões et al., 2021). In this study, their frequencies maintained 100% from before 2020 to after 2020. Though suptavumab has never been approved in China, the natural evolution of these two mutations has rendered all circulating RSV B strains in China to escape suptavumab neutralization. Thus, it is imperative to continue tracking RSV-F mutations to ensure the efficacy of current monoclonal antibodies.
Palivizumab (MEDI-493), a recombinant humanized mAb for preventing severe lower respiratory tract infections in children at a high risk of RSV infection, was approved by FDA in 1998, but has not yet been approved in China (Garegnani et al., 2021). In this study, based on published information from FDA, no mutations which were associated with resistance to palivizumab were found within or outside its binding site (AA 262–275) (Administration, 1998). This finding is consistent with the conclusion that resistance-related mutations, which are developed in vitro after the introduction of palivizumab, are not frequently produced in real-world settings, even though palivizumab is not available in China (Simões et al., 2021).
Despite its ability to prevent severe RSV infections, palivizumab is expensive, requires monthly doses throughout the RSV season, and has not been introduced in China (Administration, 1998; Mazur et al., 2023). In January 2024, nirsevimab (MEDI-8897) was approved by the National Medical Products Administration of China and is expected to be available during the 2024–2025 RSV infection season. Nirsevimab is a new mAbs targeting site Ø, with YTE mutation at its Fc terminus extending its half-life, allowing one injection of nirsevimab to protect children throughout the RSV season (Zhu et al., 2017; Keam, 2023). The binding site of nirsevimab (AA 62–69, 196–212) is highly conserved in both RSV A and RSV B, enabling it to neutralize different subgroups of RSV. However, some reports have indicated the presence of mutations associated with resistance to nirsevimab, such as E66K and K68E in RSV A, and K68N, N201S/T, Q209K/L, and S211N in RSV B (Zhu et al., 2017; Langedijk et al., 2022; Peeples and Thongpan, 2023; Rios-Guzman et al., 2024). Notably, although S211N renders only a 1.2-fold reduction in susceptibility to nirsevimab, its frequency has increased from 0% to 1.15% (3/260) (accession number: OR899386, OR899425 and EPI_ISL_6314659) in China after 2020. A BLAST search on NCBI revealed that these sequences cluster with strains isolated in the United States, Australia, and Europe (data not shown), suggesting that the increase in China may be due to the global influence of RSV B circulating in the United States after 2020 (Rios-Guzman et al., 2024). Alternatively, this mutation may result from natural evolution. Considering that nirsevimab will soon be available, the mutational frequency of S211N should be closely monitored. No other resistance-related mutations in the nirsevimab-binding site of RSV A and RSV B were found in this study.
Additionally, RSV B strains involved in this study exhibited R202Q (100%), I206M (97.72%), and Q209R (97.34%) mutations. The R202Q mutation was already present when nirsevimab was designed and has maintained a frequency of 100% from 2014 to 2021 in China (Zhu et al., 2017; Chen et al., 2018). The frequencies of the I206M and Q209R mutations were higher than global data collected from 2015 to 2021 (I206M: 68.90%, Q209R: 68.25%) (Wilkins et al., 2023). Furthermore, the frequencies of the two mutations increased after 2020, a trend consistent with global data, suggesting a worldwide prevalence (Wilkins et al., 2023; Rios-Guzman et al., 2024). Although the I206M mutation can slightly reduce nirsevimab neutralization (5.0-fold change in IC50), it often co-occurs with the Q209R mutation due to steric effects. In such cases, these RSV B strains become more susceptible to nirsevimab (0.2-fold change in IC50) (Wilkins et al., 2023). In this study, only one strain (1/305, 0.33%) (accession number: OR899555) isolated in 2017 exhibited the I206M mutation without the Q209R mutation.
Clesrovimab (MK-1654) binds to site IV (AA 426–429, 432–433, 440–441, 443, and 445–447) and contains the YTE mutation at its Fc terminus, with properties of half-life extension and broad-spectrum neutralization like nirsevimab. In vivo experiments in cotton rats and humans have shown that this mAb can inhibit RSV replication (Tang et al., 2019; Maas et al., 2021). Compared to the binding site of nirsevimab, clesrovimab's binding sites are more conserved across all RSV genotypes (Hause et al., 2017). In this study, we found that no mutations occurred in its binding site.
