Abstract
Anti-estrogen endocrine therapies greatly improve survival of estrogen receptor positive (ER + ) breast cancer. Unfortunately, about 30% of patients do not respond to endocrine therapies initially. We previously showed that deubiquitinase USP35 and ERα act in a positive feedback loop to promote the carcinogenesis of ER+ breast cancer although it is unclear whether USP35 regulates cell death in ER+ breast cancer. In this study, we uncovered that USP35 inhibited ferroptosis of ER+ breast cancer cells. Mechanistically, USP35 interacted with, deubiquitinated, and stabilized BRD4. Consequentially, BRD4 mediated USP35-induced SLC7A11 upregulation, inhibiting ferroptosis and promoting the growth of ER+ breast cancer cells. Furthermore, BRD4 inhibitor (+)-JQ-1 inhibited USP35-enhanced tumorigenesis in vivo. Our findings demonstrated that the USP35-BRD4-SLC7A11 axis contributes to the growth of ER+ breast cancer by inhibiting ferroptosis. Targeting USP35 together with ferroptosis inducer may represent a potential promising strategy for treating ER+ breast cancer that does not respond to endocrine therapies.
Subject terms: Breast cancer, Cell growth, Ubiquitylation
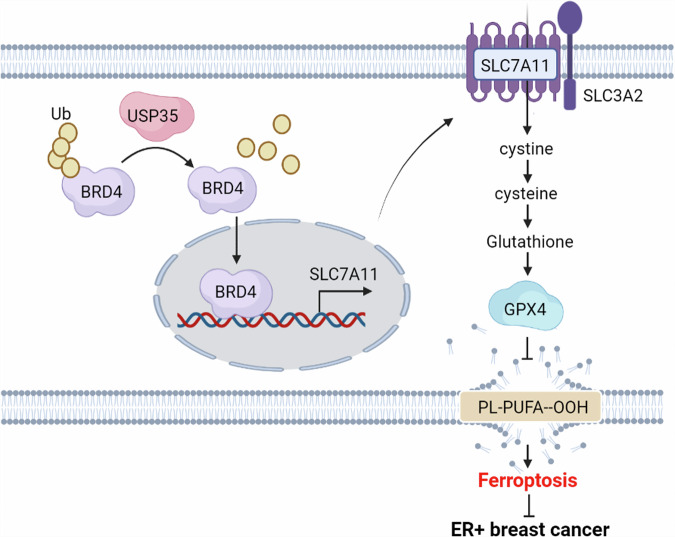
USP35 regulates ferroptosis in ER+ breast cancer. USP35 interacts with, deubiquitinates, and stabilizes BRD4, which mediates USP35-induced upregulation of SLC7A11. This process inhibits ferroptosis and promotes the growth of ER+ breast cancer cells.
Introduction
Breast cancer has surpassed lung cancer as the most commonly diagnosed cancer worldwide. Estrogen receptor positive (ER+) breast cancer accounts for 70% of breast cancer1. The initiation and progression of breast cancer, specifically ER+ breast cancer, have been found to be associated with long-term exposure to estrogen, both endogenous and exogenous2. Anti-estrogen based endocrine therapies, such as Tamoxifen, Fulvestrant, and letrozole, are widely utilized for the treatment of ER+ breast cancer. Although survival of ER+ breast cancer has been prolonged significantly largely due to the anti-estrogen based endocrine therapies3, about 30% of patients have primary resistance to endocrine therapy, and around 40% of them develop acquired resistance4. Uncovering novel mechanisms for the growth of ER+ breast cancer should help develop effective therapies for patients with ER+ breast cancer.
Ferroptosis is a type of regulated cell death that is caused by iron-dependent lipid peroxidation, which leads to the accumulation of excessive lipid peroxides (Lipid ROS) and results in the rupture of plasma membrane and cell demise5. Uptake of extracellular cystine mediated by amino-acid transporter solute carrier family 7 member 11 (SLC7A11; also known as xCT) is the primary way for most cancer cells to obtain cystine, which is the rate-limiting precursor for glutathione6. Glutathione peroxidase 4 (GPX4) inhibits ferroptosis by utilizing reduced glutathione (GSH) as a cofactor to detoxify lipid peroxidation7,8. Triple-negative breast cancer cells are more sensitive to ferroptosis inducers than ER+ breast cancer cells9. However, it is still unclear how ER+ breast cancer cells acquire resistance to ferroptotic insults. BRD4 is a member of the BET (bromodomain and extra terminal domain) protein family. It plays critical roles in cancer development through epigenetic regulation of gene transcription, which makes it a promising anti-cancer drug target10. It has been reported that BRD4 inhibitor (+)-JQ-1 or knockdown of BRD4 decreases GPX4 expression in triple-negative breast cancer cells11.
Human deubiquitylating enzymes (DUBs) are a group of isopeptidases that play crucial roles in the ubiqutin-proteasome system primarily by catalyzing the removal of ubiquitin moieties from substrate proteins, thereby regulating ubiquitylation-mediated cellular functions12. Recent evidence has implicated several deubiquitylating enzymes in the development and progression of breast cancer. For instance, USP14 has been shown to regulate cell cycle by stabilizing CDK1 in breast cancer13. USP35, the gene encoding Ubiquitin Specific Peptidase (USP35), is located on chromosome 11q14.1, where a small amplicon was amplified around 9% in breast cancer patients. USP35 plays a significant role in various types of cancers. USP35 is overexpressed in ovarian cancer with a poor prognosis. Through deubiquitination and inactivation of STING, USP35 acts as a negative regulator of the STING-TBK1-IRF3 pathway and the production of type I interferon14. Additionally, USP35 is involved in carcinogenesis of renal clear cell carcinoma by stabilizing NRF2 protein level and inhibiting cell death15.
We previously demonstrated that USP35 is upregulated in ER+ breast cancer compared with ER− breast cancer. In fact, estrogen stimulation increases USP35 protein level in ER+ breast cancer cells. USP35 promotes tumorigenesis of ER+ breast cancer via stabilizing and enhancing the transcriptional activity of ERα16. To further explore the mechanism of USP35 action in ER+ breast cancer, we investigated whether USP35 promotes the growth of ER+ breast cancer by inhibiting cell death. Our study uncovered a role of USP35 in regulating ferroptosis in ER+ breast cancer cells.
