Abstract
Infections of many viruses induce caspase activation to regulate multiple cellular pathways, including programmed cell death, immune signaling and etc. Characterizations of caspase cleavage sites and substrates are important for understanding the regulation mechanisms of caspase activation. Here, we identified and analyzed a novel caspase cleavage motif AEAD, and confirmed its caspase dependent cleavage activity in natural substrate, such as nitric oxide-associated protein 1 (NOA1). Fusing the enhanced green fluorescent protein (EGFP) with the mitochondrial marker protein Tom20 through the AEAD motif peptide localized EGFP to the mitochondria. Upon the activation of caspase triggered by Sendai virus (SeV) or herpes simplex virus type 1 (HSV-1) infection, EGFP diffusely localized to the cell due to the caspase-mediated cleavage, thus allowing visual detection of the virus-induced caspase activation. An AEAD peptide-derived inhibitor Z-AEAD-FMK were developed, which significantly inhibited the activities of caspases-1, -3, -6, -7, -8 and -9, exhibiting a broad caspase inhibition effect. The inhibitor further prevented caspases-mediated cleavage of downstream substrates, including BID, PARP1, LMNA, pro-IL-1β, pro-IL-18, GSDMD and GSDME, protecting cells from virus-induced apoptotic and pyroptotic cell death. Together, our findings provide a new perspective for the identification of novel caspase cleavage motifs and the development of new caspase inhibitors and anti-inflammatory drugs.
Keywords: Caspase, AEAD motif, Inhibitor, Virus, Cell death
Highlights
-
•
The AEAD motif is a novel caspase cleavage motif.
-
•
The AEAD peptide-derived Z-AEAD-FMK is a novel pan-caspase inhibitor.
-
•
Z-AEAD-FMK inhibits caspase-mediated cell death pathway triggered by virus infection.
1. Introduction
Members of the caspase family are proteolytic enzymes involved in multiple cell death pathways in response to damage-associated or pathogen-associated molecular patterns (DAMPs or PAMPs) (Zheng et al., 2020). Caspases are initially expressed as inactive monomeric zymogens (procaspases) that obtain catalytic activity upon activation (Chen et al., 2017). To date, 18 caspases have been identified in mammals (Chen et al., 2017). Traditionally, based on their physiological functions, caspases are grouped as apoptotic caspases (caspases-2, -3, -6, -7, -8, -9 and -10 in mammals) and inflammatory caspases (caspases-1, -4, -5 and -12 in humans and caspases-1, -11 and -12 in mice) (Shalini et al., 2015; Chen et al., 2017). The apoptotic caspases are further classified into initiator caspases (caspases-8, -9, and -10) and effector caspases (caspases-3, -6, and -7), which are responsible for the initiation and execution of apoptosis, respectively (Chen et al., 2017). The inflammatory caspases mediate the cleavage of pro-inflammatory cytokine precursors (for example, pro-IL-1β and pro-IL-18) triggering inflammatory responses (inflammation) and also involve in the inflammatory cell death known as pyroptosis (Chen et al., 2017).
Apoptosis is a form of programmed cell death mainly initiated by either the death receptor-mediated extrinsic pathway or the intracellular stress-induced, mitochondrion-mediated intrinsic pathway (Fan et al., 2005). Both extrinsic pathway-activated caspase-8 and intrinsic pathway-activated caspase-9 cleave and activate the downstream effector caspases-3, -6 and -7, which can further cleave a large number of cellular proteins, including poly(ADP-ribose)polymerase-1 (PARP1), prelamin-A/C (LMNA) and inhibitor of caspase-activated DNase (ICAD), and finally lead to cell death (Orzalli and Kagan, 2017; Zhou et al., 2017). Also, caspase-8 mediates the cleavage of BH3 interacting-domain death agonist (BID) to generate the truncated BID (tBID), which translocates to mitochondria and thus transduces apoptotic signals from cytoplasmic membrane to mitochondria triggering mitochondrial outer membrane permeabilization (MOMP) and the release of cytochrome c, and thus initiating effector caspases activation and cell death (Li et al., 1998; Zhou et al., 2017). Pyroptosis is a lytic programmed cell death pathway executed by gasdermin proteins and associated with strong inflammatory responses (Vande Walle and Lamkanfi, 2016). The activated caspases-1, -4, -5 and -11 cleave pro-IL-1β and pro-IL-18 into mature inflammatory cytokines IL-1β and IL-18 (Chen et al., 2017). Also, these activated inflammatory caspases process members of gasdermin (GSDM) family, such as GSDMD and GSDME, leading to pore formation on the plasma membrane, followed by rupture of the cell membrane and pyroptotic cell death (Tsuchiya, 2020). Many of the viruses activate caspases to cleave a number of substrates, and cause infected cells to undergo the above mentioned programmed cell death pathways (Koyama et al., 1998). Activated caspases can cleave substrate in a tetrapeptide sequence-independent cleavage way, such as active caspase-1/4/11-mediated GSDMD cleavage (Wang et al., 2020). But generally, caspases recognize a tetrapeptide motif (XXXD) in the substrate and cleave after the aspartate (D) (Crawford and Wells, 2011; Poreba et al., 2013; Wang et al., 2020). Identification of novel caspase cleavage motifs is important for the investigation of the caspase dependent cell death pathways and other caspase regulated pathways.
To identify new caspase cleavage motifs, we developed an assay for screening novel caspase cleavage motifs. With this assay we successfully identified a novel caspase cleavage motif AEAD, and confirmed its caspase sensitivity. We showed that AEAD peptide-derived Z-AEAD-FMK can inhibit the activation of multiple caspases and the cleavage of downstream substrates. The broad-spectrum inhibitory effect of Z-AEAD-FMK on caspase can protect cells against virus-induced apoptotic and pyroptotic cell death and inflammation. The novel pan-caspase inhibitor Z-AEAD-FMK can be employed to investigate the caspase-mediated cell death pathways and facilitate the development of anti-inflammatory drugs.
2. Materials and methods
2.1. Plasmids
cDNAs encoding human caspase-3 (CCDS3836.1), caspase-6 (CCDS3684.1) and caspase-7 (CCDS7581.1) were synthesized by Tsingke (Wuhan, China) and cloned into pRK plasmid with a C-terminal His tag, respectively. Plasmids, including C-terminal strep-tagged Tom20-AEADG-EGFP (Tom20, CCDS1603.1), EGFP-AEADG-Tom20 and human NOA1 (CCDS3510.1) were constructed by standard molecular biology techniques. Point mutants were generated by using the ClonExpress II One Step Cloning Kit (#C112-01, Vazyme, China) and the construct coding the wild-type proteins as template. Each construct was confirmed by sequencing.
2.2. Antibodies and reagents
The anti-strep (#A00626) antibody was purchased from GenScript (Nanjing, China). The anti-Tom20 (#sc-17764) antibody used in immunoblot analysis was purchased from Santa Cruz Biotechnology (Delaware, USA). The anti-Tom20 (#ab209606) used in immunofluorescence analysis and GSDME (#ab215191) were purchased from Abcam (Cambridge, UK). The anti-caspase-1 (#2225), anti-cleaved caspase-1 (#4199), anti-caspase-3 (#9662), anti-cleaved caspase-3 (#9661), anti-caspase-6 (#9762), anti-cleaved caspase-6 (#9761), anti-caspase-7 (#9492), anti-caspase-8 (#9746), anti-cleaved caspase-8 (#9496), anti-caspase-9 (#9502), anti-GSDMD (#96458), anti-cleaved GSDMD (#36425), anti-cleaved IL1β (#83186), anti-IL6 (#12153), anti-BID (#2002) and anti-PARP1 (#9542) antibodies were obtained from Cell Signaling Technology (Beverly, MA, USA). The anti-LMNA (#10298-1-AP), anti-IL8 (#27095-1-AP), anti-IL18 (#27095-1-AP), anti-EGFP tag (#50430-2-AP), anti-His tag (#66005-1-Ig), anti-alpha Tubulin (#14555-1-AP), goat anti-mouse (#SA00001-1) and goat anti-rabbit (#SA00001-2) IgG-horseradish peroxidase (HRP) secondary antibodies were purchased from Proteintech (Wuhan, China).
PMA (#P1585) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Z-VAD-FMK (#S7023) was obtained from Selleck (Houston, TX, USA). Z-AEAD-FMK was synthesized by GL Biochem (Shanghai, China). Staurosporine (#HY-15141) was purchased from MedChemExpress (New Jersey, USA). DAPI (#C1002) and propidium iodide (#ST511) were purchased from Beyotime (Shanghai, China).
2.3. Cell lines
The HEK293T and THP-1 cell lines were purchased from American Type Culture Collection (ATCC). HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplied with 10% fetal bovine serum (FBS; Gibco) and 1% Pen Strep antibiotics (Gibco). THP-1 cells were cultured in RPMI 1640 (Gibco) containing 10% FBS (Gibco) and 1% Pen Strep antibiotics (Gibco). All cells were cultured at 37 °C in a humidified 5% CO2 incubator.
2.4. Viruses
SeV was amplified in 9-day-old specific-pathogen-free (SPF) chicken eggs for 72 h and titrated by plaque assay. HSV-1 (F strain) was amplified in Vero cells for 48 h and titrated by plaque assay.
2.5. Cell differentiation
THP-1 cells were treated with 40 ng/mL PMA at 37 °C for 24 h to differentiate into macrophages, and then cells were cultured for 24 h without PMA.
2.6. Molecular docking
We used the program AutoDock Vina to perform the molecular docking (Trott and Olson, 2010). Crystal structure of human caspases, including caspase-1 (PDB ID code: 1rwk), caspase-3 (PDB ID code: 5i9b), caspase-4 (PDB ID code: 6nry), caspase-6 (PDB ID code: 3od5), caspase-7 (PDB ID code: 1shj), caspase-8 (PDB ID code: 1qtn) and caspase-9 (PDB ID code: 2ar9) were from Protein Data Bank. Crystal structure of human caspase-10 (AlphaFold ID code: Q92851) and caspase-14 (AlphaFold ID code: P31944) were from AlphaFold Protein Structure Database. The Z-VAD-FMK molecule was from PubChem CID: 5497174.
The Z-AEAD-FMK molecule was drawn by ChemDraw (2D structure) or Chem3D (3D structure).
All the ligands were docked 10 times. And the docking results were visualized using open-source PyMOL.
