Abstract
Histone mimicry (HM) refers to the presence of short linear motifs in viral proteins that mimic critical regions of host histone proteins. These motifs have the potential to interfere with host cell epigenome and counteract antiviral response. Recent research shows that HM is critical for the pathogenesis and transmissibility of influenza virus and coronavirus. However, the distribution, characteristics, and functions of HM in eukaryotic viruses remain obscure. Herein, we developed a bioinformatic pipeline, Histone Motif Scan (HiScan), to identify HM motifs in viral proteins and predict their functions in silico. By analyzing 592,643 viral proteins using HiScan, we found that putative HM motifs were widely distributed in most viral proteins. Among animal viruses, the ratio of HM motifs between DNA viruses and RNA viruses was approximately 1.9:1, and viruses with smaller genomes had a higher density of HM motifs. Notably, coronaviruses exhibited an uneven distribution of HM motifs, with betacoronaviruses (including most human pathogenic coronaviruses) harboring more HM motifs than other coronaviruses, primarily in the NSP3, S, and N proteins. In summary, our virome-wide screening of HM motifs using HiScan revealed extensive but uneven distribution of HM motifs in most viral proteins, with a preference in DNA viruses. Viral HM may play an important role in modulating viral pathogenicity and virus-host interactions, making it an attractive area of research in virology and antiviral medication.
Keywords: Histone mimicry, Viral proteins, Histone modification, Evolution, Coronavirus
Highlights
-
•
We developed a pipeline called HiScan to identify HM motif in viral proteins with high efficiency.
-
•
HM motifs are widely distributed across animal and plant viruses, as well as DNA and RNA viral genomes.
-
•
In Coronaviridae, HM motifs are predominantly concentrated in Betacoronaviruses, primarily located in NSP3, S and E proteins.
1. Introduction
Histone plays a crucial role in regulating gene expression and other cellular processes through various post-translation modifications (PTMs). According to the positions, histones can be classified into the core histones, including H2A, H2B, H3, H4, and the linker histone H1 (Peterson and Laniel, 2004). The N-terminus of a histone protein extends out as a “tail” which serves as the site for various PTMs, such as methylation, ubiquitination, phosphorylation, and acetylation. The patterns of histone PTMs are highly complex and dynamic with different combinations of modification types, sites, and states, which significantly alter gene expression via chromatin relaxation, transcriptional activation or transcriptional inhibition-collectively known as histone codes (Jenuwein and Allis, 2001).
Histone codes serve as carriers of epigenetic information interpreted by chromatin-binding proteins, known as “reader” (Jenuwein and Allis, 2001). Histone methylation typically occurs at lysine or arginine and is often associated with gene activation or silencing (Lee et al., 2005). Histone acetylation was the first modification found to be linked to transcriptional activation (Brownell et al., 1996), and it also contributes to the recruitment of chromatin remodeling factors (Shvedunova and Akhtar, 2022). Histone phosphorylation, primarily at Ser, Thr, or Tyr residues, plays important roles in cell cycle, growth, and signal transduction (Cheung et al., 2000; Hans and Dimitrov, 2001; Nowak and Corces, 2004). Histone ubiquitination is crucial for maintaining genome stability. It not only involves protein recruitment, such as DOT1L (Worden et al., 2019) or PRC2 (Kasinath et al., 2021), but also plays important roles in multiple key biological processes including DNA repair (Mattiroli and Penengo, 2021) and transcriptional regulation (Tamburri et al., 2020). In addition, other types of histone PTMs, such as glycosylation (Merx et al., 2022), S-nitrosylation (Yoon et al., 2021), and sumoylation (Ryu et al., 2020), also have significant effects on protein folding, conformation, and activity.
Interestingly, certain viruses have been found to possess short linear motifs on their proteins that mimic critical regions of host histone proteins, a phenomenon known as histone mimicry (HM) (Elde and Malik, 2009; Schaefer et al., 2013). For example, the influenza A H3N2 subtype possesses a human H3K4-like sequence in the tail of its non-structural protein 1 (NS1) (Marazzi et al., 2012), which can be methylated or acetylated in virus-infected cells to disrupt the normal epigenetic regulation of host cells. Similarly, the SARS-CoV-2 ORF8 has been shown to possess an HM motif (Kee et al., 2022). Although ORF8 primarily functions in the endoplasmic reticulum lumen rather than the nucleus (Liu et al., 2022), its potential role as a HM motif is not precluded, given the multifunctional nature of viral proteins. Moreover, Kee et al. demonstrated that lysine acetyltransferase 2A (KAT2A) interacts with the HM motif on ORF8, leading to KAT2A degradation and consequently disrupting the host's epigenetic regulation (Kee et al., 2022). It is possible that ORF8 sequester newly synthesized KAT2A in the cytoplasm, promoting KAT2A degradation before its nucleus translocation. Similar interactions may occur for other HMs, though direct evidence has not been reported. However, it is logical not to exclude the possibility that HM's can function out of the nucleus. Additionally, recent studies have demonstrated that adenovirus protein VII exhibits histone-like properties, interacting with host chromatin and suppressing both DNA damage response and immune signaling (Avgousti et al., 2016, 2017). Although researchers have not explicitly termed this phenomenon ‘HM’, its functional essence closely resembles that of histone modifications, capable of modulating host epigenetic regulation. Moreover, this histone-mimicking phenomenon is not confined to viral proteins. Investigations have revealed that the human histone methyltransferase G9a contains a conserved methylation motif similar to histone H3, with G9a′s auto-methylation crucial for its interaction with heterochromatin protein 1 (Sampath et al., 2007). Collectively, these findings suggest that HM may represent a ubiquitous biological mechanism found not only in viral proteins but also in host proteins. Therefore, studying HM has the potential to enhance our understanding of epigenetic regulatory mechanisms and provide new insights into viral infection and replication processes.
However, comprehensive investigations elucidating the detailed mechanisms or distributions of HM remain limited. Existing bioinformatics tools like the logistic regression model (Benveniste et al., 2014), DeepPTM (Baisya and Lonardi, 2021), and DeepHistone (Yin et al., 2019), are not designed to identify viral HM motifs, as they focus on analyzing DNA sequences rather than amino acid sequences. These tools are designed to predict histone modifications based on DNA sequence characteristics and transcription factor binding (the logistic regression model and DeepHistone) or chromatin accessibility data (DeepPTM). Thus, they are not suitable for this study. To bridge this gap, we developed a bioinformatic pipeline, HiScan, to systematically identify putative HM motifs across the viral proteome and predict their potential functions in silico.