Results
USP35 promotes the growth of ER+ breast cancer cells by inhibiting ferroptosis
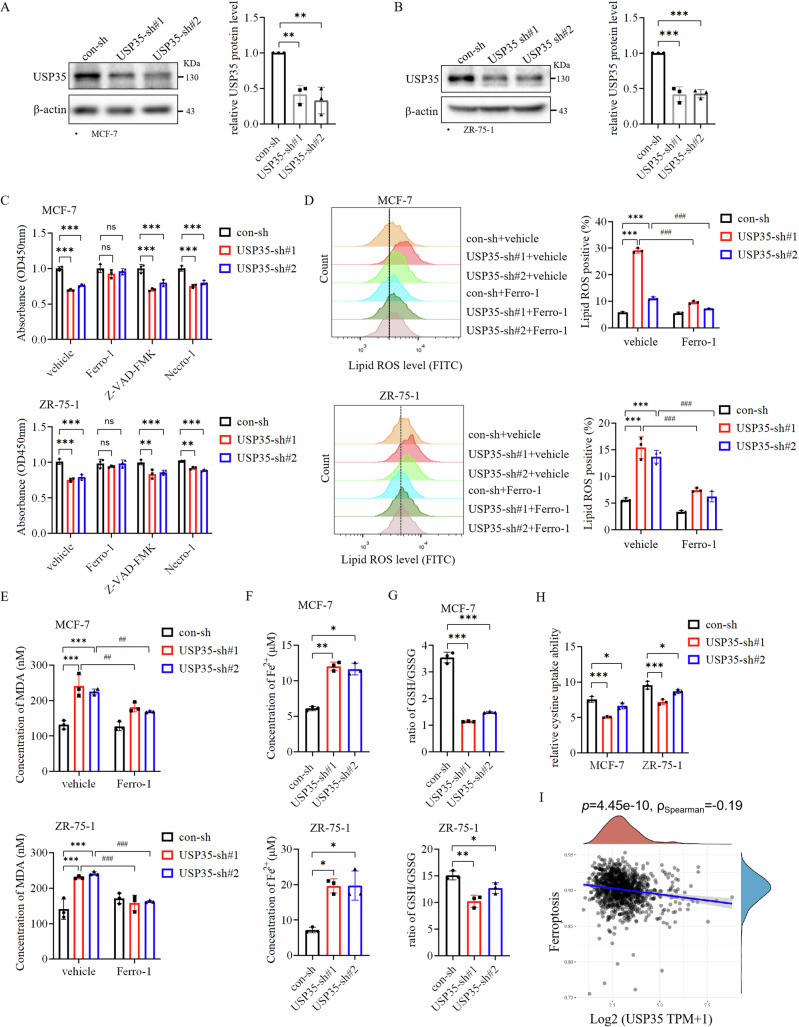
We previously showed that silencing USP35 inhibited the growth of ER+ breast cancer cells. Therefore, we investigated whether USP35 knockdown could induce cell death16. First, USP35 was knocked down by two different shRNAs (#1, #2) in two ER+ breast cancer cell lines, MCF-7 and ZR-75-1, respectively (Fig. 1A, B). Subsequently, MCF-7 and ZR-75-1 cells with USP35 knockdown were treated with inhibitors for different types of cell death including the ferroptosis inhibitor Ferrostatin-1 (Ferro-1), the apoptosis inhibitor Z-VAD-FMK, and the necrosis inhibitor Necrostatin-1. We found that only Ferro-1 could rescue cell viability from the knockdown of USP35 in both cell lines (Fig. 1C). Furthermore, silencing USP35 increased Lipid ROS levels in ER+ breast cancer cells, which were also attenuated by Ferro-1 treatment (Fig. 1D, Supplementary Fig. 1A). Knockdown of USP35 did not increase the level of cleaved PARP1, a marker of apoptosis (Supplementary Fig. 2A), nor did it affect the phosphorylation levels of RIP and MLKL (Supplementary Fig. 2B), which are the markers of necroptosis17. MDA is an end metabolite of Lipid ROS. Knockdown of USP35 also increased MDA levels, which were also decreased by Ferro-1 treatment (Fig. 1E). Iron overload from abnormal metabolism is the known trigger for ferroptosis. Interestingly, USP35 silencing enhanced Fe2+ accumulation in both breast cancer cell lines (Fig. 1F). GSH plays an indispensable role in preventing lipid peroxidation during ferroptosis8. We found that the ratio of GSH/GSSG and cystine uptake was significantly reduced in USP35 knockdown cells compared with control cells respectively (Fig. 1G, H). Consistent with the above results, our analysis of breast cancer cohort in TCGA database revealed that high expression level of USP35 was negatively correlated with ferroptosis (Fig. 1I), and it was only significant in ER+ breast cancer (Supplementary Fig. 3). Together, these results support the notion that USP35 promotes the growth of ER+ breast cancer cells by inhibiting ferroptosis.
Fig. 1. USP35 promotes the growth of ER+ breast cancer cells by inhibiting ferroptosis.
A, B USP35 was knocked down by two different shRNAs (#1, #2) in MCF-7 (A) and ZR-75-1 (B) cells (n = 3). USP35 levels were quantified and normalized to corresponding β-actin levels, which were presented as fold changes relative to the controls that were set to 1. C Ferroptosis inhibitor Ferrostatin-1 rescued the cell viability decrease induced by USP35 knockdown. MCF-7 and ZR-75-1 cells with control-shRNA and USP35-shRNAs (#1, #2) were treated with vehicle (DMSO), 5 μM Ferrostatin-1, 10 μM Z-VAD-FMK, and 2 μM Necrostatin-1 for 48 h before being subjected to CCK-8 assays (n = 3). D Ferro-1 prevented the increase in lipid peroxidation induced by USP35 knockdown. MCF-7 and ZR-75-1 cells with USP35 knockdown were seeded in twelve-well plates, treated with vehicle or 5 μM Ferro-1 for 24 h and stained with 5 μM C11-BODIPY followed by flow cytometry analysis. Quantitation and statistical analysis of the data were shown on the right (n = 3). E The same cells as (D) were subjected to analysis for the measurement of MDA concentration (n = 3). F, G Knockdown of USP35 increased Fe2+ content and decreased the ratio of GSH/GSSG in MCF-7 and ZR-75-1 cells (n = 3). H Knockdown of USP35 expression decreased cystine uptake. MCF-7 and ZR-75-1 cells with control-shRNA and USP35-shRNAs were subjected into cystine uptake assay (n = 3). I USP35 mRNA level was correlated with ferroptosis negatively in breast cancer. All experiments were performed at least three times. Data are shown as Mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ##P < 0.01, ###P < 0.001, ns no significance.