2.7. Immunofluorescence assay
THP-1PMA cells were infected or mock infected with SeV or HSV-1 (MOI = 10), or treated with staurosporine (STS), and then fixed with 4% paraformaldehyde (PFA) at 25 °C for 30 min, permeabilized with 0.3% (vol/vol) Triton X-100 for 20 min, blocked in 3% bovine serum albumin in PBS for 30 min, and incubated sequentially with Alexa Fluor® 647 anti-TOMM20 antibody (#ab209606, Abcam, Cambridge, UK) for 1 h. Cells were stained with DAPI, mounted with ProLong™ Gold antifade reagent (Invitrogen, San Diego, CA, USA), and then analyzed using confocal microscope (Leica). Images were processed by Leica.
2.8. In vitro caspase cleavage assay
HEK293T cells were transfected with the indicated plasmids expressing strep-tagged substrates for 48 h or expressing His-tagged caspases for 36 h, followed by STS treatment at 10 μmol/L for 12 h. The cells were washed twice with PBS, lysed with NP-40 buffer (#P0013F, Beyotime, Shanghai, China) and rotated for 30 min at 4 °C. After centrifugation at 16,000×g for 20 min, the whole cell lysates were incubated with streptavidin magnetic beads (#2-4090-010, Iba-Lifesciences, Germany) or Ni Sepharose beads (#17531801, Cytiva, Marlborough, MA, USA) and rotated overnight at 4 °C on a rotator. After washing three times with PBST, the streptavidin magnetic beads-pull down purified substrate was eluted by 1× BXT buffer (#2-1042-025, Iba-Lifesciences, Germany), and Ni Sepharose beads-pull down purified caspase was eluted by 500 mmol/L imidazole.
In vitro protease cleavage experiments were performed as previously described (Wang et al., 2017). Briefly, Ni Sepharose beads-pulldown purified caspase was incubated with streptavidin magnetic beads-pulldown purified substrate in cleavage buffer (100 mmol/L HEPES, 10% (w/v) sucrose, 0.1% (w/v) CHAPS, PH 7.0, 10 mmol/L DTT) at 37 °C for 5 h and then samples were applied to immunoblot analysis.
2.9. Protein detection in supernatants
Culture supernatants of THP-1PMA cells were precipitated by adding an equal volume of methanol and 0.25 volumes of chloroform. The mixtures were vortexed and then centrifuged for 10 min at 16,000×g. The upper phase was discarded and 500 μL methanol was added to the interphase. This mixture was vortexed and then centrifuged for 10 min at 16,000×g. The protein pellet was dried at 55 °C, followed by immunoblotting analysis to detect indicated protein.
2.10. Propidium iodide (PI) fluorometric assay
THP-1PMA cells were seeded in 48-well plates at a density of 200,000 cells per well in a total volume of 200 μL per well and infected or mock infected with SeV or HSV-1 (MOI = 10) for 1 h, and then media were replaced with Opti-MEM (#31985070, Gibco) with or without the treatment of Z-AEAD-FMK at 20 μmol/L. At 12 h.p.i of SeV infection or 24 h.p.i of HSV-1 infection, cells were stained with propidium iodide. The plates were imaged using Operetta (PerkinElmer) with a 10× objective in both the PI and bright filed channels. The images were analyzed using automated image analysis software (Harmony 3.5, PerkinElmer).
2.11. LDH release assay
THP-1PMA cells were treated as indicated in figure legend, and cell death was measured by a lactate dehydrogenase (LDH) assay using LDH Cytotoxicity Assay Kit (#C0017, Beyotime, Shanghai, China).
2.12. Immunoblot analysis
Cells were lysed with buffer (#P0013, Beyotime, Shanghai, China). Cell lysates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Proteins were incubated with primary antibodies overnight at 4 °C, then secondary horseradish peroxidase-conjugated goat anti-rabbit/mouse IgG. Protein bands were detected by an enhanced chemiluminescence (ECL) kit (Millipore, Billerica, MA, USA) using a Chemiluminescence Analyzer (Chemiscope600pro, Shanghai, China).
2.13. Statistical analysis
Statistical significance was determined with Student’s t test (Fig. 3) or One-way ANOVA test (Fig. 5) with P < 0.05 considered statistically significant. Data were analyzed with GraphPad Prism 9. ns, not significant; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
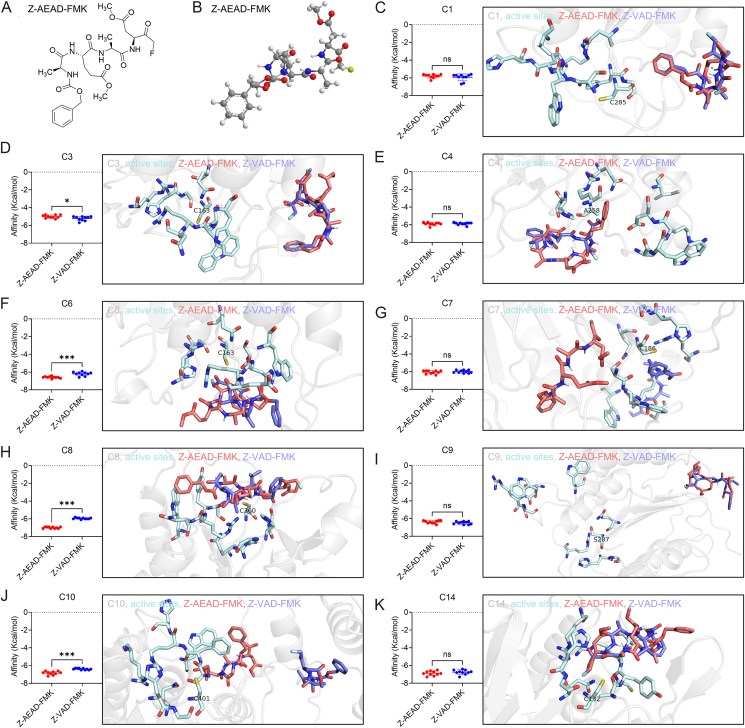
Fig. 3.
Molecular docking analysis of Z-AEAD-FMK with multiple caspases. A The 2D structure of the AEAD-derived inhibitor Z-AEAD-FMK. B The 3D structure of the AEAD-derived inhibitor Z-AEAD-FMK. C–K Molecular docking between Z-AEAD-FMK or Z-VAD-FMK and the indicated caspase. Left panel is the affinity comparison between Z-AEAD-FMK and Z-VAD-FMK with indicated caspase in 10 dockings. Right panel is the comparison of the highest-affinity conformations of Z-AEAD-FMK and Z-VAD-FMK with the indicated caspase. The Z-AEAD-FMK inhibitor is shown in deep salmon sticks, the Z-VAD-FMK inhibitor is shown in slate sticks, the caspase structure is shown in gray cartoon, the active sites of caspase is shown in pale cyan sticks. The catalytic sites (Cys residue) of caspase-4 and caspase-9 were mutated into Ala (A258) and Ser (S287), respectively. ∗P < 0.05; ∗∗∗ P < 0.001; ns, not significant (Student’s t test).
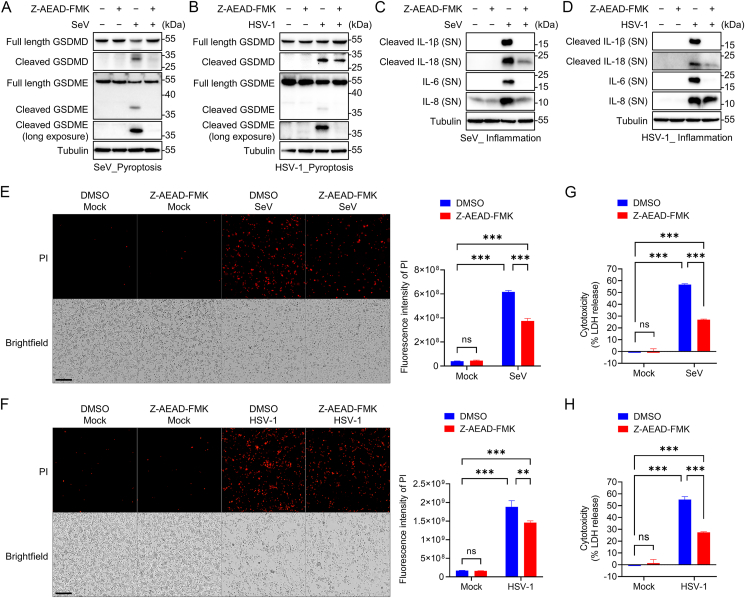
Fig. 5.
Z-AEAD-FMK inhibits caspase-mediated cell death pathway triggered by virus infection. A Immunoblot analysis of the indicated proteins in THP-1PMA cells. Cells were infected or mock infected with SeV at MOI = 10 for 1 h and then the media were replaced with fresh media and treated with Z-AEAD-FMK at 20 μmol/L for 11 h. B Immunoblot analysis of the indicated proteins in THP-1PMA cells. Cells were infected or mock infected with HSV-1 at MOI = 10 for 1 h, and then the media were replaced with fresh media and treated with Z-AEAD-FMK at 20 μmol/L for 23 h. C Immunoblot analysis of the indicated proteins in THP-1PMA cells. Cells were infected or mock infected with SeV at MOI = 10 for 1 h and then the media were replaced with fresh media and treated with Z-AEAD-FMK at 20 μmol/L for 11 h. The culture supernatants (SN) were used to extract protein, and the indicated protein in supernatants or Tubulin in lysates were determined. D Immunoblot analysis of the indicated proteins in THP-1PMA cells. Cells were infected or mock infected with HSV-1 at MOI = 10 for 1 h and then the media were replaced with fresh media and treated with Z-AEAD-FMK at 20 μmol/L for 23 h. The culture supernatants (SN) were used to extract protein, and the indicated protein in supernatants or Tubulin in lysates were determined. E Fluorescence and quantitative analysis of the propidium iodide (PI)-positive THP-1PMA cells. Cells were infected or mock infected with SeV at MOI = 10 for 1 h and then treated with Z-AEAD-FMK at 20 μmol/L for 11 h, followed by PI staining. Scale bars, 250 μm. F Fluorescence and quantitative analysis of the PI-positive THP-1PMA cells. Cells were infected or mock infected with HSV-1 at MOI = 10 for 1 h and then treated with Z-AEAD-FMK at 20 μmol/L for 23 h, followed by PI staining. Scale bars, 250 μm. G LDH release assays for the cytotoxicity of THP-1PMA cells. Cells were infected or mock infected with SeV at MOI = 10 for 1 h and then treated with Z-AEAD-FMK at 20 μmol/L for 11 h. Extracellular LDH were determined. H LDH release assays for the cytotoxicity of THP-1PMA cells. Cells were infected or mock infected with HSV-1 at MOI = 10 for 1 h and then treated with Z-AEAD-FMK at 20 μmol/L for 23 h. Extracellular LDH were determined. ∗∗P < 0.01; ∗∗∗P < 0.001; ns, not significant (One-way ANOVA test). Data are representative of three experiments with similar results (means with SD in E–H).