2. Materials and methods
2.1. Data source
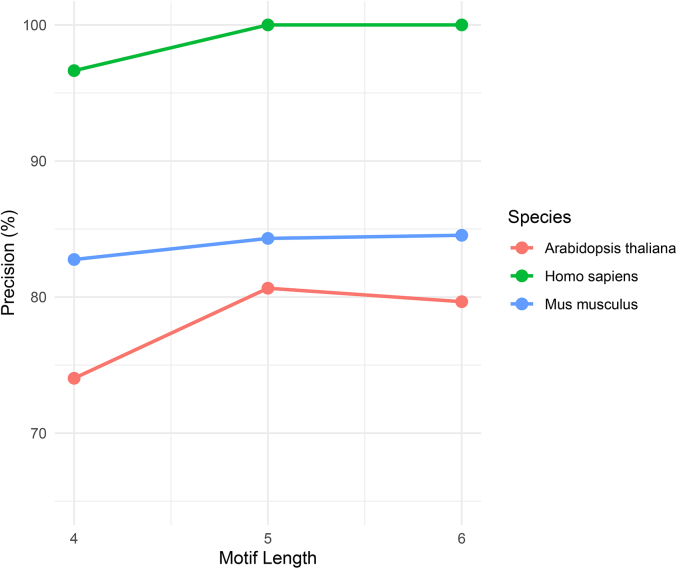
The viral protein sequence data and annotations were obtained from the NCBI Reference Sequence Database as of November 9, 2022 (https://ftp.ncbi.nlm.nih.gov/refseq/release/viral/), comprising 592, 643 viral proteins and their corresponding annotations. Since histone modification sites are highly conserved between plants and animals, and there are limited reports on plant histone modifications, this study focused on human histone datasets compiled from relevant literature and research. After preliminary testing of motif lengths (Supplementary Fig. S2, Supplementary Table S10), HM motifs are ultimately defined as 5 amino acids centered around the modification site, excluding those with fewer than 5 amino acids at the beginning or end. Each HM motif and its evidence source are provided in Supplementary Table S1. Only motifs that matched 100% with viral proteins were counted. Taxonomic information and host origin information of viral proteins were supplemented from the ICTV Virus Metadata Resource (https://ictv.global/vmr) and the NCBI Taxonomy Browser (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi). Coronaviridae sequence data were obtained from the NCBI Taxonomy Browser as of Nov 9, 2022 (https://www.ncbi.nlm.nih.gov/datasets/taxonomy/tree/?taxon=11118). Viruses without family information were excluded from subsequent analysis to ensure the accuracy of results.
2.2. HiScan
HiScan is a Python-based program that can quickly identify HM motifs in viral protein sequences across multiple operating systems. By inputting the viral protein sequence and histone motif files, HiScan can locate potential HM motifs and provide the annotation information. Additionally, HiScan can analyze the skewness distribution of amino acids in the motif and predict the potential HM motifs. Given that the motifs predicted by HiScan are putative histone mimics that may not actually possess functional significance, “HM motifs” in this study specifically denote the results obtained using HiScan. Hiscan is available at: https://github.com/xiaosheep01/hiscan.
2.3. Consistency and discrepancy analysis on HM motifs between animal and plant viruses
Since histones are unique to eukaryotic cells, the analysis focused on animal and plant viruses, with animal viruses further divided into DNA and RNA categories based on the type of their nucleic acid. HiScan was used to identify putative histone mimicry (HM) motifs in the viral protein sequences, excluding viruses without family information. In the virus family and genus annotation file, non-vertebrates and vertebrates were combined into the animal category. And viruses that infect both non-vertebrates and vertebrates were recorded only once, and viruses that infect both animals and plants were counted once for both.
2.3.1. Distribution of HM motifs modification types
The analysis result of raw result was classified according to the types of histone motif modification. The “Rate” value is the count of its HM motifs under a specific classification divided by the total number of all motifs in this classification. To verify whether there are differences in HM modification types between animal and plant viruses, and between DNA and RNA animal viruses, contingency table analysis (chi-square test) was performed using SPSS software.
2.3.2. Discrepancy analysis on HM motifs between DNA and RNA animal viruses
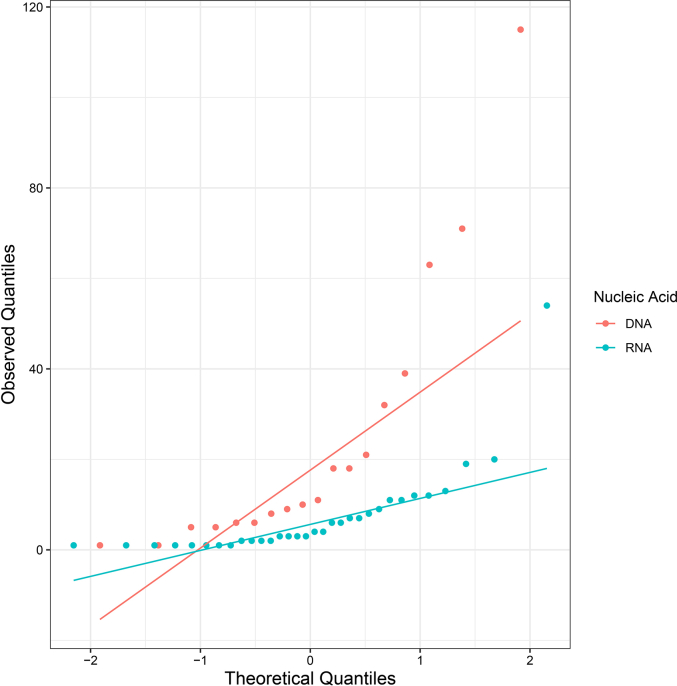
To uncover differences in HM motifs between DNA and RNA animal viruses, the Mann-Whitney U test was used to assess differences in the number of HM motifs. Additionally, the chi-square test was conducted to determine if the distributions of HM motifs across different histone subtypes varied between DNA and RNA animal viruses. Prior to the statistical tests, the normality (Shapiro-Wilk) and homogeneity of variance (Levene's test) of the datasets were analyzed in R. The Mann-Whitney U test and chi-square test were performed using SPSS.