Knockdown of USP35 enhances the susceptibility of ER+ breast cancer cells to ferroptosis inducer
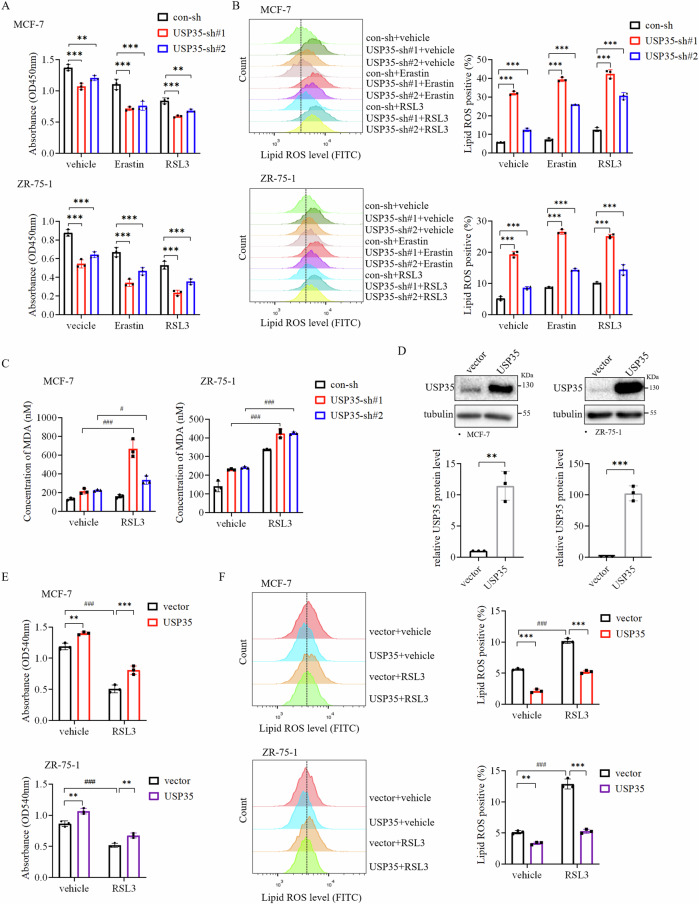
We further investigated whether USP35 was involved in regulating the sensitivity of ER+ breast cancer cells to ferroptosis inducer. Erastin and RSL3 represent the first two classes of ferroptosis-inducing compounds, inhibiting cystine uptake through the amino-acid transporter SLC7A11, and the enzyme activity of GPX4, respectively8,18. As expected, treatment with Erastin or RSL3 partially reduced cell viability in MCF-7 cells, which was enhanced significantly by silencing USP35 expression (Fig. 2A). Meanwhile, knockdown of USP35 dramatically enhanced Lipid ROS level induced by Erastin or RSL3 treatment in MCF-7 and ZR-75-1 cells (Fig. 2B, Supplementary Fig. 1B). Likewise, knockdown of USP35 also increased MDA concentration caused by RSL3 treatment (Fig. 2C). Conversely, USP35 overexpression in MCF-7 and ZR-75-1 cell (Fig. 2D) partially rescued cell growth inhibited by RSL3 treatment (Fig. 2E). Lastly, the increase in Lipid ROS induced by RSL3 treatment was dampened by USP35 overexpression (Fig. 2F, Supplementary Fig. 1C). These results indicate that USP35 silencing enhances ferroptosis and growth inhibition of ER+ breast cancer cells invoked by ferroptotic stimuli.
Fig. 2. Knockdown of USP35 enhances ferroptosis invoked by ferroptosis inducer in ER+ breast cancer cells.
A Ferroptosis inducers increased the growth inhibition of cells induced by USP35 knockdown. MCF-7 and ZR-75-1 cells with control-shRNA (sh) and USP35-shRNAs (sh#1, sh#2) were treated with Erastin (10 μM) or RSL3 (5 μM) for 2 d before being subjected to CCK-8 assay (n = 3). B Knockdown of USP35 expression enhanced lipid peroxidation caused by Erastin or RSL3 treatment. MCF-7 and ZR-75-1 cells with control-shRNA and USP35-shRNAs were treated with Erastin (10 μM) or RSL3 (5 μM) and stained with 5 μM C11-BODIPY followed by flow cytometry analysis after 24-h treatment. Quantitation and statistical analysis of the data were shown on the right (n = 3). C Knockdown of USP35 increased MDA concentration induced by RSL3 treatment (n = 3). D USP35 was overexpressed in MCF-7 and ZR-75-1 cells (n = 3). E USP35 overexpression increased cell growth inhibited by RSL3 treatment. MCF-7 and ZR-75-1 cells with USP35 overexpression were treated with RSL3 (5 μM) for 3 d before being subjected to the colony formation assay (n = 3). F USP35 overexpression reduced the elevated level of lipid peroxidation induced by RSL3 (n = 3). All experiments were performed at least three times. Data are shown as Mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.05, ###P < 0.001.
SLC7A11 mediates the effect of USP35 on ferroptosis
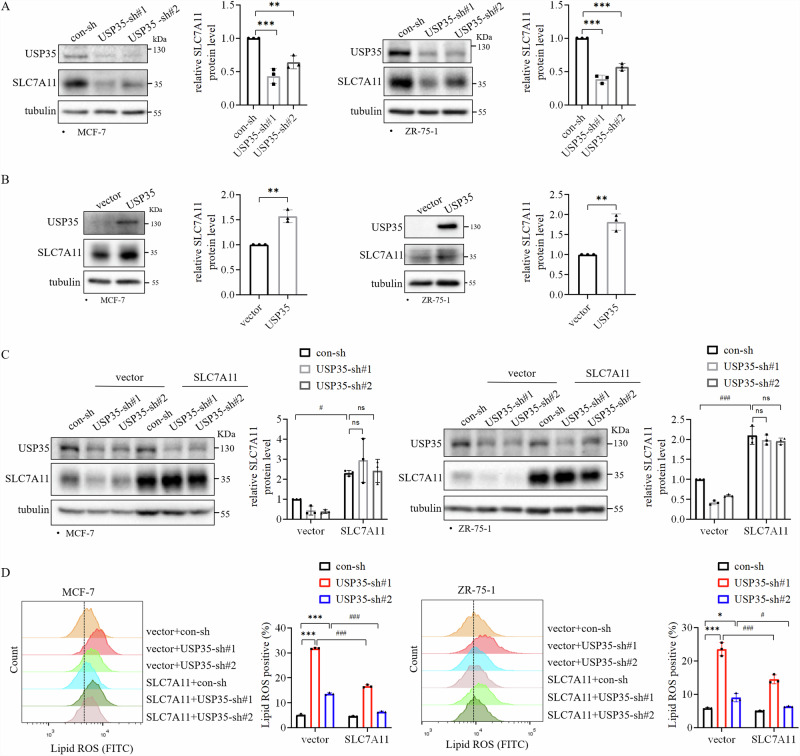
To understand the mechanism by which USP35 inhibits ferroptosis, we examined whether knockdown of USP35 affected the expressions of key ferroptosis regulators in two ER+ breast cancer cell lines. USP35 knockdown reduced the protein level of cystine/glutamate transporter SLC7A11 (Fig. 3A). Conversely, overexpression of USP35 also increased the SLC7A11 protein level (Fig. 3B), indicating that USP35 is important for SLC7A11 expression in ER+ breast cancer cells. However, MG132 (a proteasome inhibitor) treatment could not rescue the SLC7A11 protein level that was inhibited by USP35 knockdown (Supplementary Fig. 4), indicating that USP35 did not upregulate SLC7A11 through deubiquitination. To test whether SLC7A11 meditates the effect of USP35 on ferroptosis, we overexpressed exogenous SLC7A11 in USP35 knockdown cells (Fig. 3C). We found that SLC7A11 overexpression partially reduced the increase in Lipid ROS level induced by USP35 knockdown (Fig. 3D, Supplementary Fig. 1D). These results demonstrated that SLC7A11 is involved in inhibiting ferroptosis by USP35 in ER+ breast cancer cells.