In 2016, Fuentes et al. identified p27 as a new antigenic site of RSV-F and an age-dependent immunodominant epitope (Fuentes et al., 2016). Although the neutralizing activity of p27 antibodies is lower than that of antibodies targeting other sites of RSV-F in adults, mucosal p27 antibodies may play a crucial role in controlling RSV infection (Fuentes et al., 2020; Ye et al., 2020; Blunck et al., 2022). Mechanistically, p27 antibodies may protect the body through antibody-dependent cell-mediated cytotoxicity (ADCC) effect rather than directly neutralizing RSV (Lee et al., 2022; Rezende et al., 2023). Therefore, p27 may have potential values in therapy and prevention. However, the sequence of p27 is variable (Hause et al., 2017), which is also found in this study. Compared to prototype strains, the AA sequence of p27 exhibited eight and twelve changes in subgroups A and B, respectively. The frequency of three mutations in RSV A (L111I, T122A, and K123E) increased by more than 5% since 2020, suggesting a potential evolutionary trend in RSV A. BALB/c mice immunized with mutated RSV-F (N116Q) have been reported to generate an enhanced neutralizing antibody response (Leemans et al., 2018, 2019); however, this mutation was not observed in our study.
Our study has several limitations. Firstly, the sample selection for this study should be comprehensive, balanced and random in both temporal and spatial distribution. However, some samples were excluded due to sequencing failures. Secondly, we only conducted a preliminary analysis of the mutated RSV-F conformation. Extensive investigations are needed to determine the impact of variations on the conformation and biological function of RSV-F.
As RSV-F-targeted vaccines and mAbs are focal points of research, the molecular epidemiology of RSV-F deserves continued surveillance. This multi-center study sequenced and analyzed RSV-F circulating in China from 2017 to 2021, finding that the RSV F gene was relatively conserved, while some nucleotide substitutions and AA mutations were detected. Based on simple alignments of structures, the altered AAs appear to have little impact on the RSV-F conformation. However, the effects of altered AAs on RSV-F require further high-resolution structural and biological function analyses. For RSV B, K191R, I206M, and Q209R have become gradually predominant in China from before 2020 to after 2020; a nirsevimab-resistant mutation, S211N, has emerged with a low frequency in China since 2020. These results could provide foundational data from China for the development of RSV mAbs and vaccines.
5. Conclusions
The RSV F genes circulating in China from 2017 to 2021 have remained relatively conserved, although some AAs located at six specific sites and the p27 region have undergone changes. For RSV B, the K191R, I206M, and Q209R mutations have gradually become predominant in China from the time before 2020 to after 2020. Additionally, the S211N mutation has emerged at a low frequency in China since 2020.
Data availability
The datasets presented in this study can be found in GenBank and Science Data Bank. The GenBank accession number(s) can be found in the article/Supplementary Material. The datasets can be downloaded from Science Data Bank by searching DOI: https://doi.org/10.57760/sciencedb.12757.
Ethics statement
This study was performed in strict accordance with human subject protection guidance and was approved by the Ethical Review Committee of Beijing Children's Hospital. Written informed consent was obtained on the participants' behalf from their parents or guardians.
Author contributions
Yiliang Fu: methodology, software, validation, formal analysis, investigation, data curation, writing - original draft, visualization; Xiangpeng Chen: conceptualization, methodology, resources, writing - review & editing, supervision, project administration, funding acquisition; Zhengde Xie: conceptualization, methodology, resources, writing - review & editing, supervision, project administration, funding acquisition; Fei Li: validation, investigation; Yun Zhu: validation, investigation, resources; Luci Huang: validation, investigation; Qiuping Li: resources; Hanwen Zhan: validation, investigation; Lili Zhong: resources; Hailin Zhang: resources; Zhengxiu Luo: resources; Gen Lu: resources; Jikui Deng: resources; Lingfeng Cao: resources; Ying Wu: resources; Rong Jin: resources; Lei Li: resources; Lili Xu: resources.
Conflict of interest
The authors declare no competing interests.
Acknowledgements
We would like to express our sincere appreciation to our institutes and universities. This study was supported by the National Key Research and Development Program of China (grant number 2023YFC2306002), National Science and Technology Major Projects (grant number 2017ZX10104001-005-010), CAMS Innovation Fund for Medical Sciences (CIFMS) (Grant Number 2019–I2M–5–026) and Funding for Reform and Development of Beijing Municipal Health Commission.