2.4. Statistical analysis of HM density in animal viruses
All animal virus results were screened from the analysis result of raw data and then classified according to different animal virus families. The average number of HM motifs per kilobase (Kb) of nucleotide length (unit length) was calculated based on the average genome size of different animal virus families, referred to here as “HM density”. The mean genome length of each pedigree was obtained from ICTV and PubMed using the arithmetic mean. For the segmented virus, the genome size was calculated as the sum of average segment lengths. HM density was calculated using the following formula
where N is the number of matched HM motifs in viral proteins, and L is the mean genome size of the animal virus family.
2.5. Distribution of HM motifs in Coronaviridae
Representative sequences of coronavirus genera were downloaded from the NCBI database. Representative genera were not analyzed if their reference sequences were not available. The HM motifs present in Coronaviridae were identified using HiScan. The Coronaviridae genome structure was drawn based on the annotated information of the SARS-CoV-2 reference sequence, and all HM motifs matched by Coronaviridae in raw result were mapped to the genome.
3. Results
3.1. The principle and design of HiScan pipeline
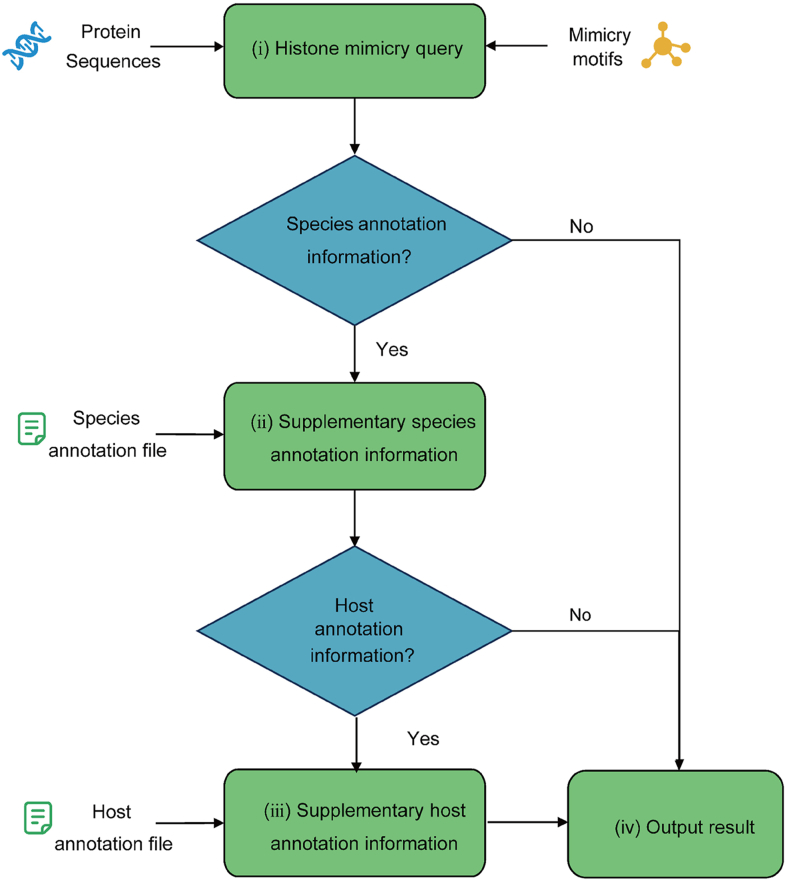
HiScan is developed using the Python programming language, and its internal workflow is illustrated in Fig. 1. The principle of HiScan is to treat each potential HM as a “probe” and the viral protein data as a library of reference sequences. The process of identifying HM motifs in viruses involves matching each probe in the HM motif library against the viral protein library. By default configuration, the algorithm requires 100% sequence identity, which means no differences in amino acid positions or types. During the matching process, HiScan employs the decision tree model and a local alignment algorithm to optimize performance. Additionally, HiScan can construct a comprehensive annotation library by integrating multiple data sources (Note: The annotation files must be in GTF or GFF format.). This allows the HM query results to be automatically paired with relevant annotation information, which facilitates efficient HM motif identification. The detailed specification of HiScan can be viewed in the software using the "-h" parameter. The general steps for analyzing data using HiScan are as follows:
-
(ⅰ)
Viral protein sequence files, preferably in “fasta” format, and their associated annotation files, typically in “gtf” format, are obtained from various databases.
-
(ⅱ)
The annotation information from the prepared protein files is used to supplement the virus classification data in the raw HM motif identification results.
-
(ⅲ)
This step involves further annotating the results from step (ii) to include the host information of the viruses and correct any discrepancies in the virus classification using the ICTV annotation file and the NCBI Taxonomy Browser to address gaps in host data.
-
(ⅳ)
Some basic statistical analysis can be performed on the raw results, such as computing the frequency of HM occurrence by the type of HM or the distribution of HM motifs across different virus families.
Fig. 1.
The overall workflow of HiScan. Note that the species annotation information is recommended to be sourced from the official website of ICTV.
It should be noted that HiScan is a versatile sequence analysis tool that can be applied not only to viral proteins but also to other sequence data.
3.2. Comprehensive view of HM motif distribution in eukaryotic viruses analyzed by HiScan
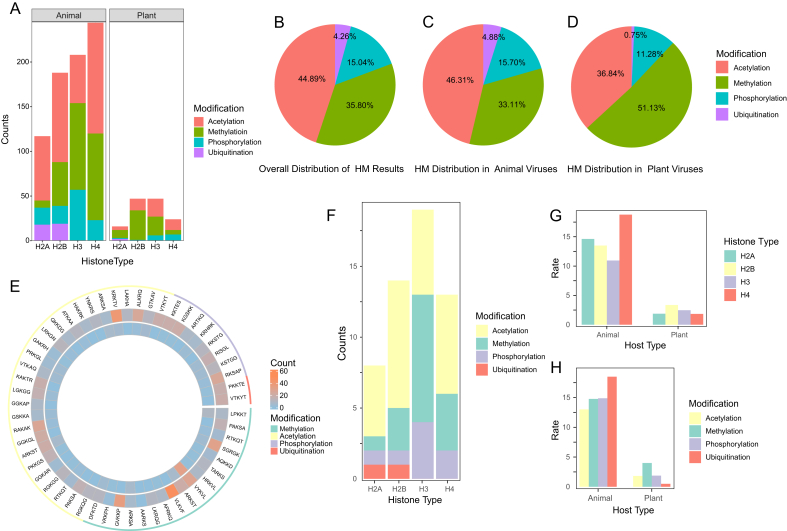
Using HiScan, 891 viral protein sequences (which can be classified into approximately 68 viral families) were predicted to have HM motifs. We identified the HM motif of four main types of modification in viral protein datasets, including methylation, acetylation, phosphorylation, and ubiquitination. The counts of these modifications and their corresponding HM motifs in each viral host are shown in Fig. 2A. According to the results, HM is a common phenomenon in both animal and plant viruses, but with distinct frequencies for the different modification types. Interestingly, animal viruses have a significantly higher number of HM motifs (758) compared to plant viruses (133). Furthermore, animal viruses also exhibit a higher number of HM motifs for each modification type, especially for methylation (251 vs. 68) and acetylation (351 vs. 49).