3. Results
3.1. Identification of the novel caspase cleavage motif
To identify novel caspase cleavage site, we performed motif screening based on the characteristic of caspases cleaving substrates at specific motifs. Generally, caspases substrates have the peptide sequence P4-P3-P2-P1↓P1', where the P4-P3-P2-P1 is the core tetrapeptide motif and the cleavage site is located between P1 and P1', represented by “↓” (Poreba et al., 2013). Although the P1 residue was believed to be exclusively Asp (D), recent evidence showed that caspases can also cleave after Glu (E) and phosphoserine (pS) residues (Seaman et al., 2016). To narrow down the scope of P1 and other positions, we performed the sites analysis through the MEROPS database, a peptidase database (http://www.merops.ac.uk) (Rawlings et al., 2002, 2010). The analysis and screening process is shown in Fig. 1A.
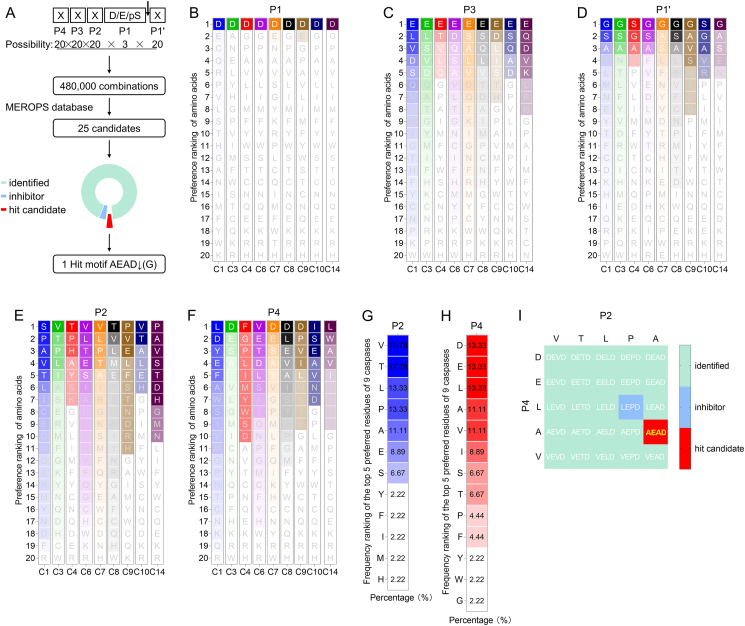
Fig. 1.
AEAD motif is a new potential caspase cleavage motif. A Analysis and screening process of caspase cleavage motifs. B Preference analysis at the P1 position of the cleavage motifs recognized by caspase-1, -3, -4, -6, -7, -8, -9, -10 and -14, respectively. C Preference analysis at the P3 position of the cleavage motifs recognized by caspase-1, -3, -4, -6, -7, -8, -9, -10 and -14, respectively. D Preference analysis at the P1' position of the cleavage motifs recognized by caspase-1, -3, -4, -6, -7, -8, -9, -10 and -14, respectively. E Preference analysis at the P2 position of the cleavage motifs recognized by caspase-1, -3, -4, -6, -7, -8, -9, -10 and -14, respectively. F Preference analysis at the P4 position of the cleavage motifs recognized by caspase-1, -3, -4, -6, -7, -8, -9, -10 and -14, respectively. G Frequency analysis of the top 5 preferred residues at the P2 position for 9 caspases (caspase-1, -3, -4, -6, -7, -8, -9, -10 and -14). H Frequency analysis of the top 5 preferred residues at the P4 position for 9 caspases (caspase-1, -3, -4, -6, -7, -8, -9, -10 and -14). I 25 candidate motifs for caspase cleavage. Single capital letters in the figures are amino acid abbreviations.
We first arranged the preference order of amino acid residues at each site in the cleavage motif of nine human caspases, including caspase-1, -3, -4, -6, -7, -8, -9, -10 and -14. Taking the analysis of the P1 site of caspase-1 as an example, based on the 189 cleavage sites mediated by caspase-1, Asp appeared 160 times, and the ratio was 160/189 × 100% ≈ 84.66%. The percentages of other amino acids were also calculated in the same way as Asp. Then ordering from large to small, we can obtain the preference list of P1 site of caspase-1. It was found that all caspases consistently showed exquisite selectivity for D in the P1 position (Fig. 1B) and a preference for E in P3 position (Fig. 1C). Compared with caspase-4 and caspase-10, the other caspases-1, -3, -6, -7, -8, -9 and -14 showed a preference for Gly (G) rather than E at the P1' position (Fig. 1D). More caspases seem to prefer Val (V) at P2 (Fig. 1E) and D at P4 (Fig. 1F), respectively, and the generated motif combination DEVD has been shown to be a classical motif recognized by caspase-3 and has a derived inhibitor Z-DEVD-FMK.
Therefore, we selected the top 5 amino acid residues preferred by each caspase at P2 position or P4 position for combination, and analyzed the frequency of occurrence of each amino acid. Taking the analysis of the P2 site of caspases as an example (Fig. 1E), top 5 preferred residues for caspase-1 (C1) were Ser, Pro, Ala, Val and Thr; top 5 preferred residues for C3 were Val, Thr, Pro, Leu and Ile; top 5 preferred residues for C4 were Thr, Pro, His, Ala and Tyr; top 5 preferred residues for C6 were Val, Leu, Thr, Glu and Ser; top 5 preferred residues for C7 were Val, Leu, Pro, Thr and Ala; top 5 preferred residues for C8 were Thr, Val, Leu, Met and Glu; top 5 preferred residues for C9 were Pro, Val, Glu, Leu and Phe; top 5 preferred residues for C10 were Val, Thr, Ala, Leu and Glu; top 5 preferred residues for C14 were Pro, Ala, Val, Ser and Thr. By calculating the frequency of each amino acid, we obtained a ranking table, as shown in Fig. 1G. The analysis of the P4 position was also performed in the same way as P2 position (Fig. 1H). Altogether, these analyses showed that the top 5 amino acid residues preferred by theses caspases at the P2 position were Val (V), Thr (T), Leu (L), Pro (P) and Ala (A) (Fig. 1G), and the top 5 residues preferred by caspases at the P4 position were Asp (D), Glu (E), Leu (L), Ala (A) and Val (V) (Fig. 1H).
We further performed a combined matrix analysis of the preferred amino acid residues selected at each position, and thus obtained 25 candidate motifs (Fig. 1I). Among them, 23 motifs have been verified by previous studies, as shown in Table 1, and one motif LEPD has been used to develop caspase inhibitors Ac-LEPD-CHO and Ac-LEPD-FMK (Kelotra et al., 2014), which supports the relevance of our analysis. Based on this analysis the AEAD motif appears as a novel caspase cleavage motif (Fig. 1I).
Table 1.
List of identified caspase cleavage motifs and their derived inhibitors.
| Number | Caspase cleavage motifs | Caspase inhibitor | Substrate (UniProt entry) | Position of motifs | References |
|---|---|---|---|---|---|
| 1 | DEVD | Z-DEVD-FMK (caspase-3 inhibitor) | PARP1_HUMAN | DEVD (214) | Lazebnik et al. (1994) |
| CXA7_HUMAN | DEVD (367) | Yin et al. (2001) | |||
| PRKDC_HUMAN | DEVD (2713) | Casciola-Rosen et al. (1995) | |||
| IF4H_HUMAN | DEVD (92) | Van Damme et al. (2005) | |||
| ITPR1_HUMAN | DEVD (1838) | Hirota et al. (1999) | |||
| KPCT_HUMAN | DEVD (354) | Datta et al. (1997) | |||
| RFC1_HUMAN | DEVD (722) | Rheaume et al. (1997) | |||
| SGG_DROME | DEVD (300) | Kanuka et al. (2005) | |||
| SPTB1_HUMAN | DEVD (1457) | Wang et al. (1998) | |||
| SPTB2_HUMAN | DEVD (1457) | Vanags et al. (1996) | |||
| 2 | EEVD | Non | DHX9_HUMAN | EEVD (167) | Takeda et al. (1999) |
| 3 | LEVD | Z-LEVD-FMK (caspase-4 inhibitor) | CFLAR_HUMAN | LEVD (376) | Irmler et al. (1997) |
| TRAF1_HUMAN | LEVD (163) | Irmler et al. (2000) | |||
| 4 | AEVD | Z-AEVD-FMK (caspase-10 Inhibitor) | IF2A_HUMAN | AEVD (301) | Satoh et al. (1999) |
| 5 | VEVD | Z-VEVD-FMK | APLP1_HUMAN | VEVD (620) | Galvan et al. (2002) |
| A4_HUMAN | VEVD (739) | Galvan et al. (2002), Gervais et al. (1999) | |||
| K1C18_HUMAN | VEVD (237) | Ku et al. (1997) | |||
| 6 | DETD | Non | IF4B_HUMAN | DETD (45) | Bushell et al. (2000) |
| IF4G2_HUMAN | DETD (790) | Henis-Korenblit et al. (2000) | |||
| DFFA_MOUSE | DETD (117) | Liu et al. (1997), Enari et al. (1998), Yakovlev et al. (2001) | |||
| ROCK1_HUMAN | DETD (1113) | Coleman et al. (2001), Sebbagh et al. (2001), Chang et al. (2006) | |||
| SPTA2_HUMAN | DETD (1185) | Martin et al. (1996) | |||
| TADBP_HUMAN | DETD (89) | Van Damme et al. (2005) | |||
| TIAM1_HUMAN | DETD (993) | Qi et al. (2001) | |||
| 7 | EETD | Z-EETD-FMK; Ac-EETD- pNA (caspase-8 inhibitor) |
KPCZ_HUMAN | EETD (210) | Frutos et al. (1999) |
| 8 | LETD | Z-LETD-FMK Ac-LETD-CHO |
RPB1_HUMAN | LETD (1339) | Lu et al. (2002) |
| 9 | AETD | Non | STK24_HUMAN | AETD (313) | Huang et al. (2002) |
| 10 | VETD | Non | CASP8_HUMAN | VETD (374) | Boldin et al. (1996) |
| 11 | DELD | Non | ACINU_HUMAN | DELD (1093) | Sahara et al. (1999), Joselin et al. (2006) |
| PA24A_HUMAN | DELD (522) | Adam-Klages et al. (1998) | |||
| ITB4_HUMAN | DELD (1109) | Werner et al. (2007) | |||
| STK3_HUMAN | DELD (322) | Lee et al. (2001) | |||
| NFAC2_HUMAN | DELD (29) | Wu et al. (2006) | |||
| GDIS_HUMAN | DELD (19) | Na et al. (1996), Danley et al. (1996) | |||