Footnotes
Peer review under the responsibility of Editorial Board of Virologica Sinica.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2024.09.002.
Contributor Information
Xiangpeng Chen, Email: chenxp1111@163.com.
Zhengde Xie, Email: xiezhengde@bch.com.cn.
Appendix A. .Supplementary data
The following are the Supplementary data to this article:
Supplementary Figure S1.
Phylogenetic analysis of the fusion gene of RSV A (n = 245) and RSV B (n = 306). Phylogenetic trees of RSV A (A) and RSV B (B) were generated using the maximum likelihood method, with bootstrap values of ≥ 70% indicated. Strains marked with red (RSV A) and blue (RSV B) dots in front of their labels were obtained by sequencing, while those without these markings were acquired from GenBank and GISAID. Different clusters were distinctly colored and noted accordingly.
References
- Ackerson B., Tseng H.F., Sy L.S., Solano Z., Slezak J., Luo Y., Fischetti C.A., Shinde V. Severe morbidity and mortality associated with respiratory syncytial virus versus influenza infection in hospitalized older adults. Clin. Infect. Dis. 2019;69:197–203. doi: 10.1093/cid/ciy991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Administration U.F.a.D. Synagis® (palivizumab) injection, for intramuscular use initial u.S. Approval: 1998. 1998. https://www.accessdata.fda.gov/drugsatfda_docs/label/2014/103770s5185lbl.pdf/
- Blunck B.N., Aideyan L., Ye X., Avadhanula V., Ferlic-Stark L., Zechiedrich L., Gilbert B.E., Piedra P.A. Antibody responses of healthy adults to the p27 peptide of respiratory syncytial virus fusion protein. Vaccine. 2022;40:536–543. doi: 10.1016/j.vaccine.2021.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu B., Guo J., Li C., An S., Zhou Y., Chen A., Deng L., Fu Z., Zhu Y., Liu C., Xu L., Wang W., Shen K., Xie Z. Genetic variations in the fusion protein of respiratory syncytial virus isolated from children hospitalized with community-acquired pneumonia in China. Sci. Rep. 2018;8:4491. doi: 10.1038/s41598-018-22826-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou C., Abidi S.H., Harker J., Stevenson N.J. Revisiting respiratory syncytial virus's interaction with host immunity, towards novel therapeutics. Cell. Mol. Life Sci. 2020;77:5045–5058. doi: 10.1007/s00018-020-03557-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes S., Coyle E.M., Beeler J., Golding H., Khurana S. Antigenic fingerprinting following primary rsv infection in young children identifies novel antigenic sites and reveals unlinked evolution of human antibody repertoires to fusion and attachment glycoproteins. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes S., Hahn M., Chilcote K., Chemaly R.F., Shah D.P., Ye X., Avadhanula V., Piedra P.A., Golding H., Khurana S. Antigenic fingerprinting of respiratory syncytial virus (rsv)-a-infected hematopoietic cell transplant recipients reveals importance of mucosal anti-rsv g antibodies in control of rsv infection in humans. J. Infect. Dis. 2020;221:636–646. doi: 10.1093/infdis/jiz608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garegnani L., Styrmisdóttir L., Roson Rodriguez P., Escobar Liquitay C.M., Esteban I., Franco J.V. Palivizumab for preventing severe respiratory syncytial virus (rsv) infection in children. Cochrane Database Syst. Rev. 2021;11 doi: 10.1002/14651858.CD013757.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacking D., Hull J. Respiratory syncytial virus--viral biology and the host response. J. Infect. 2002;45:18–24. doi: 10.1053/jinf.2002.1015. [DOI] [PubMed] [Google Scholar]
- Hause A.M., Henke D.M., Avadhanula V., Shaw C.A., Tapia L.I., Piedra P.A. Sequence variability of the respiratory syncytial virus (rsv) fusion gene among contemporary and historical genotypes of rsv/a and rsv/b. PLoS One. 2017;12 doi: 10.1371/journal.pone.0175792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keam S.J. Nirsevimab: first approval. Drugs. 2023;83:181–187. doi: 10.1007/s40265-022-01829-6. [DOI] [PubMed] [Google Scholar]