Fig. 3. SLC7A11 mediates the effect of USP35 on ferroptosis.
Knockdown of USP35 decreased (A) and USP35 overexpression enhanced (B) SLC7A11 expression in ER+ breast cancer cell lines MCF-7 and ZR-75-1 (n = 3). C USP35 and SLC7A11 levels in the indicated MCF-7 and ZR-75-1 stable cell lines with vector and SLC7A11 overexpression together with control-sh or two different USP35-sh (#1, #2). Quantitation and statistical analysis of the data were shown on the left. SLC7A11 levels were quantified and normalized to corresponding β-actin levels, which were presented as fold changes relative to the controls that were set to 1 (n = 3). D Cells as indicated in (C) were stained with 5 μM C11-BODIPY followed by flow cytometry analysis (n = 3). All experiments were performed at least three times. Data are shown as Mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.05, ###P < 0.001.
USP35 regulates BRD4 protein level by interacting and deubiquitinating BRD4
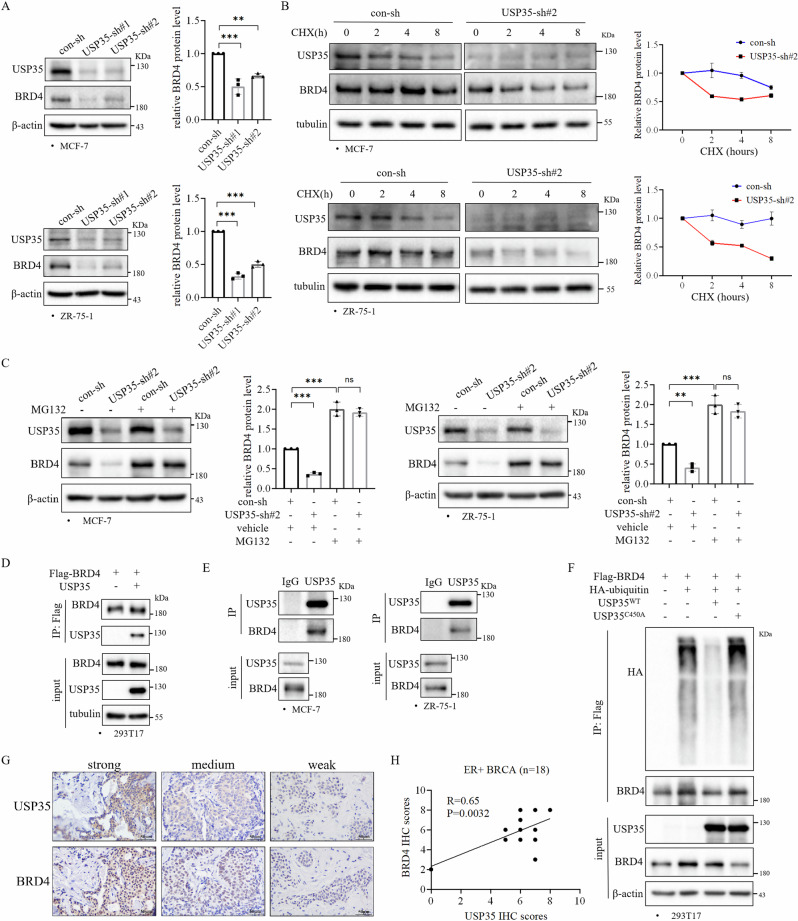
Sui et al. reported that BRD4 knockdown decreased the expression of SLC7A1111. Therefore, we investigated whether USP35 regulates BRD4 to affect SLC7A11 level. Knockdown of USP35 significantly decreased BRD4 protein level in ER+ breast cancer cells (Fig. 4A). However, knockdown of USP35 did not affect BRD4 levels in TNBC cell lines (Supplementary Fig. 5A). Considering that USP35 is a deubiquitinase, we suspected that USP35 may regulate the stability of the BRD4 protein. We treated the cells with cycloheximide, a protein synthesis inhibitor to block new protein synthesis. Knockdown of USP35 with shRNA accelerated BRD4 protein turnover in comparison to control-shRNA (Fig. 4B). Furthermore, MG132 treatment prevented the decrease of BRD4 protein induced by USP35 knockdown in both MCF-7 and ZR-75-1 cells (Fig. 4C).
Fig. 4. USP35 interacts with and deubiquitinates BRD4.