| TWST1_MOUSE | DELD (173) | Demontis et al. (2006) | |||
| 12 | EELD | Non | MCL1_HUMAN | EELD (127) | Weng et al. (2005), Herrant et al. (2004) |
| NF2L2_HUMAN | EELD (366) | Ohtsubo et al. (1999) | |||
| 13 | LELD | Non | GSDMC_HUMAN | LELD (365) | Hou et al. (2020) |
| 14 | AELD | Non | GGA3_HUMAN | AELD (333) | Tesco et al. (2007) |
| 15 | VELD | Non | GFAP_HUMAN | VELD (225) | Chen et al. (2013) |
| 16 | DEPD | Non | SRBP2_HUMAN | DEPD (486) | Wang et al. (1995) |
| 17 | EEPD | Non | PAWR_HUMAN | EEPD (131) | Chaudhry et al. (2012) |
| 18 | AEPD | Non | PLCG1_HUMAN | AEPD (770) | Bae et al. (2000) |
| 19 | VEPD | Non | GANP_HUMAN | VEPD (1023) | Wickramasinghe et al. (2011) |
| 20 | DEAD | Non | RB_HUMAN | DEAD (886) | Janicke et al. (1996) |
| 21 | EEAD | Non | RAD9A_HUMAN | EEAD (187) | Lee et al. (2003) |
| TRAF3_HUMAN | EEAD (348) | Lee et al. (2001) | |||
| 22 | LEAD | Non | CASP5_HUMAN | LEAD (330) | Munday et al. (1995) |
| 23 | VEAD | Z-VEAD-FMK | STAP2_HUMAN | VEAD (260) | Sekine et al. (2012) |
| GRAB_HUMAN | VEAD (192) | Casciola-Rosen et al. (2007) | |||
| 24 | LEPD | Ac-LEPD-FMK; Ac-LEPD-CHO |
Kelotra et al. (2014) |
3.2. The AEAD motif is a novel caspase cleavage motif
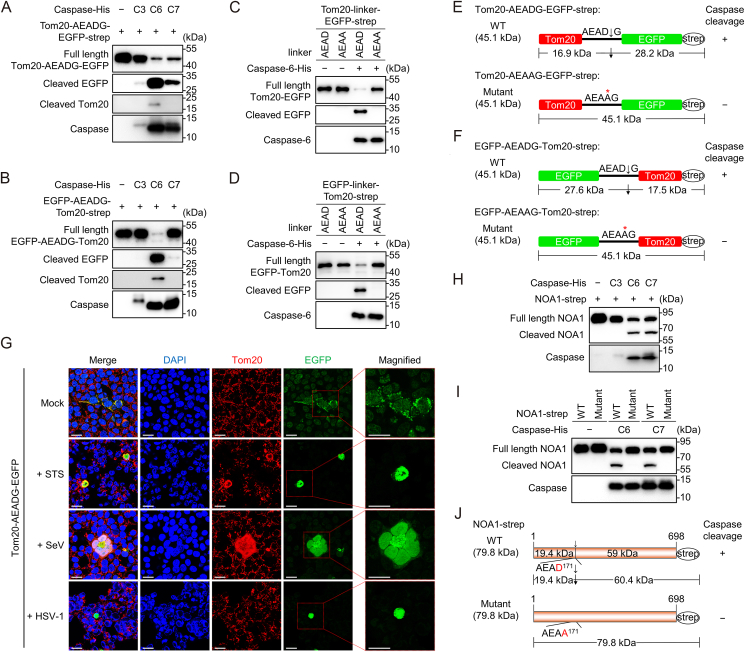
To analyze whether activated caspases can cleave substrates at the AEAD motif, we fused EGFP to the mitochondrial marker protein Tom20 using the AEADG peptide as a linker to express a fusion protein Tom20-AEADG-EGFP or EGFP-AEADG-Tom20, which may allow the fusion protein to localize to mitochondria. We first performed an in vitro caspase cleavage assay to verify whether the Tom20-AEADG-EGFP or EGFP-AEADG-Tom20 can be cleaved by activated caspases. Purified C-terminal strep-tagged Tom20-AEADG-EGFP or EGFP-AEADG-Tom20 was incubated with active C-terminal His-tagged human caspase-3, -6, or -7 protein (C3, C6 or C7). Immunoblotting analysis showed that caspases-3, -6 and -7 cleaved Tom20-AEADG-EGFP to produce a ∼30 kDa cleaved band, which was recognized by the anti-EGFP antibody and corresponds to the EGFP-strep (Fig. 2A and E). And caspase-6 can cleave Tom20-AEADG-EGFP to produce a ∼17 kDa fragment, which was recognized by the anti-Tom20 antibody and corresponds to the Tom20 (Fig. 2A and E). But we did not detect the ∼17 kDa cleaved band for caspase-3 and caspase-7 mediated Tom20-AEADG-EGFP cleavage, and one of the possible reasons is that these caspases may also cleave Tom20 at other sites, making it difficult to be recognized by anti-Tom20 antibody (Fig. 2A). At the same time, we found that caspase-6 and caspase-7 cleaved EGFP-AEADG-Tom20 to generate a ∼30 kDa cleaved band, which was recognized by the anti-EGFP antibody and corresponds to the EGFP (Fig. 2B and F). And caspase-6 can cleave EGFP-AEADG-Tom20 to produce a ∼17 kDa fragment, which was recognized by the anti-Tom20 antibody and corresponds to the Tom20-strep (Fig. 2B and F). However, unlike caspase-3 mediated Tom20-AEADG-EGFP cleavage, we did not detect the cleavage of EGFP-AEADG-Tom20 by caspase-3. One of the possible reasons is that the conformation of EGFP-AEADG-Tom20 hinders the recognition and cleavage of caspase-3 at the AEAD motif.
Fig. 2.
Caspases can cleave substrates at the AEAD motif. A, B Immunoblot analysis of Tom20-AEADG-EGFP (A) or EGFP-AEADG-Tom20 (B) cleavage by the in vitro cleavage assay. The C-terminal strep-tagged Tom20-AEADG-EGFP protein or EGFP-AEADG-Tom20 was expressed and purified from HEK293T cells, and subjected to the in vitro cleavage assay with his-tagged caspase-3, 6 or 7 protein, followed by immunoblot analysis. Anti-EGFP and anti-Tom20 antibodies were used to determine the cleavage of Tom20-AEADG-EGFP or EGFP-AEADG-Tom20. Anti-His antibody was used to identify caspases. C, D Immunoblot analysis of the cleavage of Tom20-AEADG-EGFP and its mutant Tom20-AEAAG-EGFP (C) or the cleavage of EGFP-AEADG-Tom20 and its mutant EGFP-AEAAG-Tom20 (D) by the in vitro cleavage assay. The indicated C-terminal strep-tagged substrate was expressed and purified from HEK293T cells, and subjected to the in vitro cleavage assay with his-tagged caspase-6 protein, followed by immunoblot analysis. Anti-EGFP and anti-Tom20 antibodies were used to determine the cleavage of Tom20-AEADG-EGFP, Tom20-AEAAG-EGFP, EGFP-AEADG-Tom20 or EGFP-AEAAG-Tom20. Anti-His antibody was used to identify active caspase-6. E, F Schematic of the cleavage of Tom20-AEADG-EGFP and Tom20-AEAAG-EGFP (E) or EGFP-AEADG-Tom20 and EGFP-AEAAG-Tom20 (F). G Immunofluorescence analysis of Tom20-AEADG-EGFP cleavage. HEK293T cells were transfected with vector expressing Tom20-AEADG-EGFP for 24 h and then infected or mock infected with SeV or HSV-1 at MOI = 10 for 24 h, or treated with STS at 10 μmol/L for 12 h. Mitochondria were labeled with anti-Tom20 antibody (red), green represents EGFP signals and Nuclei were stained with DAPI (blue). Scale bars, 20 μm. H Immunoblot analysis of NOA1 cleavage by the in vitro cleavage assay as described in (A, B). Anti-strep antibody was used to determine the cleavage of NOA1. Anti-His antibody was used to identify caspases. I Immunoblot analysis of the cleavage of NOA1 and its mutant NOA1D171A by the in vitro cleavage assay as described in (C, D). Anti-strep antibody was used to determine the cleavage of NOA1 and its mutant NOA1D171A. Anti-His antibody was used to identify caspases. J Schematic of the cleavage of wild type and mutant NOA1. Data are representative of three experiments with similar results.
To further verify whether Tom20-AEADG-EGFP or EGFP-AEADG-Tom20 is cleaved by caspase at AEAD motif, we substituted the D of AEAD motif with A to construct the mutant, Tom20-AEAAG-EGFP or EGFP-AEAAG-Tom20. The in vitro caspase cleavage assay showed that the Tom20-AEAAG-EGFP or EGFP-AEAAG-Tom20 mutant was resistant to caspase-6 cleavage (Fig. 2C–F), confirming that caspase can specifically recognize and cleave Tom20-AEADG-EGFP or EGFP-AEADG-Tom20 at the AEAD motif. Next, we found that Tom20-AEADG-EGFP indeed localized to mitochondria in the absence of caspase activation (Fig. 2G). When intracellular caspases were activated upon SeV/HSV-1 infection, or with STS treatment, the EGFP signals were diffusely distributed in the cell (Fig. 2G), suggesting that virus infection induced caspase-mediated Tom20-AEADG-EGFP cleavage at the AEAD motif. Thus, these results confirmed that the AEAD motif is a caspase cleavage motif.
To explore the possibility of caspase cleavage of natural substrate proteins containing AEAD motif, we used ‘AEADG’ as a query sequence, to perform protein blast on the NCBI website and obtained multiple candidates. Among that, 20 candidate proteins are shown in Table 2. We cloned and constructed a plasmid expressing the C-terminal strep-tagged NOA1. The in vitro caspase cleavage assay showed that caspase-6 and caspase-7 can cleave NOA1 (Fig. 2H and J). Additionally, incubation of caspase-3 with NOA1 decreased the abundance of full-length NOA1 (Fig. 2H), suggesting that caspase-3 cleaved NOA1. We failed to detect the cleaved NOA1 by the anti-strep tag antibody. This is potentially due to less efficient cleavage of NOA1 by caspase-3 resulting in less cleaved NOA1 band which is below the detection limit. We then substituted the D of AEAD171 motif with A to construct mutants, NOA1D171A, and the in vitro caspase cleavage assay showed that mutant NOA1D171A was resistant to caspase-6 and caspase-7 mediated cleavage (Fig. 2I and J), confirming that caspase can recognize and cleave NOA1 at the AEAD motif. Thus, active caspase can cleave natural substrates at the AEAD motif, showing that AEAD is indeed a caspase cleavage motif.