Fig. 2.
Distribution of HM motifs in animal and plant viruses. A Distribution of HM motif numbers. Different colors represent different types of histone modifications: acetylation (red), methylation (green), phosphorylation (blue), and ubiquitination (purple). B–D Distribution of different modification types of HM motifs. Panels (B–D) represent the overall distribution of identification results (B), animal virus results (C) and plant virus results (D), respectively. E Abundance of each HM motif in animal and plant viruses. The narrowest outer ring represents the different modification types of HM motifs, with green, yellow, purple and red representing methylation, acetylation, phosphorylation, and ubiquitination, respectively. The innermost ring shows plant virus results, and the outer ring shows animal virus results. F The distribution of all histone motifs used in this study. Different colors represent different modification types. G, H The normalized distribution of HM motifs across different histone subunits and different modifications.
To investigate the variation in HM types among different eukaryotic viruses, we analyzed the ratios of four HM types in animal and plant viruses. As shown in Fig. 2B, acetylation (44.89%) and methylation (35.80%) are the most common HM motif types, followed by phosphorylation (15.04%) and ubiquitination (4.26%). Similar trends were observed in the animal virus results (Fig. 2C), with acetylation being the most abundant (46.31%). However, plant viruses showed some divergence (Fig. 2D), with methylation as the dominant category (51.13%), while acetylation accounted for only 36.84% and ubiquitination a mere 0.75%. Contingency table analysis (Supplementary Table S2) confirmed that the distribution of different HM modification types differs significantly between animal and plant viruses. Since the quantity of motifs (encompassing both modification and histone subunit types) can significantly influence the outcomes, we conducted a ratio analysis of the results across different categories (Fig. 2F–H). Unlike the distribution pattern of the utilized motifs (Fig. 2F), our research indicates that the HM motifs in animal viruses are actually significantly concentrated on the H4 subunit and ubiquitination (Fig. 2G and H), rather than methylation. The HM motifs of plant viruses are still mainly distributed on the H2B subunit and methylation (Fig. 2G and H). And by comparing Fig. 2A and F, we found that the distribution of viral HM motifs on H3 histones shows a high degree of similarity.
To further explore the internal heterogeneity of HM motifs, the sequence differences within each modification type were analyzed and the result was visualized using a circular heatmap (Fig. 2E, Supplementary Table S3). This analysis revealed that certain HM motifs, such as VLKVF, KRKTV, and GVKKP, were more abundant in animal viruses than in plant viruses, while VYKVL was found to be relatively common in both (29 vs. 32).
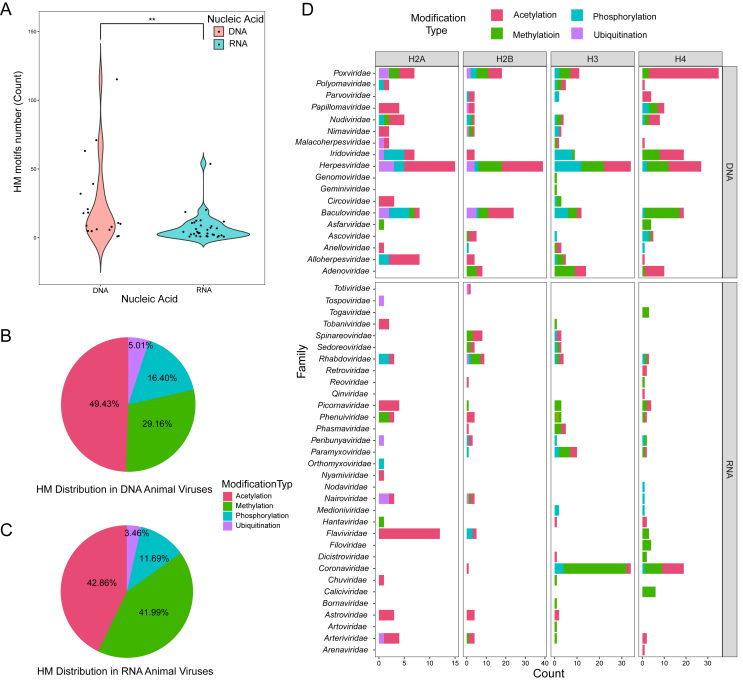
3.3. Discrepancy of HM motifs in DNA and RNA animal viruses
DNA and RNA animal viruses exhibit different strategies and characteristics during viral evolution, which are closely related to their biological properties. To investigate the distribution pattern of HM motifs in animal viruses, the number of HM motifs in both DNA and RNA animal viruses was statistically analyzed (Fig. 3A). By evaluating sample characteristics (Supplementary Tables S4–6, Supplementary Fig. S1), the differences in HM motif distribution between DNA and RNA animal viruses were analyzed using the Mann-Whitney U test. The P-value of 0.006 indicated significant statistical differences in the distribution of HM motifs between DNA and RNA animal viruses, further suggesting that DNA animal viruses are more likely to exhibit HM phenomena. Additionally, when studying the relationship between HM motifs of different histone subunits and animal virus nucleic acid types, we found no significant association, indicating that HM motifs from a particular subunit do not preferentially occur in either DNA or RNA animal viruses (Supplementary Table S7).
Fig. 3.
Distribution of HM motifs in DNA and RNA viruses. A Distribution of HM motif numbers in DNA and RNA animal viruses. Red and blue represent DNA and RNA animal viruses, respectively. Significant difference, ∗∗P < 0.01. B, C Distribution of different modification types of HM motifs. “B” represents the DNA virus results, and “C” represents the RNA virus results. D Distribution of HM motifs in different animal virus families. Different colors represent different types of histone modifications, with red, green, blue, and purple representing the acetylation, methylation, phosphorylation, and ubiquitination, respectively.