A Knockdown of USP35 decreased BRD4 protein level in ER+ breast cancer cells (n = 3). B Knockdown of USP35 accelerated BRD4 degradation. MCF-7 and ZR-75-1 cells expressing con-sh and USP35-sh#2 were treated with cycloheximide (CHX, 10 μM) for the indicated times (n = 3). C MG132 rescued the decreased BRD4 level caused by knockdown of USP35. MCF-7 and ZR-75-1 cells with con-sh or USP35-sh#2 were treated with MG132 (10 μM) for 8 h, and then were subjected to western blot analysis (n = 3). D USP35 interacted with BRD4. 293T17 cells were transiently cotransfected with vector or Flag-BRD4 plasmid together with vector or USP35 plasmid. Cell lysates were subjected to immunoprecipitation with anti-Flag antibody beads followed by immunoblotting with the indicated antibodies. E USP35 interacted with BRD4 in ER+ breast cancer cells. MCF-7 and ZR-75-1 cell lysates were subjected to immunoprecipitation with anti-USP35 antibodies and rabbit IgG (negative control). F USP35 promoted BRD4 deubiquitination. 293T17 cells were transiently cotransfected with Flag-BRD4 or HA-ubiquitin plasmid together with USP35WT or USP35C450A plasmid, and then were treated with MG132 (10 μM) for 8 h. Cell lysates were incubated with anti-Flag antibody and immunoblotted with the indicated antibodies. BRD4 levels were quantified and normalized to corresponding β-actin levels, which were presented as fold changes relative to the controls that were set to 1. G, H Immunohistochemistry analysis showed a positive correlation between USP35 and BRD4 protein levels in ER+ breast tumors (n = 18) (H). The representative images are shown on the right (G). Scale bar = 50 μm. All experiments were performed at least three times. Data are shown as Mean ± SEM. **P < 0.01, ***P < 0.001, ns no significance.
To further examine how USP35 regulates BRD4 protein level, we performed immunoprecipitation analysis to examine whether there is an interaction between USP35 and BRD4. Immunoprecipitation data showed that Flag-tagged BRD4 interacted with USP35 in transiently transfected 293T17 cells (Fig. 4D). In addition, results from immunoprecipitation experiments using anti-USP35 antibodies revealed that the endogenous USP35 and BRD4 can be coimmunoprecipitated in ER+ breast cancer cells (Fig. 4E). To test whether USP35 deubiquitinates BRD4, HA-ubiquitin and Flag-BRD4 plasmids were cotransfected with vector or vector expressing USP35WT or USP35C450A (the catalytic dead USP35 mutant) into 293T17 cells. Compared to vector control, USP35WT dramatically reduced the ubiquitination level of BRD4, whereas USP35C450A failed to affect ubiquitination of BRD4 (Fig. 4F). Lastly, we investigated the correlation between USP35 and BRD4 protein levels in ER+ breast cancer patients. Immunohistochemistry staining demonstrated that USP35 protein levels were positively associated with BRD4 protein levels in ER+ breast tumors (Fig. 4G, H). The examples of each staining scoring category (0/1/2/3) were shown in Supplementary Fig. 6. Interestingly, there was a trend towards a positive correlation between USP35 and BRD4 protein level in the CPTAC database, although it did not reach statistical significance (Supplementary Fig. 7A), considering that samples in the CPTAC database consist of different subtypes of breast cancer. Together, our results demonstrate that USP35 interacts with BRD4, and regulates BRD4 protein level by increasing BRD4 protein stability through deubiquitination in ER+ breast cancer cells.
BRD4 mediates USP35 upregulating SLC7A11 level and inhibiting ferroptosis in ER+ breast cancer cells
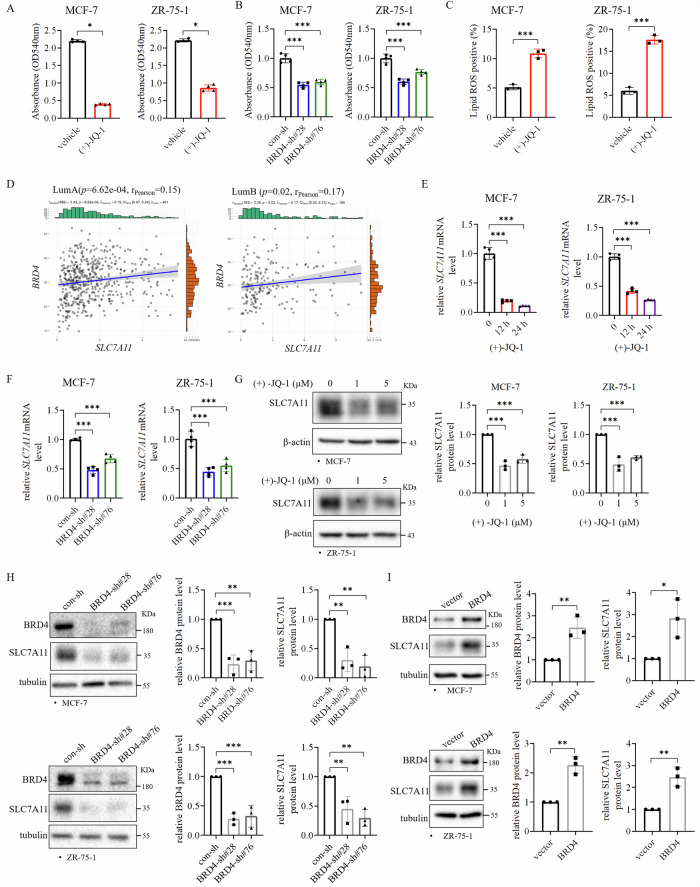
Since it is not known whether BRD4 regulates ferroptosis in ER+ breast cancer cells, we examined whether BRD4 inhibition affects ferroptosis in ER+ breast cancer cells. First, we examined the effects of BRD4 inhibitor (+)-JQ-1 or BRD4 knockdown on cell growth and Lipid ROS level in ER+ breast cancer cells. The results showed that (+)-JQ-1 inhibited the growth of MCF-7 and ZR-75-1 cells (Fig. 5A). Likewise, knockdown of BRD4 inhibited the cell growth of these two cell lines (Fig. 5B). In addition, (+)-JQ-1 treatment also increased the Lipid ROS levels in these cells (Fig. 5C, Supplementary Fig. 1E). Furthermore, knockdown of BRD4 enhanced ferroptosis of ER+ breast cancer cells induced by RSL3 (Supplementary Fig. 8, 1H). To understand how BRD4 regulates ferroptosis, we analyzed the correlation of BRD4 level with genes important in regulating ferroptosis in breast cancer cohort in TCGA database. Our analysis showed that BRD4 mRNA level was positively correlated with SLC7A11 mRNA level in luminal A and luminal B subtypes of breast cancer (Fig. 5D). Importantly, (+)-JQ-1 treatment or knockdown of BRD4 decreased SLC7A11 mRNA levels in MCF-7 and ZR-75-1 cells (Fig. 5E, F). Accordingly, inhibition of BRD4 with (+)-JQ-1 or BRD4 knockdown also decreased SLC7A11 protein level (Fig. 5G, H), whereas BRD4 overexpression enhanced SLC7A11 protein levels in these two cell lines (Fig. 5I). These results indicate that BRD4 regulates SLC7A11 mRNA level in ER+ breast cancer cells. SLC7A11 may mediate the effect of BRD4 on ferroptosis.