Table 2.
Potential natural substrates in human containing AEAD↓(G) motif for caspase cleavage.
| Number | Uniprot entry name | Length (aa) | Mass (kDa) | Amino acid position of caspase cleavage motif |
Fragments |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| P4 (A) | P3 (E) | P2 (A) | P1 (D) | P1' (G) | N (kDa) | C (kDa) | ||||
| 1 | MYH7B_HUMAN | 1983 | 225.85 | 418 | 419 | 420 | 421 | 422 | 47.95 | 177.90 |
| 2 | SNPC4_HUMAN | 1469 | 159.43 | 1165 | 1166 | 1167 | 1168 | 1169 | 126.76 | 32.67 |
| 3 | SOGA1_HUMAN | 1423 | 159.76 | 1053 | 1054 | 1055 | 1056 | 1057 | 118.56 | 41.20 |
| 4 | PHC1_HUMAN | 1004 | 105.53 | 339 | 340 | 341 | 342 | 343 | 35.95 | 69.59 |
| 5 | ARMC3_HUMAN | 872 | 96.41 | 395 | 396 | 397 | 398 | 399 | 44.00 | 52.40 |
| 6 | CFA58_HUMAN | 872 | 103.42 | 590 | 591 | 592 | 593 | 594 | 70.33 | 33.09 |
| 7 | MILK1_HUMAN | 863 | 93.44 | 265 | 266 | 267 | 268 | 269 | 29.02 | 64.42 |
| 8 | IL31R_HUMAN | 732 | 82.95 | 239 | 240 | 241 | 242 | 243 | 27.42 | 55.53 |
| 9 | PLPR3_HUMAN | 718 | 76.04 | 613 | 614 | 615 | 616 | 617 | 65.24 | 10.80 |
| 10 | NOA1_HUMAN | 698 | 78.46 | 168 | 169 | 170 | 171 | 172 | 19.22 | 59.24 |
| 11 | ZN653_HUMAN | 615 | 67.24 | 423 | 424 | 425 | 426 | 427 | 46.57 | 20.66 |
| 12 | CENPT_HUMAN | 561 | 60.42 | 377 | 378 | 379 | 380 | 381 | 40.93 | 19.49 |
| 13 | MOT3_HUMAN | 504 | 52.32 | 220 | 221 | 222 | 223 | 224 | 23.15 | 29.17 |
| 14 | PDCD7_HUMAN | 485 | 54.70 | 198 | 199 | 200 | 201 | 202 | 22.67 | 32.03 |
| 15 | IGA2_HUMAN | 455 | 48.93 | 448 | 449 | 450 | 451 | 452 | 48.50 | 0.43 |
| 16 | PLIN3_HUMAN | 434 | 47.08 | 6 | 7 | 8 | 9 | 10 | 0.98 | 46.10 |
| 17 | NAGK_HUMAN | 344 | 37.38 | 28 | 29 | 30 | 31 | 32 | 3.37 | 34.01 |
| 18 | IGHA2_HUMAN | 340 | 36.59 | 333 | 334 | 335 | 336 | 337 | 36.16 | 0.43 |
| 19 | CGRE1_HUMAN | 318 | 33.46 | 212 | 213 | 214 | 215 | 216 | 22.62 | 10.84 |
| 20 | MPU1_HUMAN | 247 | 26.64 | 3 | 4 | 5 | 6 | 7 | 0.65 | 25.99 |
3.3. Molecular docking analysis of Z-AEAD-FMK with multiple caspases
Next, we designed a peptide derived from the AEAD motif to analyze its potential as a caspase inhibitor. The peptide was developed by linking the AEAD peptide to the benzyloxycarbonyl (Z) group and fluoromethylketone (FMK) group, named as Z-AEAD-FMK (Fig. 3A and B). To analyze the engagement of the inhibitor with caspases, a crystal structure of the catalytic domain of human caspases, including caspases-1, -3, -4, -6, -7, -8, -9, -10 and -14, were docked with Z-AEAD-FMK respectively. The Z-VAD-FMK, a well-known pan-caspase inhibitor, was included as a positive control.
A higher negative Autodock Vina score represents a strong binding affinity (Trott and Olson, 2010). We found that the affinity between Z-AEAD-FMK and caspase-1, -4, -9 or -14 was similar to the affinity between Z-VAD-FMK and caspase-1, -4, -9 or -14, and for each caspase, the highest-affinity conformation of Z-AEAD-FMK almost overlapped with the highest-affinity conformation of Z-VAD-FMK (Fig. 3C, E, I and K). The caspase-3-binding affinity of Z-AEAD-FMK is slightly lower than the caspase-3-binding affinity of Z-VAD-FMK, but the highest caspase-3-binding affinity conformation of Z-AEAD-FMK and Z-VAD-FMK almost overlapped and both conformations relatively far from the active pocket of caspase-3 (Fig. 3D). The caspase-7-binding affinity of Z-AEAD-FMK is not significantly different from the caspase-7-binding affinity of Z-VAD-FMK; however, the highest-affinity conformation of Z-AEAD-FMK seems to be slightly further away from the active pocket of caspase-7, compared with Z-VAD-FMK (Fig. 3G).
The highest caspase-6-binding affinity conformations of Z-AEAD-FMK and Z-VAD-FMK nearly overlapped, but Z-AEAD-FMK has a higher binding affinity with caspase-6 (Fig. 3F). The highest-affinity conformations of Z-AEAD-FMK and Z-VAD-FMK are very close to the active pocket of caspase-8, but we noticed that the more stretched Z-AEAD-FMK is tightly lying on the active pocket of caspase-8 and has higher binding affinity with caspase-8 (Fig. 3H). The caspase-10-binding affinity of Z-AEAD-FMK is higher and the highest-affinity conformation of Z-AEAD-FMK is closer to the active pocket of caspase-10, compared with Z-VAD-FMK (Fig. 3J).
Together, these results suggest that Z-AEAD-FMK may be a potential pan-caspase inhibitor similar to Z-VAD-FMK.
3.4. Z-AEAD-FMK is a novel pan-caspase inhibitor
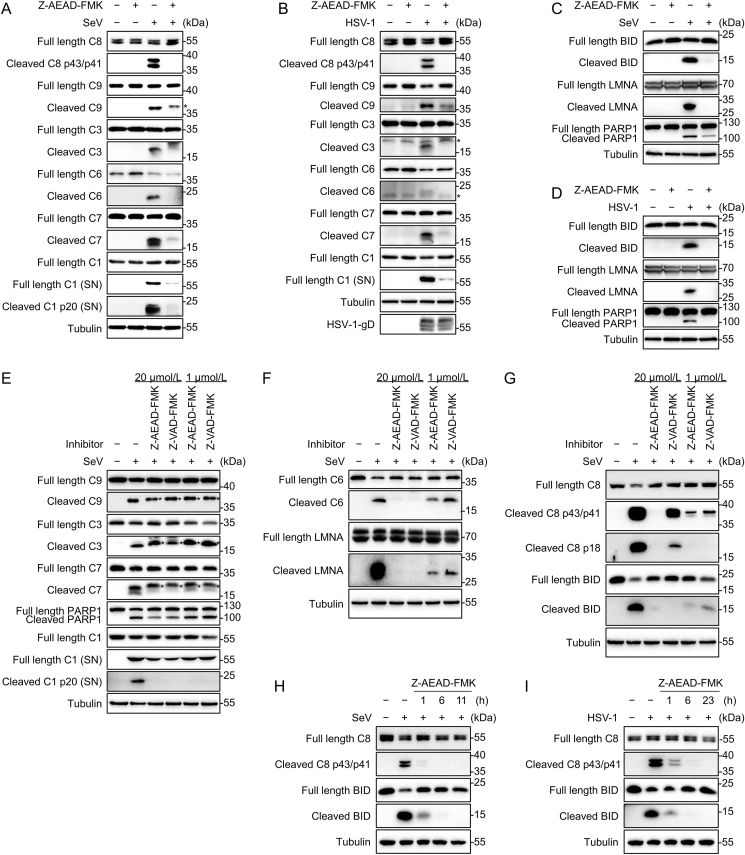
Next, we investigated the effect of Z-AEAD-FMK on caspase activity during virus infection. THP-1PMA cells were infected with SeV or HSV-1 and the activation of multiple caspases, including caspases-1, -3, -6, -7, -8 and -9 were observed (Fig. 4A and B). The SeV or HSV-1 induced activation of multiple caspases were inhibited by Z-AEAD-FMK treatment (Fig. 4A and B). These results indicate that Z-AEAD-FMK has a pan inhibitory effect on activation of multiple caspases, in line with the results of molecular docking. Immunoblot analysis showed that SeV or HSV-1 infection induced obvious cleavage of BID, LMNA and PARP1 (the downstream substrates of caspase-8, -6 and -3/7 respectively) (Fig. 4C and D). Z-AEAD-FMK treatment significantly inhibited the cleavage of these caspase substrates (Fig. 4C and D).
Fig. 4.
Broad inhibition of Z-AEAD-FMK on virus-induced activation of multiple caspases. A Immunoblot analysis of caspases activation in THP-1PMA cells. Cells were infected or mock infected with SeV at MOI = 10 for 1 h, and then the media were replaced with fresh media and treated with Z-AEAD-FMK at 20 μmol/L for 11 h. The culture supernatants (SN) were used to extract protein, and the indicated protein in supernatants or in lysates were determined. The asterisk (∗) represents inactive cleaved band of caspase. B Immunoblot analysis of caspases activation in THP-1PMA cells. Cells were infected or mock infected with HSV-1 at MOI = 10 for 1 h, and then the media were replaced with fresh media and treated with Z-AEAD-FMK at 20 μmol/L for 23 h. The culture supernatants (SN) were used to extract protein, and the indicated protein in supernatants or in lysates were determined. The asterisk (∗) represents non-specific bands. C Immunoblot analysis of the indicated proteins cleavage in THP-1PMA cells. The process was described as (A). D Immunoblot analysis of the indicated proteins cleavage in THP-1PMA cells. The process was described as (B). E–G Immunoblot analysis of the indicated proteins in THP-1PMA cells. Cells were infected or mock infected with SeV at MOI = 10 for 1 h, and then the media were replaced with fresh media and treated with the Z-AEAD-FMK or Z-VAD-FMK at indicated concentration for 11 h. The asterisks (∗) represent inactive cleaved bands of caspases. H, I Immunoblot analysis of the indicated proteins in THP-1PMA cells. Cells were infected or mock infected with SeV (H) or HSV-1 (I) at MOI = 10 for 1 h, and then the media were replaced with fresh media, followed by Z-AEAD-FMK treatment at 20 μmol/L for the indicated times. Data are representative of three experiments with similar results.