Analyzing the distribution of different HM modification types in DNA and RNA animal viruses revealed distinct patterns (Fig. 3B and C). Among DNA animal viruses (Fig. 3B), acetylation is the most common, accounting for 49.43%, followed by methylation (29.16%), phosphorylation (16.40%), and ubiquitination (5.01%). In RNA animal viruses (Fig. 3C), acetylation and methylation account for similar proportions (42.86% and 41.99%, respectively), with phosphorylation at 11.69% and ubiquitination at 3.46%. Contingency table analysis (Supplementary Table S8) confirmed that DNA and RNA animal viruses exhibit significant differences in the types of modifications present in HM motifs.
Furthermore, we investigated the variations in histone subunits for HM motifs across different animal virus families (Fig. 3D). The H2B, H3, and H4 histone subunits accounted for the majority of the matched HM motifs. However, certain virus families such as Poxviridae, Herpesviridae, Coronaviridae, Baculoviridae, and Iridoviridae, displayed a significantly higher number of HM motifs compared to other families. Some virus families exhibited a notable abundance of specific types of HM motifs, such as H4 acetylation in Poxviridae and H3 methylation in Coronaviridae. Most virus families have only one type of HM motif with a relatively low proportion. For example, Tospoviridae, Reoviridae, Orthomyxoviridae, and Genomoviridae only contain HM motifs of ubiquitination, acetylation, phosphorylation, and methylation, respectively. Only a few virus families, including Poxviridae, Iridoviridae, Herpesviridae, and Baculoviridae, have a widespread distribution across all four types of HM motif modifications.
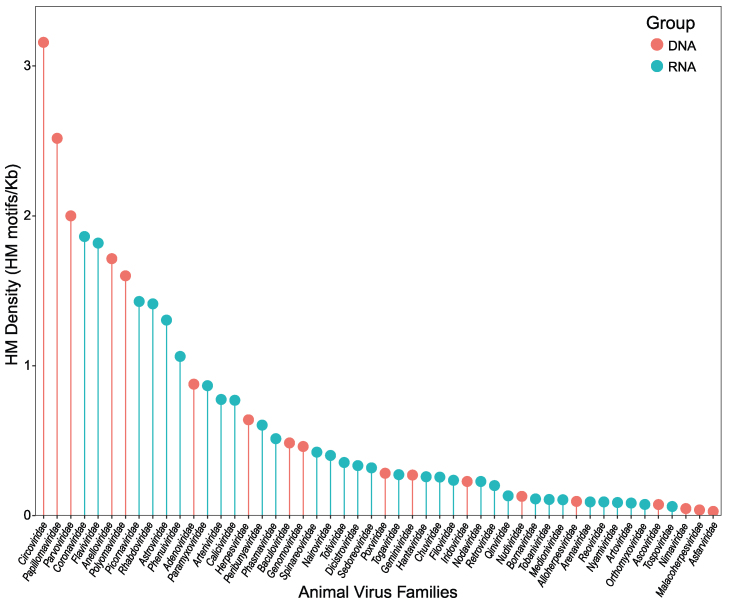
3.4. Different density of HM motifs in animal viruses
The abundance of HM can be significantly influenced by the length of the viral genome. To account for this, the density of HM is used as a measure of how frequently HM occurs in each virus family, rather than solely relying on the genome length of the virus itself. Fig. 4 shows that 11 virus families (5 DNA and 6 RNA animal virus families) have an HM density exceeding 1.00 HM motifs/Kb. Among these families, Circoviridae has the highest HM density at 3.16 HM motifs/Kb. However, none of the other families analyzed reached a HM density of 1.00 HM motifs/Kb. It is worth noting that, despite some families such as Herpesviridae, Baculoviridae, and Poxviridae matching a large number of HM motifs, their relatively large genome sizes contribute to a higher probability of successful matches, rather than indicating a preference for specific HM motifs or types.
Fig. 4.
Statistics of HM density in animal virus families. Red and blue represent DNA and RNA animal viruses, respectively.
3.5. Conservation and localization of HM motifs in Coronaviridae
Coronaviridae is a virus family containing many pathogenic and epidemic human viruses, including SARS-CoV-2. Our analysis focused on the HM states within the Coronaviridae family. The results revealed that the majority of these viruses belonged to the Betacoronavirus genus. A total of 19 viruses were identified, of which 15 belonged to the Betacoronavirus genus and 2 belonged to the Alphacoronavirus genus (Scotophilus bat coronavirus 512 and Swine enteric coronavirus). Two other coronaviruses had not been clearly classified yet (Rat coronavirus Parker and Bat coronavirus).
To further verify this phenomenon, we analyzed the distribution of HM motifs in different genera of Coronaviridae (Supplementary Table S9). As expected, the identified HM motifs were concentrated in the Betacoronavirus genus. Out of the 12 selected reference strains, 9 harbored HM motifs, with 6 of them located in ORF1ab. In contrast, in the Alphacoronavirus genus, only 3 reference strains have HM motifs. No HM motif was identified in the reference strains of Gammacoronavirus and Deltacoronavirus. These findings indicate that the HM motifs are primarily concentrated in the Betacoronavirus genus and may have evolved alongside these highly pathogenic viruses.
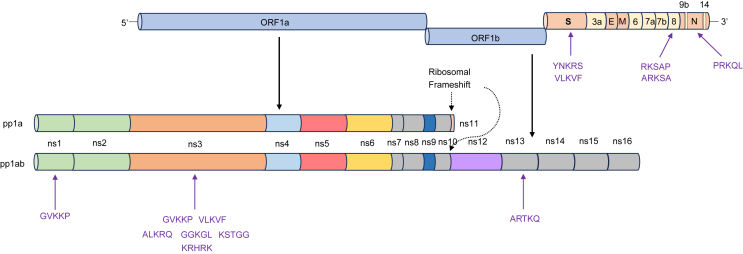
In Supplementary Table S9, we observed that most of the HM motifs were concentrated in ORF1ab and structural proteins. Specifically, 87.04% of the HM motifs were located in ORF1ab, with 31.92% residing in the NSP3 protein (Fig. 5). It is important to note that we detected two HM motifs (ALKRQ and GVKKP, respectively) on NSP3 in both MERS-CoV and Human coronavirus OC43, both of which belong to the Betacoronavirus genus. Furthermore, we also identified HM motifs on the S and N structural proteins (YNKRS, VLKVF, and PRKQL) but not on the E and M proteins.