- Korsten K., Adriaenssens N., Coenen S., Butler C., Ravanfar B., Rutter H., Allen J., Falsey A., Pirçon J.Y., Gruselle O., Pavot V., Vernhes C., Balla-Jhagjhoorsingh S., Öner D., Ispas G., Aerssens J., Shinde V., Verheij T., Bont L., Wildenbeest J. Burden of respiratory syncytial virus infection in community-dwelling older adults in europe (resceu): an international prospective cohort study. Eur. Respir. J. 2021;57 doi: 10.1183/13993003.02688-2020. [DOI] [PubMed] [Google Scholar]
- Krzyzaniak M.A., Zumstein M.T., Gerez J.A., Picotti P., Helenius A. Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the f protein. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langedijk A.C., Bont L.J. Respiratory syncytial virus infection and novel interventions. Nat. Rev. Microbiol. 2023;21:734–749. doi: 10.1038/s41579-023-00919-w. [DOI] [PubMed] [Google Scholar]
- Langedijk A.C., Harding E.R., Konya B., Vrancken B., Lebbink R.J., Evers A., Willemsen J., Lemey P., Bont L.J. A systematic review on global rsv genetic data: identification of knowledge gaps. Rev. Med. Virol. 2022;32 doi: 10.1002/rmv.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Lee Y., Klenow L., Coyle E.M., Tang J., Ravichandran S., Golding H., Khurana S. Protective antigenic sites identified in respiratory syncytial virus fusion protein reveals importance of p27 domain. EMBO Mol. Med. 2022;14 doi: 10.15252/emmm.202013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans A., Boeren M., Van der Gucht W., Martinet W., Caljon G., Maes L., Cos P., Delputte P. Characterization of the role of n-glycosylation sites in the respiratory syncytial virus fusion protein in virus replication, syncytium formation and antigenicity. Virus Res. 2019;266:58–68. doi: 10.1016/j.virusres.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Leemans A., Boeren M., Van der Gucht W., Pintelon I., Roose K., Schepens B., Saelens X., Bailey D., Martinet W., Caljon G., Maes L., Cos P., Delputte P. Removal of the n-glycosylation sequon at position n116 located in p27 of the respiratory syncytial virus fusion protein elicits enhanced antibody responses after DNA immunization. Viruses. 2018;10:426. doi: 10.3390/v10080426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive tree of life (itol) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–w296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang X., Blau D.M., Caballero M.T., Feikin D.R., Gill C.J., Madhi S.A., Omer S.B., Simões E.A.F., Campbell H., Pariente A.B., Bardach D., Bassat Q., Casalegno J.S., Chakhunashvili G., Crawford N., Danilenko D., Do L.A.H., Echavarria M., Gentile A., Gordon A., Heikkinen T., Huang Q.S., Jullien S., Krishnan A., Lopez E.L., Markić J., Mira-Iglesias A., Moore H.C., Moyes J., Mwananyanda L., Nokes D.J., Noordeen F., Obodai E., Palani N., Romero C., Salimi V., Satav A., Seo E., Shchomak Z., Singleton R., Stolyarov K., Stoszek S.K., von Gottberg A., Wurzel D., Yoshida L.M., Yung C.F., Zar H.J., Nair H. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: a systematic analysis. Lancet. 2022;399:2047–2064. doi: 10.1016/S0140-6736(22)00478-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas B.M., Lommerse J., Plock N., Railkar R.A., Cheung S.Y.A., Caro L., Chen J., Liu W., Zhang Y., Huang Q., Gao W., Qin L., Meng J., Witjes H., Schindler E., Guiastrennec B., Bellanti F., Spellman D.S., Roadcap B., Kalinova M., Fok-Seang J., Catchpole A.P., Espeseth A.S., Stoch S.A., Lai E., Vora K.A., Aliprantis A.O., Sachs J.R. Forward and reverse translational approaches to predict efficacy of neutralizing respiratory syncytial virus (rsv) antibody prophylaxis. EBioMedicine. 2021;73 doi: 10.1016/j.ebiom.2021.103651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorov V.N., Crippen G.M. Significance of root-mean-square deviation in comparing three-dimensional structures of globular proteins. J. Mol. Biol. 1994;235:625–634. doi: 10.1006/jmbi.1994.1017. [DOI] [PubMed] [Google Scholar]