Furthermore, Z-VAD-FMK is an established pan caspase inhibitor, and we then compared the inhibitory effects on caspase activation of Z-AEAD-FMK and Z-VAD-FMK during SeV infection (Fig. 4E–G). Immunoblot analysis showed that Z-AEAD-FMK and Z-VAD-FMK had similar inhibitory effects on the activation of caspases-1, 3, -7 and -9 at concentrations of 20 μmol/L and 1 μmol/L (Fig. 4E). At a concentration of 20 μmol/L, Z-AEAD-FMK and Z-VAD-FMK may have similar inhibitory effects on caspase-6 activation (Fig. 4F). At the concentration of 1 μmol/L, Z-AEAD-FMK still had a good inhibitory effect on caspase-6 activation, showing a dose-dependent effect, while Z-VAD-FMK no longer inhibited the activation of caspase-6 (Fig. 4F). Notably, either at the concentration of 20 μmol/L or 1 μmol/L, Z-AEAD-FMK has a profoundly inhibition on caspase-8 activation compared with Z-VAD-FMK, exhibiting a dose-dependent effect (Fig. 4G). Additionally, we tested the effect of short-time (e.g., 1 h or 6 h) treatment of Z-AEAD-FMK on caspase-8 activation and BID cleavage. Immunoblot analysis showed that Z-AEAD-FMK treatment at 20 μmol/L for 1 h, 6 h or 11 h significantly inhibited the SeV-induced cleavage of caspase-8 and BID in a time-dependent manner (Fig. 4H). Similarly, Z-AEAD-FMK treatment at 20 μmol/L for 1 h, 6 h or 23 h significantly inhibited HSV-1-induced cleavage of caspase-8 and BID in a time-dependent manner (Fig. 4I).
Together, these findings suggest that Z-AEAD-FMK can broadly inhibit multiple caspases, including caspases-1, -3, -6, -7, -8 and -9, and inhibit caspase cleavage of their substrates during virus infection.
3.5. Z-AEAD-FMK inhibits caspase-mediated cell death pathway triggered by virus infection
To analyze the effects of Z-AEAD-FMK on caspase-mediated pathway, we assessed the effects of the inhibitor on caspase-mediated apoptosis, pyroptosis and inflammatory responses induced by viruses.
Infection of SeV or HSV-1 can induce pyroptosis in THP-1PMA cells with the cleavage of GSDMD and GSDME, whereas these cleavages were concurrently attenuated by Z-AEAD-FMK (Fig. 5A and B). As a form of inflammatory programmed cell death, pyroptosis can result in cellular lysis and release of inflammatory intracellular contents (Bergsbaken et al., 2009). Consistently, SeV or HSV-1 infection induced inflammation in THP-1PMA cells with the release of multiple inflammatory factors, including IL-1β, IL-18, IL-6 and IL-8 (Fig. 5C and D). The secretion of inflammatory mediators was decreased by Z-AEAD-FMK (Fig. 5C and D). These results indicated that Z-AEAD-FMK could not only inhibit the cleavage of inflammatory substrates IL-1β and IL-18 by inhibiting the activation of inflammatory caspases, but also prevent the release of inflammatory cytokines IL-1β, IL-18, IL-6 and IL-8 by inhibiting pyroptosis.
Furthermore, we used a propidium iodide (PI) fluorometric assay to assess the effect of Z-AEAD-FMK on virus-induced cell death. We observed that SeV or HSV-1 infection can cause a large number of THP-1PMA cells staining positive for PI, whereas Z-AEAD-FMK treatment significantly reduces the proportion of PI-positive cells (Fig. 5E and F). Also, the potency of Z-AEAD-FMK against virus-induced cytotoxicity was measured by lactate dehydrogenase (LDH) release assays. The results showed that Z-AEAD-FMK potently inhibited SeV or HSV-1-induced LDH release (Fig. 5G and H). These results indicate that Z-AEAD-FMK potently inhibits caspase-dependent cell death triggered by the virus infection.
Taken together, our data indicate that Z-AEAD-FMK broadly inhibits the activation of multiple caspases triggered by virus infection, thereby potently inhibiting caspase-mediated apoptotic and pyroptotic cell death and inflammatory response.
4. Discussion
Caspases are a family of cysteine proteases participating in regulating multiple biological processes including programmed cell death and innate immune signaling pathways. Virus infection often triggers caspases activation. Identification of caspase substrates is important for analyzing the intracellular pathways regulated by virus activated caspases. Here, we identified a novel caspase cleavage motif AEAD, and demonstrated its susceptibility to caspase cleavage. We fused EGFP with Tom20 using the AEADG peptide as a linker, which allowed EGFP to localize to mitochondria. Upon caspase activation either through virus infection or drug treatment, EGFP diffused into the cells due to caspase-dependent cleavage at the AEAD site. Thus, the AEAD motif can be employed to detect caspase activation in living cells. Furthermore, we developed an AEAD peptide-derived inhibitor Z-AEAD-FMK, which significantly inhibited the activities of multiple caspases, showing a broad inhibitory effect on caspase activation. The caspase inhibitor suppressed cleavage of multiple caspases substrates, such as BID, PARP1, LMNA, pro-IL-1β, pro-IL-18, GSDMD and GSDME, protecting cells from virus-induced apoptotic and pyroptotic cell death and inflammatory response.
Here, we developed an assay of identification of novel caspase cleavage motifs. The assay is based on the characteristic of caspases cleavage, that is, caspases always recognize substrates at the highly specific tetrapeptide motifs (denoted as P4-P3-P2-P1) and cleave substrates after P1 residue. We collected the cleavage sites of nine caspases, including caspases-1, -3, -4, -6, -7, -8, -9, -10 and -14, from the MEROPS peptidase database. Based on the characteristics of caspase cleavage motifs, we analyzed the preference of each caspase at each position of the motif and obtained 25 tetrapeptide motifs. Strikingly, 24 of the 25 motifs have been verified and the novel motif AEAD was verified to be a functional caspase cleavage site. Further analysis is needed to analyze whether all the proteins contain the AEAD motif are susceptible for caspase cleavage and which functionalities are involved upon these cleavage events.
In general, synthetic caspase inhibitors are classified as peptide-based and non-peptide based compounds (Dhani et al., 2021). Both Z group and FMK group are commonly used modification groups for peptide-based caspase inhibitors. Among that, Z group is a key protecting group for amines in peptide synthesis and could enhance the hydrophobicity of the caspase inhibitor, thereby being cell-permeant. The FMK group is a commonly used C-terminal modification group for caspase inhibitors. The Asp (D) was modified with the reactive electrophilic FMK group, enabling the inhibitor Z-AEAD-FMK to covalently link with the nucleophilic active thiol site of caspase in an irreversible way (Dhani et al., 2021). Collectively, the Z and FMK modifications of AEAD peptide make Z-AEAD-FMK become a cell-permeant and irreversible caspase inhibitor. The binding affinity between Z-AEAD-FMK and purified caspase proteins should be tested in the future research.
Regulation of caspase activity may also find potential clinical application. For example, the reversible caspase-1 inhibitor VX-765 (belnacasan) was used to treat the inflammatory disease psoriasis in a phase II clinical trial (clinical trial: NCT00205465) (Dhani et al., 2021). VX-740 (pralnacasan), another caspase-1 inhibitor, was orally used in a phase II clinical trial for the treatment of rheumatoid arthritis (RA) and osteoarthritis (OA) (Rudolphi et al., 2003; Dhani et al., 2021). Nevertheless, clinical application of caspase inhibitors faces the challenges, namely an inadequate efficacy, poor target specificity or adverse side effects. Development of the caspase inhibitor Z-AEAD-FMK provides an alternative backbone that can be further developed to analyze its potential of being applied for therapeutic purposes such as anti-inflammation treatment.
5. Conclusions
In conclusion, we developed an assay for identifying novel caspase cleavage motifs. With this assay we identified a novel caspase cleavage motif AEAD, and confirmed its presence in cellular proteins and susceptibility to caspase cleavage. Furthermore, we showed that AEAD peptide-derived Z-AEAD-FMK is a novel pan caspase inhibitor, which broadly inhibits multiple caspases during virus infection, protecting cells from virus-induced apoptotic and pyroptotic cell death and inflammation. Z-AEAD-FMK can be further developed to investigate its potential for clinical therapeutic purposes such as anti-inflammation therapy.
Data availability
All the data generated during the current study are included in the manuscript.
Ethics statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author contributions
Yujie Fang: conceptualization, resources, investigation, formal analysis, data curation, visualization, writing-original draft. Zhou Gong: software, formal analysis. Miaomiao You: resources. Ke Peng: conceptualization, funding acquisition, project administration, writing-review & editing, supervision.
Conflict of interest
The authors declare no conflict of interest. The authors have patented the Z-AEAD-FMK as a pan-caspase inhibitor.
Acknowledgements
This work was supported by the National Key R&D Program of China (2021YFC2300700), National Science and Technology Major Project (No. 2018ZX10101004001005), National Natural Science Foundation of China (numbers 32070179). We thank Dr. Qinxue Hu (Wuhan Institute of Virology) and Dr. Yuchen Xia (Wuhan University) for help with materials. We thank Ding Gao from Center for Instrumental Analysis and Metrology at Wuhan Institute of Virology for his help with the Leica confocal microscope and the Operetta.
Footnotes
Peer review under the responsibility of Editorial Board of Virologica Sinica.