Fig. 5. BRD4 inhibits ferroptosis and regulates SLC7A11 expression in ER+ breast cancer cells.
A, B BRD4 inhibition decreased the growth of ER+ breast cancer cells. MCF-7 and ZR-75-1 cells treated with BRD4 inhibitor (+)-JQ-1 (5 μM) (A) for 3 d and expressing BRD4-shRNAs (B) were subjected to the colony formation assay (n = 4). C (+)-JQ-1 increased lipid peroxidation in ER+ breast cancer cells. MCF-7 and ZR-75-1 cells were treated with (+)-JQ-1 (5 μM) for 24 h and stained with 5 μM C11-BODIPY before being subjected to flow cytometry analysis (n = 3). D BRD4 mRNA level was positively correlated with SLC7A11 mRNA level in ER+ breast cancer. Analysis of breast cancer data in TCGA showed that BRD4 mRNA level was positively associated with SLC7A11 mRNA level in luminal A and B subtypes of breast cancer. E, F (+)-JQ-1 or knockdown of BRD4 decreased SLC7A11 mRNA level. MCF-7 and ZR-75-1 cells were treated with (+)-JQ-1 (5 μM) (E) for the indicated times, or knocked down with BRD4-shRNAs (sh#28 and sh#76) (F), and subjected to quantitative RT-PCR (n = 4). G, H BRD4 inhibition decreased SLC7A11 protein level. Cells treated with (+)-JQ-1 or with BRD4 knockdown were subjected to western blot analysis (n = 3). I BRD4 overexpression increased SLC7A11 level. MCF-7 and ZR-75-1 cells with vector alone and BRD4 overexpression were subjected to western blot analysis. BRD4 and SLC7A11 levels were quantified and normalized to corresponding β-actin levels, which were presented as fold changes relative to the controls that were set to 1 (n = 3). All experiments were performed at least three times. A Mann–Whitney test, (C) unpaired t test. Data are shown as Mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
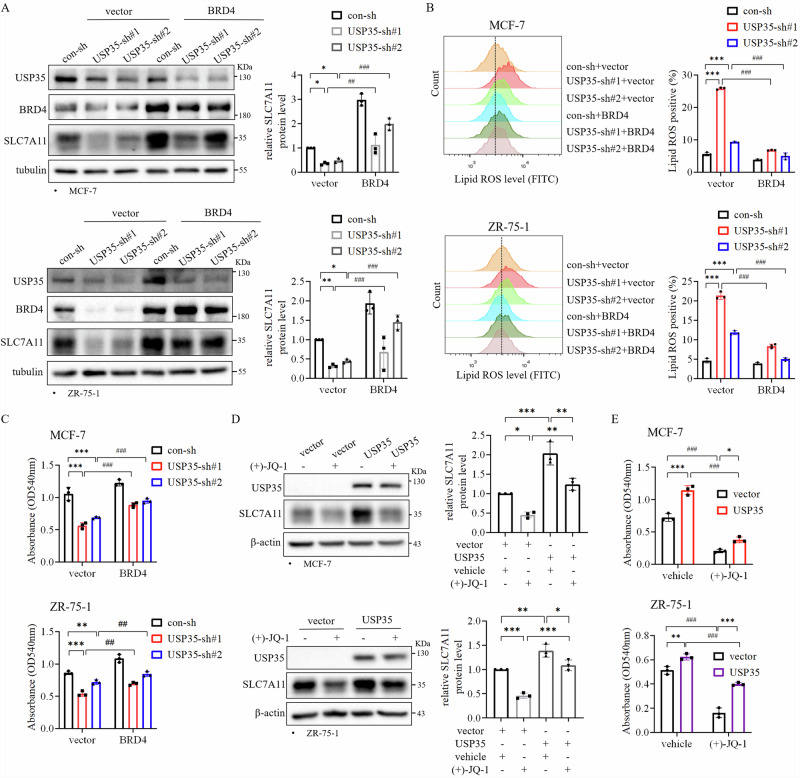
Considering that BRD4 interacts with USP35, and regulates SLC7A11 level, we hypothesized that USP35 via BRD4 regulates SLC7A11 level and ferroptosis in ER+ breast cancer cells. First, we examined whether overexpression of BRD4 can rescue the effects induced by USP35 knockdown. Consequently, we found that BRD4 overexpression rescued the decrease in SLC7A11 protein level induced by USP35 knockdown (Fig. 6A). In addition, BRD4 overexpression dramatically decreased Lipid ROS level induced by USP35 knockdown in two cell lines (Fig. 6B, Supplementary Fig. 1F). Furthermore, BRD4 overexpression also partially rescued the cell growth inhibition induced by USP35 knockdown (Fig. 6C). Conversely, although USP35 overexpression increased SLC7A11 protein level compared with vector control, BRD4 inhibitor (+)-JQ-1 treatment reduced the increase in SLC7A11 protein level induced by USP35 overexpression (Fig. 6D). Accordingly, (+)-JQ-1 treatment blocked the increase in cell growth induced by USP35 overexpression (Fig. 6E). These results indicated that USP35 regulates ferroptosis through the BRD4-SLC7A11 axis in ER+ breast cancer cells.
Fig. 6. USP35 via BRD4 regulates SLC7A11 level and ferroptosis in ER+ breast cancer cells.
A BRD4 overexpression rescued the decrease in SLC7A11 protein level induced by USP35 knockdown. MCF-7 and ZR-75-1 cells with vector alone and BRD4 overexpression together with control or USP35 knockdown were subjected to western blot analysis (n = 3). B BRD4 overexpression reduced the elevated level of lipid peroxides induced by USP35 knockdown. Cells as indicated in (A) were subjected to flow cytometry analysis. Quantitation and statistical analysis of the data were shown on the right (n = 3). C BRD4 overexpression rescued cell growth inhibited by USP35 knockdown. Cells as indicated in (A) were subjected to colony formation assay (n = 3). D BRD4 inhibitor reduced SLC7A11 level elevated by USP35 overexpression. Cells with USP35 overexpression were treated with (+)-JQ-1 (5 μM) for 24 h before being subjected to western blot analysis (n = 3). E BRD4 inhibitor dampened cell growth elevated by USP35 overexpression. MCF-7 and ZR-75-1 cells with vector alone and USP35 overexpression were treated with (+)-JQ-1 (5 μM) for 3 d before being subjected to the colony formation assay (n = 3). All experiments were performed at least three times. Data are shown as Mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ##P < 0.01, ###P < 0.001.