- Mazur N.I., Terstappen J., Baral R., Bardají A., Beutels P., Buchholz U.J., Cohen C., Crowe J.E., Jr., Cutland C.L., Eckert L., Feikin D., Fitzpatrick T., Fong Y., Graham B.S., Heikkinen T., Higgins D., Hirve S., Klugman K.P., Kragten-Tabatabaie L., Lemey P., Libster R., Löwensteyn Y., Mejias A., Munoz F.M., Munywoki P.K., Mwananyanda L., Nair H., Nunes M.C., Ramilo O., Richmond P., Ruckwardt T.J., Sande C., Srikantiah P., Thacker N., Waldstein K.A., Weinberger D., Wildenbeest J., Wiseman D., Zar H.J., Zambon M., Bont L. Respiratory syncytial virus prevention within reach: the vaccine and monoclonal antibody landscape. Lancet Infect. Dis. 2023;23:e2–e21. doi: 10.1016/S1473-3099(22)00291-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangesti K.N.A., Abd El Ghany M., Walsh M.G., Kesson A.M., Hill-Cawthorne G.A. Molecular epidemiology of respiratory syncytial virus. Rev. Med. Virol. 2018;28 doi: 10.1002/rmv.1968. [DOI] [PubMed] [Google Scholar]
- Peeples M.E., Thongpan I. Nirsevimab-resistant respiratory syncytial virus strains are rare but there. Lancet Infect. Dis. 2023;23:773–774. doi: 10.1016/S1473-3099(23)00137-8. [DOI] [PubMed] [Google Scholar]
- Qin X., Zhang C., Zhao Y., Zhao X. Genetic variability of subgroup a and b respiratory syncytial virus strains circulating in southwestern China from 2009 to 2011. Arch. Virol. 2013;158:1487–1495. doi: 10.1007/s00705-012-1552-z. [DOI] [PubMed] [Google Scholar]
- Rezende W., Neal H.E., Dutch R.E., Piedra P.A. The rsv f p27 peptide: current knowledge, important questions. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1219846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Guzman E., Simons L.M., Dean T.J., Agnes F., Pawlowski A., Alisoltanidehkordi A., Nam H.H., Ison M.G., Ozer E.A., Lorenzo-Redondo R., Hultquist J.F. Deviations in rsv epidemiological patterns and population structures in the United States following the covid-19 pandemic. Nat. Commun. 2024;15:3374. doi: 10.1038/s41467-024-47757-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossey I., McLellan J.S., Saelens X., Schepens B. Clinical potential of prefusion rsv f-specific antibodies. Trends Microbiol. 2018;26:209–219. doi: 10.1016/j.tim.2017.09.009. [DOI] [PubMed] [Google Scholar]
- Shi T., McAllister D.A., O'Brien K.L., Simoes E.A.F., Madhi S.A., Gessner B.D., Polack F.P., Balsells E., Acacio S., Aguayo C., Alassani I., Ali A., Antonio M., Awasthi S., Awori J.O., et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet. 2017;390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes E.A. Respiratory syncytial virus infection. Lancet. 1999;354:847–852. doi: 10.1016/S0140-6736(99)80040-3. [DOI] [PubMed] [Google Scholar]
- Simões E.A.F., Forleo-Neto E., Geba G.P., Kamal M., Yang F., Cicirello H., Houghton M.R., Rideman R., Zhao Q., Benvin S.L., Hawes A., Fuller E.D., Wloga E., Pizarro J.M.N., Munoz F.M., Rush S.A., McLellan J.S., Lipsich L., Stahl N., Yancopoulos G.D., Weinreich D.M., Kyratsous C.A., Sivapalasingam S. Suptavumab for the prevention of medically attended respiratory syncytial virus infection in preterm infants. Clin. Infect. Dis. 2021;73:e4400–e4408. doi: 10.1093/cid/ciaa951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Wang H., Ng T.I., Cui A., Zhu S., Huang Y., Sun L., Yang Z., Yu D., Yu P., Zhang H., Zhang Y., Xu W. Sequence analysis of the fusion protein gene of human respiratory syncytial virus circulating in China from 2003 to 2014. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-35894-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni A., Kabra S.K., Lodha R. Respiratory syncytial virus infection: an update. Indian J. Pediatr. 2023;90:1245–1253. doi: 10.1007/s12098-023-04613-w. [DOI] [PubMed] [Google Scholar]