References
- Adam-Klages S., Schwandner R., Luschen S., Ussat S., Kreder D., Kronke M. Caspase-mediated inhibition of human cytosolic phospholipase A2 during apoptosis. J. Immunol. 1998;161:5687–5694. [PubMed] [Google Scholar]
- Bae S.S., Perry D.K., Oh Y.S., Choi J.H., Galadari S.H., Ghayur T., Ryu S.H., Hannun Y.A., Suh P.G. Proteolytic cleavage of phospholipase C-gamma1 during apoptosis in Molt-4 cells. FASEB J. 2000;14:1083–1092. doi: 10.1096/fasebj.14.9.1083. [DOI] [PubMed] [Google Scholar]
- Bergsbaken T., Fink S.L., Cookson B.T. Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldin M.P., Goncharov T.M., Goltsev Y.V., Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- Bushell M., Wood W., Clemens M.J., Morley S.J. Changes in integrity and association of eukaryotic protein synthesis initiation factors during apoptosis. Eur. J. Biochem. 2000;267:1083–1091. doi: 10.1046/j.1432-1327.2000.01101.x. [DOI] [PubMed] [Google Scholar]
- Casciola-Rosen L., Garcia-Calvo M., Bull H.G., Becker J.W., Hines T., Thornberry N.A., Rosen A. Mouse and human granzyme B have distinct tetrapeptide specificities and abilities to recruit the bid pathway. J. Biol. Chem. 2007;282:4545–4552. doi: 10.1074/jbc.M606564200. [DOI] [PubMed] [Google Scholar]
- Casciola-Rosen L.A., Anhalt G.J., Rosen A. DNA-dependent protein kinase is one of a subset of autoantigens specifically cleaved early during apoptosis. J. Exp. Med. 1995;182:1625–1634. doi: 10.1084/jem.182.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Xie M., Shah V.R., Schneider M.D., Entman M.L., Wei L., Schwartz R.J. Activation of Rho-associated coiled-coil protein kinase 1 (ROCK-1) by caspase-3 cleavage plays an essential role in cardiac myocyte apoptosis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14495–14500. doi: 10.1073/pnas.0601911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry P., Singh M., Parent S., Asselin E. Prostate apoptosis response 4 (Par-4), a novel substrate of caspase-3 during apoptosis activation. Mol. Cell. Biol. 2012;32:826–839. doi: 10.1128/MCB.06321-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Ning X., Jiang Z. Caspases control antiviral innate immunity. Cell. Mol. Immunol. 2017;14:736–747. doi: 10.1038/cmi.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.H., Hagemann T.L., Quinlan R.A., Messing A., Perng M.D. Caspase cleavage of GFAP produces an assembly-compromised proteolytic fragment that promotes filament aggregation. ASN Neuro. 2013;5 doi: 10.1042/AN20130032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M.L., Sahai E.A., Yeo M., Bosch M., Dewar A., Olson M.F. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 2001;3:339–345. doi: 10.1038/35070009. [DOI] [PubMed] [Google Scholar]
- Crawford E.D., Wells J.A. Caspase substrates and cellular remodeling. Annu. Rev. Biochem. 2011;80:1055–1087. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- Danley D.E., Chuang T.H., Bokoch G.M. Defective Rho GTPase regulation by IL-1 beta-converting enzyme-mediated cleavage of D4 GDP dissociation inhibitor. J. Immunol. 1996;157:500–503. [PubMed] [Google Scholar]
- Datta R., Kojima H., Yoshida K., Kufe D. Caspase-3-mediated cleavage of protein kinase C theta in induction of apoptosis. J. Biol. Chem. 1997;272:20317–20320. doi: 10.1074/jbc.272.33.20317. [DOI] [PubMed] [Google Scholar]
- Demontis S., Rigo C., Piccinin S., Mizzau M., Sonego M., Fabris M., Brancolini C., Maestro R. Twist is substrate for caspase cleavage and proteasome-mediated degradation. Cell Death Differ. 2006;13:335–345. doi: 10.1038/sj.cdd.4401744. [DOI] [PubMed] [Google Scholar]
- Dhani S., Zhao Y., Zhivotovsky B. A long way to go: caspase inhibitors in clinical use. Cell Death Dis. 2021;12:949. doi: 10.1038/s41419-021-04240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enari M., Sakahira H., Yokoyama H., Okawa K., Iwamatsu A., Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- Fan T.J., Han L.H., Cong R.S., Liang J. Caspase family proteases and apoptosis. Acta Biochim. Biophys. Sin. (Shanghai) 2005;37:719–727. doi: 10.1111/j.1745-7270.2005.00108.x. [DOI] [PubMed] [Google Scholar]
- Frutos S., Moscat J., Diaz-Meco M.T. Cleavage of zetaPKC but not lambda/iotaPKC by caspase-3 during UV-induced apoptosis. J. Biol. Chem. 1999;274:10765–10770. doi: 10.1074/jbc.274.16.10765. [DOI] [PubMed] [Google Scholar]
- Galvan V., Chen S., Lu D., Logvinova A., Goldsmith P., Koo E.H., Bredesen D.E. Caspase cleavage of members of the amyloid precursor family of proteins. J. Neurochem. 2002;82:283–294. doi: 10.1046/j.1471-4159.2002.00970.x. [DOI] [PubMed] [Google Scholar]
- Gervais F.G., Xu D., Robertson G.S., Vaillancourt J.P., Zhu Y., Huang J., Leblanc A., Smith D., Rigby M., Shearman M.S., Clarke E.E., Zheng H., Van Der Ploeg L.H., Ruffolo S.C., Thornberry N.A., Xanthoudakis S., Zamboni R.J., Roy S., Nicholson D.W. Involvement of caspases in proteolytic cleavage of Alzheimer's amyloid-beta precursor protein and amyloidogenic A beta peptide formation. Cell. 1999;97:395–406. doi: 10.1016/s0092-8674(00)80748-5. [DOI] [PubMed] [Google Scholar]
- Henis-Korenblit S., Strumpf N.L., Goldstaub D., Kimchi A. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol. Cell. Biol. 2000;20:496–506. doi: 10.1128/mcb.20.2.496-506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrant M., Jacquel A., Marchetti S., Belhacene N., Colosetti P., Luciano F., Auberger P. Cleavage of Mcl-1 by caspases impaired its ability to counteract Bim-induced apoptosis. Oncogene. 2004;23:7863–7873. doi: 10.1038/sj.onc.1208069. [DOI] [PubMed] [Google Scholar]
- Hirota J., Furuichi T., Mikoshiba K. Inositol 1,4,5-trisphosphate receptor type 1 is a substrate for caspase-3 and is cleaved during apoptosis in a caspase-3-dependent manner. J. Biol. Chem. 1999;274:34433–34437. doi: 10.1074/jbc.274.48.34433. [DOI] [PubMed] [Google Scholar]
- Hou J., Zhao R., Xia W., Chang C.W., You Y., Hsu J.M., Nie L., Chen Y., Wang Y.C., Liu C., Wang W.J., Wu Y., Ke B., Hsu J.L., Huang K., Ye Z., Yang Y., Xia X., Li Y., Li C.W., Shao B., Tainer J.A., Hung M.C. PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis. Nat. Cell Biol. 2020;22:1264–1275. doi: 10.1038/s41556-020-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.Y., Wu Y.M., Hsu C.Y., Lee W.S., Lai M.D., Lu T.J., Huang C.L., Leu T.H., Shih H.M., Fang H.I., Robinson D.R., Kung H.J., Yuan C.J. Caspase activation of mammalian sterile 20-like kinase 3 (Mst3). Nuclear translocation and induction of apoptosis. J. Biol. Chem. 2002;277:34367–34374. doi: 10.1074/jbc.M202468200. [DOI] [PubMed] [Google Scholar]
- Irmler M., Steiner V., Ruegg C., Wajant H., Tschopp J. Caspase-induced inactivation of the anti-apoptotic TRAF1 during Fas ligand-mediated apoptosis. FEBS Lett. 2000;468:129–133. doi: 10.1016/s0014-5793(00)01206-0. [DOI] [PubMed] [Google Scholar]
- Irmler M., Thome M., Hahne M., Schneider P., Hofmann K., Steiner V., Bodmer J.L., Schroter M., Burns K., Mattmann C., Rimoldi D., French L.E., Tschopp J. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- Janicke R.U., Walker P.A., Lin X.Y., Porter A.G. Specific cleavage of the retinoblastoma protein by an ICE-like protease in apoptosis. EMBO J. 1996;15:6969–6978. [PMC free article] [PubMed] [Google Scholar]
- Joselin A.P., Schulze-Osthoff K., Schwerk C. Loss of Acinus inhibits oligonucleosomal DNA fragmentation but not chromatin condensation during apoptosis. J. Biol. Chem. 2006;281:12475–12484. doi: 10.1074/jbc.M509859200. [DOI] [PubMed] [Google Scholar]
- Kanuka H., Kuranaga E., Takemoto K., Hiratou T., Okano H., Miura M. Drosophila caspase transduces Shaggy/GSK-3beta kinase activity in neural precursor development. EMBO J. 2005;24:3793–3806. doi: 10.1038/sj.emboj.7600822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelotra S., Jain M., Kelotra A., Jain I., Bandaru S., Nayarisseri A., Bidwai A. An in silico appraisal to identify high affinity anti-apoptotic synthetic tetrapeptide inhibitors targeting the mammalian caspase 3 enzyme. Asian Pac. J. Cancer Prev. 2014;15:10137–10142. doi: 10.7314/apjcp.2014.15.23.10137. [DOI] [PubMed] [Google Scholar]
- Koyama A.H., Irie H., Fukumori T., Hata S., Iida S., Akari H., Adachi A. Role of virus-induced apoptosis in a host defense mechanism against virus infection. J. Med. Invest. 1998;45:37–45. [PubMed] [Google Scholar]
- Ku N.O., Liao J., Omary M.B. Apoptosis generates stable fragments of human type I keratins. J. Biol. Chem. 1997;272:33197–33203. doi: 10.1074/jbc.272.52.33197. [DOI] [PubMed] [Google Scholar]
- Lazebnik Y.A., Kaufmann S.H., Desnoyers S., Poirier G.G., Earnshaw W.C. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- Lee K.K., Ohyama T., Yajima N., Tsubuki S., Yonehara S. MST, a physiological caspase substrate, highly sensitizes apoptosis both upstream and downstream of caspase activation. J. Biol. Chem. 2001;276:19276–19285. doi: 10.1074/jbc.M005109200. [DOI] [PubMed] [Google Scholar]
- Lee M.W., Hirai I., Wang H.G. Caspase-3-mediated cleavage of Rad9 during apoptosis. Oncogene. 2003;22:6340–6346. doi: 10.1038/sj.onc.1206729. [DOI] [PubMed] [Google Scholar]
- Lee Z.H., Lee S.E., Kwack K., Yeo W., Lee T.H., Bae S.S., Suh P.G., Kim H.H. Caspase-mediated cleavage of TRAF3 in FasL-stimulated Jurkat-T cells. J. Leukoc. Biol. 2001;69:490–496. [PubMed] [Google Scholar]