BRD4 inhibitor impairs the growth of ER+ breast tumors enhanced by USP35 overexpression
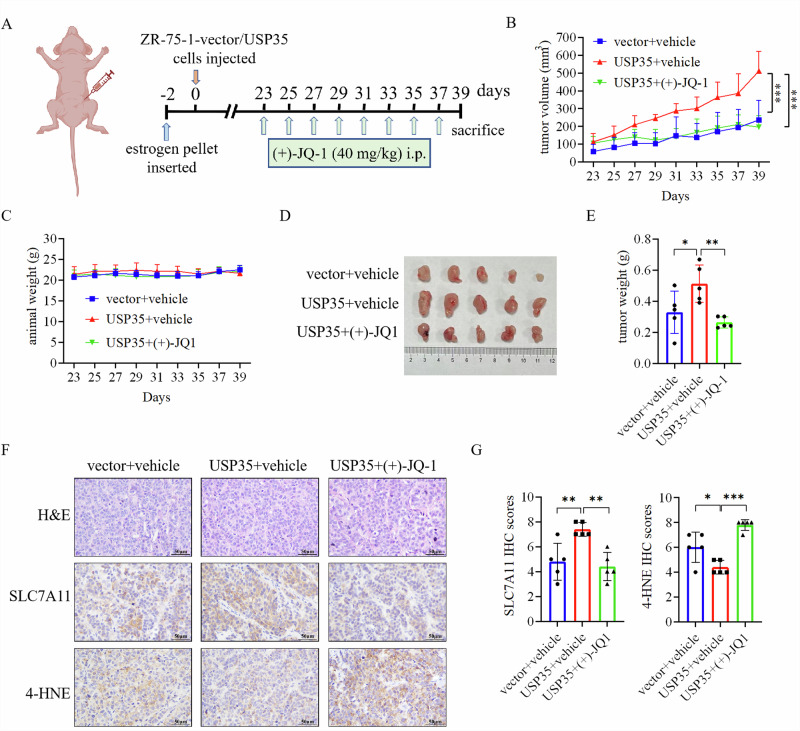
Since BRD4 mediates the effects of USP35 on inhibiting ferroptosis in ER+ breast cancer cells (Fig. 6), we tested the effect of BRD4 inhibition on USP35-enhanced growth of ER+ breast cancer cells in vivo. The efficacy of BRD4 inhibitor (+)-JQ-1 was evaluated in the ZR-75-1 xenograft model (Fig. 7A). Consistent with our previous finding16, USP35 overexpression promoted the growth of ER+ breast tumors, whereas (+)-JQ-1 inhibited the tumor growth increased by USP35 overexpression (Fig. 7B, D, E). (+)-JQ-1 treatment did not affect the weight of mice (Fig. 7C). Furthermore, immunohistochemistry analysis revealed that USP35 increased the levels of SLC7A11, whereas (+)-JQ-1 treatment significantly reduced the levels of SLC7A11, and increased the level of 4-HNE, a marker of ferroptosis, in ER+ breast tumors (Fig. 7F, G). These data support that BRD4 mediates the effects of USP35 in promoting tumor growth and inhibiting ferroptosis in ER+ breast tumors.
Fig. 7. BRD4 inhibitor impairs the growth of ER+ breast tumors enhanced by USP35 overexpression.
A ZR-75-1 cells stably expressing with vector or USP35 was injected orthotopically into the 4th mammary fat pad of the BABL/C nude female mice, which were treated with vehicle or BRD4 inhibitor (+)-JQ-1 at the indicated time points (n = 5). B The growth of the ZR-75-1 tumors in response to treatment with (+)-JQ-1 over the course of the experiment. The volumes of the tumors were recorded every 2 days, and the tumor growth curves were plotted. C The weights of mice were not different in three groups. Tumors were dissected out from the euthanized mice at the end of the experiment, photographed (D), and weighted (E) (n = 5). F Iamges depicting Hematoxylin and Eosin (H&E) staining and immunostaining for SLC7A11 and 4-HNE in ZR-75-1 tumors with the indicated treatments. Scale bar = 50 μm. G SLC7A11 and 4-HNE IHC staining intensities were quantified and scored (n = 5). Data are shown as Mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Our study uncovers a novel mechanism by which USP35 inhibits ferroptosis in ER+ breast cancer. Our results demonstrated that USP35 inhibits ferroptosis via the BRD4-SLC7A11 axis, maintaining glutathione level in ER+ breast cancer cells. Despite the fact that no ferroptosis inducer has been approved by the FDA for cancer treatment, published papers demonstrated that ER+ breast cancer cell lines are more resistant to ferroptosis inducers FIN56, Erastin, and RSL39,19. A recent study has begun to provide some mechanistic insight, showing that estrogen receptor (ER) transcriptionally upregulated the expression of the phospholipid-modifying enzyme MBOAT1, a suppressor of ferroptosis, in ER+ breast cancer cells. Ferroptosis induction combined with ER antagonist significantly inhibited the growth of ER+ breast tumors, even when tumors were resistant to single-agent endocrine therapy20. These studies together with our finding support the notion of targeting ferroptosis as a promising option for the treatment of ER+ breast cancer.
Extrinsic and intrinsic pathways contribute to the induction of ferroptosis. The extrinsic pathway is triggered by the inhibition of cell membrane cystine/glutamate transporter SLC7A11, resulting in the reduced intracellular level of glutathione21. Depleting glutathione can sensitize cells to ferroptosis22. A published report suggests that higher glutathione level in ER+ breast cancer may contribute to resistance to ferroptotic therapy23. TNBC is particularly susceptible to ferroptosis compared with other subtypes of breast cancer, with TNBC cells being more sensitive to ferroptosis inducers9. Despite this, the knockdown of USP35 resulted in increased Lipid ROS levels and inhibited the growth of TNBC cells MDA-MB-231 and SUM159PT (Supplementary Fig. 5B, 1G, 5C), whereas USP35 overexpression partially inhibited ferroptosis induced by Erastin in MDA-MB-231 cells (Supplementary Fig. 9). However, the knockdown of USP35 did not affect BRD4 protein level in TNBC cells (Supplementary Fig. 5A). Furthermore, USP35 did not interact with BRD4 in TNBC cells (Supplementary Fig. 5D), suggesting that USP35 inhibited ferroptosis independent of the BRD4-SLC7A11 axis in TNBC although BRD4 and SLC7A11 mRNA levels were positively correlated (Supplementary Fig. 7B), which was different from in ER+ breast cancer (Fig. 4). Our published report has demonstrated that USP35 is overexpressed especially in ER+ breast cancer. The results from this study suggest that USP35 overexpression in ER+ breast cancer may confer resistance to ferroptotic insult by elevating GSH level via the BRD4-SLC7A11 axis.