Fig. 5.
Localization of the HM motifs matched by Coronaviridae. Note that the Coronaviridae genome structure is based on SARS-CoV-2 genome annotation information.
4. Discussion
In nature, viral mimicry of host proteins is a common strategy that facilitates viral replication or immune evasion (Elde and Malik, 2009). HM, a recently discovered form of viral mimicry, is likely the result of a long-term co-evolution between viruses and hosts. This adaptation allows viruses to effectively utilize cellular resources, enhancing their survival, replication, and propagation in host cells. HM motifs exist extensively in both animal and plant viruses (Schaefer et al., 2013; Tarakhovsky and Prinjha, 2018). However, it is still an important challenge to rapidly screen and identify the HM motifs in viruses. In response to this challenge we present HiScan, a software designed to address this problem.
HiScan is a bioinformatics pipeline developed to identify HM motifs in viral proteins. It utilizes a decision-tree model and a local comparison algorithm for optimization. HiScan has successfully predicted viral HM motifs that have been experimentally verified, such as those found in the influenza H3N2 subtype (Marazzi et al., 2012) and SARS-CoV-2 (Kee et al., 2022). However, due to the limited number of reported viral HM motifs, it is challenging to comprehensively evaluate the accuracy of HiScan and improve its performance by expanding the reference dataset. Therefore, if more relevant studies are reported in the future, different algorithms and models will be employed for optimization.
Using HiScan, we discovered a higher prevalence of HM motifs in animal viruses compared to plant viruses (Supplementary Table S2). One possible reason for this phenomenon is that the hosts of animal viruses are closer to humans in evolutionary distance. Since humans are mammals, their histone sequences are more similar to those of other animals. Consequently, the presence of human histone motifs in animal viruses may facilitate viral transmission between different species.
In addition, Fig. 2A and F show a strong similarity in the distribution of viral HM motifs on the H3 subunit. This similarity may be attributed to various factors, such as the evolutionary conservation of H3 its regulatory significance, chromatin accessibility, and viral manipulation strategies. However, further empirical investigation is required to validate these hypotheses and elucidate the underlying mechanisms. Furthermore, Fig. 2H reveals a significant difference in the distribution of HM motifs with different modification types in animal and plant viruses. In animal viruses, ubiquitination is dominant, whereas in plant viruses, methylation is distributed in more than half of the cases. This discrepancy could be due to variations in the in the host histone modification process of viruses. For example, although methylation occurs in both plant and animal cell proteins, the methyltransferases involved in the protein methylation process may differ greatly (Cheng et al., 2020; Jacob and Michaels, 2009). Additionally, we cannot exclude the possibility of bias resulting from the different functions of the same histone modification in animals and plants. For example, while H3K9me2 is involved in chromatin stabilization and gene silences in both animals and plants, in plants it primarily silences transposons (Jackson et al., 2004), whereas in animals, it also participates in certain antiviral responses like against HIV-1 (Suzuki et al., 2008). Therefore, these findings suggest that the distribution of viral HMs does not merely reflect the frequency of modification sites in host histones but rather indicates a potential evolutionary adaptation of viruses to target specific histone modifications or subunits. Moreover, animal and plant viruses may employ different strategies for manipulating the host's epigenetic machinery.
This study reveals that DNA animal viruses have a significantly higher abundance of HM motifs compared to RNA viruses (Fig. 3A). Additionally, the proportion of HM modification types is also different. This elevated HM in DNA animal viruses may be attributed to the replication characteristics of the virus itself. Unlike most RNA viruses that replicate exclusively in the cytoplasm, DNA viruses generally direct their DNA genomes into the host cell nucleus. In the nucleus, they can utilize the host cell's DNA replication machinery or other nuclear proteins to synthesize new viral nucleic acids (Challberg and Kelly, 1989). As a result, DNA viral proteins have more opportunities to interact with host nuclear histones.
From Fig. 3D, we found the preference of HM types among several virus families, including Poxviridae and Coronaviridae. These families harbor significantly high ratios of acetylation and methylation HM motifs. This suggests that certain HM motifs may play a role in viral life activities.
Fig. 4 illustrates that HM is a relatively common phenomenon in animal viruses. Interestingly, viruses with smaller genomes tend to have higher densities of HMs. Viruses have simpler genome lengths and structures compared to eukaryotes, and they employ various strategies to maximize their coding potential. For example, some viruses can generate multiple mRNAs from a single genome (Pellett et al., 2014), while others utilize programmed ribosomal frameshifting to produce multiple proteins from a single mRNA (Champagne et al., 2022). Similarly, the observed higher HM density in viruses with smaller genomes may represent an additional strategy to promote their replication.
Finally, our study focused on coronaviruses, specifically those that have caused recurring pandemics, like SARS-CoV-2, which is highly pathogenic. We discovered that the majority of HM motifs within the SARS-CoV-2 proteome were found in the ORF1ab, particularly in NSP3 (Fig. 5). NSP3 contains a crucial papain-like protease essential for the maturation of viral proteins and evasion of host immune responses (Moustaqil et al., 2021; Yuan et al., 2022). A smaller number of HM motifs were also located in structural proteins, such as the S and N proteins that are abundant in coronavirus particles (Yang and Rao, 2021). It is possible that these histone motifs help facilitate the folding of these two proteins to ensure the proper assembly of progeny virions. Additionally, a study has shown that certain SARS-CoV-2 structural proteins (S and N proteins) and non-structural proteins (NSP1 and NSP3) can traffic to the host cell nucleus, potentially disrupting the transport of host proteins in the nuclear pore (Lafon-Hughes, 2023). Moreover, HM motifs enriched in NSP3, S, and N proteins may also induce epigenetic changes downstream by mediating unfolded protein response similar to ORF8 of SARS-CoV-2.
When comparing the HM motifs distribution across different genera in Coronaviridae, we observed that HM motifs are mainly concentrated in Betacoronavirus, with a few in Alphacoronavirus, but none in Gammacoronavirus and Deltacoronavirus (Supplementary Table S9). This pattern may be associated with the mammalian host tropism displayed by Alphacoronavirus and Betacoronavirus (Cui et al., 2019). However, the exceptionally high abundance of HM motifs, particularly in highly pathogenic Betacoronavirus like SARS-CoV-2 and MERS-CoV, necessitates further experimental investigation to elucidate the underlying mechanisms.