- Sun Y.P., Lei S.Y., Wang Y.B., Wang Y.Z., Qiang H.S., Yin Y.F., Jiang Z.M., Zhu M., Chen X.L., Ye H.M., Zheng Z.Z., Xia N.S. Molecular evolution of attachment glycoprotein (g) and fusion protein (f) genes of respiratory syncytial virus on1 and ba9 strains in xiamen, China. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.02083-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Chen Z., Cox K.S., Su H.P., Callahan C., Fridman A., Zhang L., Patel S.B., Cejas P.J., Swoyer R., Touch S., Citron M.P., Govindarajan D., Luo B., Eddins M., Reid J.C., Soisson S.M., Galli J., Wang D., Wen Z., Heidecker G.J., Casimiro D.R., DiStefano D.J., Vora K.A. A potent broadly neutralizing human rsv antibody targets conserved site iv of the fusion glycoprotein. Nat. Commun. 2019;10:4153. doi: 10.1038/s41467-019-12137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. Swiss-model: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–w303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins D., Langedijk A.C., Lebbink R.J., Morehouse C., Abram M.E., Ahani B., Aksyuk A.A., Baraldi E., Brady T., Chen A.T., Chi H., Choi E.H., Cohen R., Danilenko D.M., Gopalakrishnan V., Greenough A., Heikkinen T., Hosoya M., Keller C., Kelly E.J., Kragten-Tabatabaie L., Martinón-Torres F., de Los Santos A.H.M., Nunes M.C., Palomino M.A., Papenburg J., Pernica J.M., Richmond P., Stein R.T., Tuffy K.M., Verwey C., Esser M.T., Tabor D.E., Bont L.J. Nirsevimab binding-site conservation in respiratory syncytial virus fusion glycoprotein worldwide between 1956 and 2021: an analysis of observational study sequencing data. Lancet Infect. Dis. 2023;23:856–866. doi: 10.1016/S1473-3099(23)00062-2. [DOI] [PubMed] [Google Scholar]
- Xia Q., Zhou L., Peng C., Hao R., Ni K., Zang N., Ren L., Deng Y., Xie X., He L., Tian D., Wang L., Huang A., Zhao Y., Zhao X., Fu Z., Tu W., Liu E. Detection of respiratory syncytial virus fusion protein variants between 2009 and 2012 in China. Arch. Virol. 2014;159:1089–1098. doi: 10.1007/s00705-013-1870-9. [DOI] [PubMed] [Google Scholar]
- Ye X., Cabral de Rezende W., Iwuchukwu O.P., Avadhanula V., Ferlic-Stark L.L., Patel K.D., Piedra F.A., Shah D.P., Chemaly R.F., Piedra P.A. Antibody response to the furin cleavable twenty-seven amino acid peptide (p27) of the fusion protein in respiratory syncytial virus (rsv) infected adult hematopoietic cell transplant (hct) recipients. Vaccines. 2020;8:192. doi: 10.3390/vaccines8020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.Y., Liao H.Y., Fan X.J., Lu L., Pei X.F., Xu X. [gene sequence and antigenic epitope variation within the f protein of respiratory syncytial virus in sichuan, China] Sichuan Da Xue Xue Bao Yi Xue Ban. 2013;44:175–178. [PubMed] [Google Scholar]
- Zhang Y., Yuan L., Zhang Y., Zhang X., Zheng M., Kyaw M.H. Burden of respiratory syncytial virus infections in China: systematic review and meta-analysis. J. Glob. Health. 2015;5 doi: 10.7189/jogh.05.020417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., McLellan J.S., Kallewaard N.L., Ulbrandt N.D., Palaszynski S., Zhang J., Moldt B., Khan A., Svabek C., McAuliffe J.M., Wrapp D., Patel N.K., Cook K.E., Richter B.W.M., Ryan P.C., Yuan A.Q., Suzich J.A. A highly potent extended half-life antibody as a potential rsv vaccine surrogate for all infants. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aaj1928. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Xu B., Li C., Chen Z., Cao L., Fu Z., Shang Y., Chen A., Deng L., Bao Y., Sun Y., Ning L., Yu S., Gu F., Liu C., Yin J., Shen A., Xie Z., Shen K. A multicenter study of viral aetiology of community-acquired pneumonia in hospitalized children in Chinese mainland. Virol. Sin. 2021;36:1543–1553. doi: 10.1007/s12250-021-00437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in GenBank and Science Data Bank. The GenBank accession number(s) can be found in the article/Supplementary Material. The datasets can be downloaded from Science Data Bank by searching DOI: https://doi.org/10.57760/sciencedb.12757.