- Li H., Zhu H., Xu C.J., Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- Liu X., Zou H., Slaughter C., Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- Lu Y., Luo Z., Bregman D.B. RNA polymerase II large subunit is cleaved by caspases during DNA damage-induced apoptosis. Biochem. Biophys. Res. Commun. 2002;296:954–961. doi: 10.1016/s0006-291x(02)02028-4. [DOI] [PubMed] [Google Scholar]
- Martin S.J., Finucane D.M., Amarante-Mendes G.P., O'brien G.A., Green D.R. Phosphatidylserine externalization during CD95-induced apoptosis of cells and cytoplasts requires ICE/CED-3 protease activity. J. Biol. Chem. 1996;271:28753–28756. doi: 10.1074/jbc.271.46.28753. [DOI] [PubMed] [Google Scholar]
- Munday N.A., Vaillancourt J.P., Ali A., Casano F.J., Miller D.K., Molineaux S.M., Yamin T.T., Yu V.L., Nicholson D.W. Molecular cloning and pro-apoptotic activity of ICErelII and ICErelIII, members of the ICE/CED-3 family of cysteine proteases. J. Biol. Chem. 1995;270:15870–15876. doi: 10.1074/jbc.270.26.15870. [DOI] [PubMed] [Google Scholar]
- Na S., Chuang T.H., Cunningham A., Turi T.G., Hanke J.H., Bokoch G.M., Danley D.E. D4-GDI, a substrate of CPP32, is proteolyzed during Fas-induced apoptosis. J. Biol. Chem. 1996;271:11209–11213. doi: 10.1074/jbc.271.19.11209. [DOI] [PubMed] [Google Scholar]
- Ohtsubo T., Kamada S., Mikami T., Murakami H., Tsujimoto Y. Identification of NRF2, a member of the NF-E2 family of transcription factors, as a substrate for caspase-3(-like) proteases. Cell Death Differ. 1999;6:865–872. doi: 10.1038/sj.cdd.4400566. [DOI] [PubMed] [Google Scholar]
- Orzalli M.H., Kagan J.C. Apoptosis and necroptosis as host defense strategies to prevent viral infection. Trends Cell Biol. 2017;27:800–809. doi: 10.1016/j.tcb.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poreba M., Strozyk A., Salvesen G.S., Drag M. Caspase substrates and inhibitors. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Juo P., Masuda-Robens J., Caloca M.J., Zhou H., Stone N., Kazanietz M.G., Chou M.M. Caspase-mediated cleavage of the TIAM1 guanine nucleotide exchange factor during apoptosis. Cell Growth Differ. 2001;12:603–611. [PubMed] [Google Scholar]
- Rawlings N.D., Barrett A.J., Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N.D., O'brien E., Barrett A.J. MEROPS: the protease database. Nucleic Acids Res. 2002;30:343–346. doi: 10.1093/nar/30.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheaume E., Cohen L.Y., Uhlmann F., Lazure C., Alam A., Hurwitz J., Sekaly R.P., Denis F. The large subunit of replication factor C is a substrate for caspase-3 in vitro and is cleaved by a caspase-3-like protease during Fas-mediated apoptosis. EMBO J. 1997;16:6346–6354. doi: 10.1093/emboj/16.21.6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolphi K., Gerwin N., Verzijl N., Van Der Kraan P., Van Den Berg W. Pralnacasan, an inhibitor of interleukin-1beta converting enzyme, reduces joint damage in two murine models of osteoarthritis. Osteoarthritis Cartilage. 2003;11:738–746. doi: 10.1016/s1063-4584(03)00153-5. [DOI] [PubMed] [Google Scholar]
- Sahara S., Aoto M., Eguchi Y., Imamoto N., Yoneda Y., Tsujimoto Y. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature. 1999;401:168–173. doi: 10.1038/43678. [DOI] [PubMed] [Google Scholar]
- Satoh S., Hijikata M., Handa H., Shimotohno K. Caspase-mediated cleavage of eukaryotic translation initiation factor subunit 2alpha. Biochem. J. 1999;342(Pt 1):65–70. [PMC free article] [PubMed] [Google Scholar]
- Seaman J.E., Julien O., Lee P.S., Rettenmaier T.J., Thomsen N.D., Wells J.A. Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues. Cell Death Differ. 2016;23:1717–1726. doi: 10.1038/cdd.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebbagh M., Renvoize C., Hamelin J., Riche N., Bertoglio J., Breard J. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat. Cell Biol. 2001;3:346–352. doi: 10.1038/35070019. [DOI] [PubMed] [Google Scholar]
- Sekine Y., Yamamoto C., Kakisaka M., Muromoto R., Kon S., Ashitomi D., Fujita N., Yoshimura A., Oritani K., Matsuda T. Signal-transducing adaptor protein-2 modulates Fas-mediated T cell apoptosis by interacting with caspase-8. J. Immunol. 2012;188:6194–6204. doi: 10.4049/jimmunol.1103467. [DOI] [PubMed] [Google Scholar]
- Shalini S., Dorstyn L., Dawar S., Kumar S. Old, new and emerging functions of caspases. Cell Death Differ. 2015;22:526–539. doi: 10.1038/cdd.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y., Caudell P., Grady G., Wang G., Suwa A., Sharp G.C., Dynan W.S., Hardin J.A. Human RNA helicase A is a lupus autoantigen that is cleaved during apoptosis. J. Immunol. 1999;163:6269–6274. [PubMed] [Google Scholar]
- Tesco G., Koh Y.H., Kang E.L., Cameron A.N., Das S., Sena-Esteves M., Hiltunen M., Yang S.H., Zhong Z., Shen Y., Simpkins J.W., Tanzi R.E. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron. 2007;54:721–737. doi: 10.1016/j.neuron.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya K. Inflammasome-associated cell death: pyroptosis, apoptosis, and physiological implications. Microbiol. Immunol. 2020;64:252–269. doi: 10.1111/1348-0421.12771. [DOI] [PubMed] [Google Scholar]
- Van Damme P., Martens L., Van Damme J., Hugelier K., Staes A., Vandekerckhove J., Gevaert K. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat. Methods. 2005;2:771–777. doi: 10.1038/nmeth792. [DOI] [PubMed] [Google Scholar]
- Vanags D.M., Porn-Ares M.I., Coppola S., Burgess D.H., Orrenius S. Protease involvement in fodrin cleavage and phosphatidylserine exposure in apoptosis. J. Biol. Chem. 1996;271:31075–31085. doi: 10.1074/jbc.271.49.31075. [DOI] [PubMed] [Google Scholar]
- Vande Walle L., Lamkanfi M. Pyroptosis. Curr. Biol. 2016;26:R568–R572. doi: 10.1016/j.cub.2016.02.019. [DOI] [PubMed] [Google Scholar]
- Wang K., Sun Q., Zhong X., Zeng M., Zeng H., Shi X., Li Z., Wang Y., Zhao Q., Shao F., Ding J. Structural mechanism for GSDMD targeting by autoprocessed caspases in pyroptosis. Cell. 2020;180:941–955.e920. doi: 10.1016/j.cell.2020.02.002. [DOI] [PubMed] [Google Scholar]
- Wang K.K., Posmantur R., Nath R., Mcginnis K., Whitton M., Talanian R.V., Glantz S.B., Morrow J.S. Simultaneous degradation of alphaII- and betaII-spectrin by caspase 3 (CPP32) in apoptotic cells. J. Biol. Chem. 1998;273:22490–22497. doi: 10.1074/jbc.273.35.22490. [DOI] [PubMed] [Google Scholar]
- Wang X., Pai J.T., Wiedenfeld E.A., Medina J.C., Slaughter C.A., Goldstein J.L., Brown M.S. Purification of an interleukin-1 beta converting enzyme-related cysteine protease that cleaves sterol regulatory element-binding proteins between the leucine zipper and transmembrane domains. J. Biol. Chem. 1995;270:18044–18050. doi: 10.1074/jbc.270.30.18044. [DOI] [PubMed] [Google Scholar]
- Wang Y., Ning X., Gao P., Wu S., Sha M., Lv M., Zhou X., Gao J., Fang R., Meng G., Su X., Jiang Z. Inflammasome activation triggers caspase-1-mediated cleavage of cGAS to regulate responses to DNA virus infection. Immunity. 2017;46:393–404. doi: 10.1016/j.immuni.2017.02.011. [DOI] [PubMed] [Google Scholar]
- Weng C., Li Y., Xu D., Shi Y., Tang H. Specific cleavage of Mcl-1 by caspase-3 in tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in Jurkat leukemia T cells. J. Biol. Chem. 2005;280:10491–10500. doi: 10.1074/jbc.M412819200. [DOI] [PubMed] [Google Scholar]
- Werner M.E., Chen F., Moyano J.V., Yehiely F., Jones J.C., Cryns V.L. Caspase proteolysis of the integrin beta4 subunit disrupts hemidesmosome assembly, promotes apoptosis, and inhibits cell migration. J. Biol. Chem. 2007;282:5560–5569. doi: 10.1074/jbc.M603669200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe V.O., Mcmurtrie P.I., Marr J., Amagase Y., Main S., Mills A.D., Laskey R.A., Takei Y. MCM3AP is transcribed from a promoter within an intron of the overlapping gene for GANP. J. Mol. Biol. 2011;406:355–361. doi: 10.1016/j.jmb.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Misra R.S., Russell J.Q., Flavell R.A., Rincon M., Budd R.C. Proteolytic regulation of nuclear factor of activated T (NFAT) c2 cells and NFAT activity by caspase-3. J. Biol. Chem. 2006;281:10682–10690. doi: 10.1074/jbc.M511759200. [DOI] [PubMed] [Google Scholar]
- Yakovlev A.G., Di X., Movsesyan V., Mullins P.G., Wang G., Boulares H., Zhang J., Xu M., Faden A.I. Presence of DNA fragmentation and lack of neuroprotective effect in DFF45 knockout mice subjected to traumatic brain injury. Mol. Med. 2001;7:205–216. [PMC free article] [PubMed] [Google Scholar]
- Yin X., Gu S., Jiang J.X. The development-associated cleavage of lens connexin 45.6 by caspase-3-like protease is regulated by casein kinase II-mediated phosphorylation. J. Biol. Chem. 2001;276:34567–34572. doi: 10.1074/jbc.M106073200. [DOI] [PubMed] [Google Scholar]
- Zheng M., Karki R., Vogel P., Kanneganti T.D. Caspase-6 is a key regulator of innate immunity, inflammasome activation, and host defense. Cell. 2020;181:674–687 e613. doi: 10.1016/j.cell.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Jiang W., Liu Z., Liu S., Liang X. Virus infection and death receptor-mediated apoptosis. Viruses. 2017;9:316. doi: 10.3390/v9110316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data generated during the current study are included in the manuscript.