As intracellular parasites, viruses are unable to independently encode all the necessary enzymes and proteins required for replication and propagation process (Louten, 2016; Summers, 2009). Therefore, during viral evolution, HM is a complex and ingenious evolutionary strategy for viruses (Schaefer et al., 2013). By HM, viruses can more effectively utilize host cellular resources and evade immune responses to facilitate their survival and replication (Yu et al., 2021). Understanding the mechanisms of viral HM will provide critical insights into viral infection and replication, offering important clues for developing more effective antiviral therapies. Furthermore, recent research has suggested that viruses may possess their own nucleosomes, hinting at the possibility that, through a long-term interaction with host cells, virus have gradually developed a sophisticated mechanism for packaging genetic material, similar to that of host cell (Bryson et al., 2022). Thus, we anticipate applying HiScan to identify potential viral histones. While this study has revealed intriguing viral HM distribution patterns and hypotheses regarding viral HM, numerous questions persist regarding the molecular mechanisms and evolutionary origins of this phenomenon. Further experimental validation using techniques like mutagenesis, structural biology, and comparative genomics, is essential for fully elucidating this intricate virus-host interaction and leveraging this knowledge for novel antiviral interventions.
5. Conclusions
Through the development and application of the bioinformatic tool HiScan, we performed a virome-wide screening to gain a comprehensive understanding of the distribution, characteristics, and potential roles of HM in viral proteins. Our findings provide valuable insights into the mechanisms of host-virus interactions and co-evolution, suggesting that viral HM may represent an important strategy for viruses to modulate the host cell epigenome and counteract antiviral responses. Further experimental validation is required to fully elucidate the functional implications of the identified HM motifs. Nonetheless, the development of HiScan and the virome-wide analysis of viral HM presented in this study offer a promising starting point for further exploration of this intriguing phenomenon.
Data availability
The source code of and releases of HiScan is available at https://github.com/xiaosheep01/hiscan.
A tutorial video of HiScan is presented in the Supplementary Video. The raw data in this research is available at Science Data Bank (https://doi.org/10.57760/sciencedb.12215).
Ethics statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author contributions
Yang Xiao: resources, data curation, formal analysis, investigation, methodology, software, validation, visualization, writing-original draft, writing-review & editing. Shuofeng Yuan: methodology, writing-review & editing. Ye Qiu: methodology, formal analysis, validation, writing-original draft, writing-review & editing. Xingyi Ge: conceptualization, funding acquisition, project administration, resources, supervision, validation, writing-review & editing.
Conflict of interest
All authors declare that there are no competing interests.
Acknowledgements
This study was jointly funded by the National Natural Science Foundation of China (No. U2002218, 32270170 and 81902070), the Fund of Hunan University (521119400156), and the Science and Technology Innovation Program of Hunan Province (2024RC1028).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2024.09.004.
Contributor Information
Ye Qiu, Email: qiuye@hnu.edu.cn.
Xing-Yi Ge, Email: xyge@hnu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Figure S1.
Quantile-Quantile (Q-Q) Plot of DNA and RNA animal viruses.
Supplementary Figure S2.
The tutorial video of HiScan.
The tutorial video of HiScan.
References
- Avgousti D.C., Della Fera A.N., Otter C.J., Herrmann C., Pancholi N.J., Weitzman M.D. Adenovirus core protein VII downregulates the DNA damage response on the host genome. J. Virol. 2017;91 doi: 10.1128/JVI.01089-17. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgousti D.C., Herrmann C., Kulej K., Pancholi N.J., Sekulic N., Petrescu J., Molden R.C., Blumenthal D., Paris A.J., Reyes E.D., et al. A core viral protein binds host nucleosomes to sequester immune danger signals. Nature. 2016;535:173–177. doi: 10.1038/nature18317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baisya D.R., Lonardi S. Prediction of histone post-translational modifications using deep learning. Bioinformatics. 2021;36:5610–5617. doi: 10.1093/bioinformatics/btaa1075. [DOI] [PubMed] [Google Scholar]
- Benveniste D., Sonntag H.J., Sanguinetti G., Sproul D. Transcription factor binding predicts histone modifications in human cell lines. Proc. Natl. Acad. Sci. U.S.A. 2014;111:13367–13372. doi: 10.1073/pnas.1412081111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell J.E., Zhou J., Ranalli T., Kobayashi R., Edmondson D.G., Roth S.Y., Allis C.D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Bryson T.D., De Ioannes P., Valencia-Sánchez M.I., Henikoff J.G., Talbert P.B., Lee R., La Scola B., Armache K.J., Henikoff S. A giant virus genome is densely packaged by stable nucleosomes within virions. Mol. Cell. 2022;82:4458–4470.e5. doi: 10.1016/j.molcel.2022.10.020. [DOI] [PubMed] [Google Scholar]
- Challberg M.D., Kelly T.J. Animal virus DNA replication. Annu. Rev. Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- Champagne J., Mordente K., Nagel R., Agami R. Slippy-Sloppy translation: a tale of programmed and induced-ribosomal frameshifting. Trends Genet. 2022;38:1123–1133. doi: 10.1016/j.tig.2022.05.009. [DOI] [PubMed] [Google Scholar]
- Cheng K., Xu Y., Yang C., Ouellette L., Niu L., Zhou X., Chu L., Zhuang F., Liu J., Wu H., et al. Histone tales: lysine methylation, a protagonist in Arabidopsis development. J. Exp. Bot. 2020;71:793–807. doi: 10.1093/jxb/erz435. [DOI] [PubMed] [Google Scholar]
- Cheung P., Tanner K.G., Cheung W.L., Sassone-Corsi P., Denu J.M., Allis C.D. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde N.C., Malik H.S. The evolutionary conundrum of pathogen mimicry. Nat. Rev. Microbiol. 2009;7:787–797. doi: 10.1038/nrmicro2222. [DOI] [PubMed] [Google Scholar]
- Hans F., Dimitrov S. Histone H3 phosphorylation and cell division. Oncogene. 2001;20:3021–3027. doi: 10.1038/sj.onc.1204326. [DOI] [PubMed] [Google Scholar]
- Jackson J.P., Johnson L., Jasencakova Z., Zhang X., PerezBurgos L., Singh P.B., Cheng X., Schubert I., Jenuwein T., Jacobsen S.E. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma. 2004;112:308–315. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- Jacob Y., Michaels S.D. H3K27me1 is E(z) in animals, but not in plants. Epigenetics. 2009;4:366–369. doi: 10.4161/epi.4.6.9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kasinath V., Beck C., Sauer P., Poepsel S., Kosmatka J., Faini M., Toso D., Aebersold R., Nogales E. JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Science. 2021;371 doi: 10.1126/science.abc3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee J., Thudium S., Renner D.M., Glastad K., Palozola K., Zhang Z., Li Y., Lan Y., Cesare J., Poleshko A., et al. SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry. Nature. 2022;610:381–388. doi: 10.1038/s41586-022-05282-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon-Hughes L. Towards understanding long COVID: SARS-CoV-2 strikes the host cell nucleus. Pathogens. 2023;12:806. doi: 10.3390/pathogens12060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.Y., Teyssier C., Strahl B.D., Stallcup M.R. Role of protein methylation in regulation of transcription. Endocr. Rev. 2005;26:147–170. doi: 10.1210/er.2004-0008. [DOI] [PubMed] [Google Scholar]
- Liu P., Wang X., Sun Y., Zhao H., Cheng F., Wang J., Yang F., Hu J., Zhang H., Wang C.-C., et al. SARS-CoV-2 ORF8 reshapes the ER through forming mixed disulfides with ER oxidoreductases. Redox Biol. 2022;54 doi: 10.1016/j.redox.2022.102388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louten J. Essential Human Virology. Elsevier Inc.; 2016. Virus Replication; pp. 49–70. [Google Scholar]
- Marazzi I., Ho J.S.Y., Kim J., Manicassamy B., Dewell S., Albrecht R.A., Seibert C.W., Schaefer U., Jeffrey K.L., Prinjha R.K., et al. Suppression of the antiviral response by an influenza histone mimic. Nature. 2012;483:428–433. doi: 10.1038/nature10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F., Penengo L. Histone ubiquitination: an integrative signaling platform in genome stability. Trends Genet. 2021;37:566–581. doi: 10.1016/j.tig.2020.12.005. [DOI] [PubMed] [Google Scholar]
- Merx J., Hintzen J.C.J., Proietti G., Elferink H., Wang Y., Porzberg M.R.B., Sondag D., Bilgin N., Park J., Mecinović J., et al. Investigation of in vitro histone H3 glycosylation using H3 tail peptides. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-21883-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustaqil M., Ollivier E., Chiu H.-P., Van Tol S., Rudolffi-Soto P., Stevens C., Bhumkar A., Hunter D.J.B., Freiberg A.N., Jacques D., et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg. Microb. Infect. 2021;10:178–195. doi: 10.1080/22221751.2020.1870414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak S.J., Corces V.G. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Pellett P.E., Mitra S., Holland T.C. Basics of virology. Handb. Clin. Neurol. 2014;123:45–66. doi: 10.1016/B978-0-444-53488-0.00002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C.L., Laniel M.A. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Ryu H.-Y., Zhao D., Li J., Su D., Hochstrasser M. Histone sumoylation promotes Set3 histone-deacetylase complex-mediated transcriptional regulation. Nucleic Acids Res. 2020;48:12151–12168. doi: 10.1093/nar/gkaa1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath Srihari C., Marazzi I., Yap K.L., Sampath Srinath C., Krutchinsky A.N., Mecklenbräuker I., Viale A., Rudensky E., Zhou M.-M., Chait B.T., et al. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol. Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Schaefer U., Ho J.S.Y., Prinjha R.K., Tarakhovsky A. The “histone mimicry” by pathogens. Cold Spring Harbor Symp. Quant. Biol. 2013;78:81–90. doi: 10.1101/sqb.2013.78.020339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvedunova M., Akhtar A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2022;23:329–349. doi: 10.1038/s41580-021-00441-y. [DOI] [PubMed] [Google Scholar]
- Summers W.C. third ed. Elsevier Inc.; 2009. Virus Infection. Encyclopedia of Microbiology; p. 546. [Google Scholar]
- Suzuki K., Juelich T., Lim H., Ishida T., Watanebe T., Cooper D.A., Rao S., Kelleher A.D. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J. Biol. Chem. 2008;283:23353–23363. doi: 10.1074/jbc.M709651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburri S., Lavarone E., Fernández-Pérez D., Conway E., Zanotti M., Manganaro D., Pasini D. Histone H2AK119 mono-ubiquitination is essential for polycomb-mediated transcriptional repression. Mol. Cell. 2020;77:840–856.e5. doi: 10.1016/j.molcel.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarakhovsky A., Prinjha R.K. Drawing on disorder: how viruses use histone mimicry to their advantage. J. Exp. Med. 2018;215:1777–1787. doi: 10.1084/jem.20180099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden E.J., Hoffmann N.A., Hicks C.W., Wolberger C. Mechanism of cross-talk between H2B ubiquitination and H3 methylation by Dot1L. Cell. 2019;176:1490–1501.e12. doi: 10.1016/j.cell.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Rao Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021;19:685–700. doi: 10.1038/s41579-021-00630-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q., Wu M., Liu Q., Lv H., Jiang R. DeepHistone: a deep learning approach to predicting histone modifications. BMC Genom. 2019;20:193. doi: 10.1186/s12864-019-5489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Kim M., Lee H., Kang G., Bedi K., Margulies K.B., Jain R., Nam K.-I., Kook H., Eom G.H. S-nitrosylation of histone deacetylase 2 by neuronal nitric oxide synthase as a mechanism of diastolic dysfunction. Circulation. 2021;143:1912–1925. doi: 10.1161/CIRCULATIONAHA.119.043578. [DOI] [PubMed] [Google Scholar]
- Yu Y., Wen H., Shi X. Histone mimics: more tales to read. Biochem. J. 2021;478:2789–2791. doi: 10.1042/BCJ20210357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Gao X., Tang K., Cai J.P., Hu M., Luo P., Wen L., Ye Z.W., Luo C., Tsang J.O., et al. Targeting papain-like protease for broad-spectrum coronavirus inhibition. Protein Cell. 2022;13:940–953. doi: 10.1007/s13238-022-00909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The tutorial video of HiScan.
Data Availability Statement
The source code of and releases of HiScan is available at https://github.com/xiaosheep01/hiscan.
A tutorial video of HiScan is presented in the Supplementary Video. The raw data in this research is available at Science Data Bank (https://doi.org/10.57760/sciencedb.12215).