Our published work indicates that USP35 promotes the growth of ER+ breast cancer cells in vitro and in vivo, at least in part by affecting the G1 phase of cell cycle16. In this study, we revealed that USP35 regulates breast cancer cell growth at least in part through the inhibition of ferroptosis (Figs. 1, 2). What remains unclear is how knockdown of USP35 resulted in increased Fe2+ level (Fig. 1F). It has been reported that USP35 inhibits ferroptosis in lung cancer cells by promoting stability of ferroportin, an iron exporter24. However, we did not observe that USP35 affects the levels of ferroportin in ER+ breast cancer cells, which excludes the possibility that USP35 regulates cellular iron content through ferroportin.
Acting as an epigenetic reader, BRD4 recognizes acetylated lysine residues in histone tails of chromatin through its bromodomains and promotes gene transcription by recruiting positive transcription elongation factor b (P-TEFb), RNA polymerase II, and transcription factors to the promoter regions25–27. In addition, BRD4 can also enhance transcription by binding to the acetylated regions in enhancer and super enhancer. JQ-1, the first reported and most studied bromodomain inhibitor (BETi), interferes gene transcription by binding competitively to the acetyl-lysine recognition pocket in bromodomains28. Inhibition of BRD4 by the pan-bromodomain inhibitor such as JQ-1 has revealed the important roles of BRD4 in different cancer models28. Our results demonstrated that BRD4 inhibition with (+)-JQ-1 treatment significantly inhibited the ER+ breast tumors enhanced by USP35 overexpression, along with decrease of SLC7A11 protein level, and increase of 4-HNE protein levels (Fig. 7), supporting that BRD4 mediated USP35 inhibition of ferroptosis in ER+ breast tumors.
Reported studies have shown SLC7A11 expression can be regulated at translational and post-translational levels29–31. Our analysis showed that BRD4 regulated SLC7A11 mRNA level (Fig. 5). Interestingly, a published study showed that BRD4 is important for ERα−regulated gene transcription by occupying the estrogen response elements enriched for H3K27ac26. Our previous study demonstrated that USP35 via interaction with ERα enhances ERα mediated gene transcription in ER+ breast cancer cells16. Since USP35 also interacted with BRD4 (Fig. 4D, E), it is possible that BRD4 together with USP35 and/or ERα regulate the transcription of SLC7A11, conferring resistance to ferroptosis in ER+ breast cancer cells, which could explain why BRD4 partially rescued SLC7A11 expression in USP35 knockdown ER+ breast cancer cells (Fig. 6A). Future study is certainly required to address these questions. Our data also showed that USP35 inhibits ferroptosis independent of BRD4 in TNBC cells (Supplementary Fig. 5). USP35 may target one of the many other key proteins regulating ferroptosis in some subtypes of TNBC, which certainly deserve further investigation in the future.
Conclusions
Our study uncovers that USP35 interacts with and deubiquitinates BRD4, and enhances SLC7A11 expression in ER+ breast cancer. USP35 overexpression may contribute to the ferroptotic resistance of ER+ breast cancer cells (Fig. 8). Combining USP35 targeting with ferroptosis inducers and BET inhibitor may represent a potential therapy for ER+ breast cancer.
Fig. 8. A working model of USP35 regulating ferroptosis in ER+ breast cancer tumors.
USP35 interacts with and deubiquitinates BRD4, stabilizing the protein level of BRD4 that regulates SLC7A11. This interaction leads to USP35-invoked upregulating SLC7A11 expression, inhibiting ferroptosis, and promoting cell growth in ER+ breast cancer.
Materials and methods
Cell lines and reagents
The human breast cancer cell lines (MCF-7, ZR-75-1, and MDA-MB-231) and HEK293T-17 were obtained from the American Type Culture Collection (Maryland, USA). SUM159PT cells were obtained from Asterand Bioscience (Detroit, Michigan, USA). MCF-7 and HEK293T-17 cells were cultured in DMEM medium (Gibco, California, USA) with 5% FBS (ExCell Bio, Suzhou, China), supplemented with 1% penicillin/streptomycin (Beyotime Biotechnology, Jiangsu, China). ZR-75-1 cells were were cultured in DMEM with 10% FBS, supplemented with 1% penicillin/streptomycin. MDA-MB-231 cells were cultured in MEM medium (Gibco) containing 10% FBS, 1% penicillin/streptomycin, and 1.8 μg mL−1 insulin (Solarbio, Beijing, China). SUM159PT cells were cultured in DMEM/F-12 medium with 5% FBS, 1% penicillin/streptomycin, 5 μg mL−1 insulin, 1 μg mL−1 hydrocortisone. MCF-7, ZR-75-1, and HEK293T-17 cells were incubated in tissue culture incubators with an atmosphere of 7.5% CO2 at 37 °C, while MDA-MB-231 and SUM159PT cells were cultured with an atmosphere of 5% CO2. All cell lines were characterized by DNA fingerprinting and isozyme detection.
MG-132 was purchased from Selleck (Texas, USA). Cycloheximide was purchased from Sigma (Darmstadt, Germany).
Plasmids
pBABE-puro-USP35 and pLKO.1-shUSP35 plasmids were described in the previous study16. pcDNA4.0-Flag-BRD4 plasmid was a gift from Dr. Wenyu Wang, the Sixth Affiliated Hospital of Sun Yat-sen University, Guangzhou, China32. pBABE-puro-Flag-BRD4 plasmid was generated from pcDNA4.0-Flag-BRD4 by the seamless cloning kit (Vazyme, Jiangsu, China). DNA oligos for BRD4-shRNA#28 and BRD4-shRNA#76 (sequence information from Sigma) were subcloned into EcoRI and AgeI-digested pLKO.1 vector. BRD4-shRNA#28 sequence: 5′-CCAGAGTGATCTATTGTCAAT-3′, BRD4-shRNA#76 sequence: 5′-GCCAAATGTCTACACAGTATA-3′. DNA oligo for SLC7A11-sh#24 was subcloned into EcoRI and AgeI-digested pLKO.1 vector. SLC7A11 shRNA#24 sequence: 5′-GCACCCTTTGACAATGATAAT-3′. The sequences of the generated plasmids were verified by DNA sequencing (Qingke, Hangzhou, China).
Retrovirus/lentivirus production and viral infection
The production of retroviruses and lentiviruses in 293T17 cells and viral infection of breast cancer cells were performed as described33. ER+ breast cancer cells were incubated with retrovirus or lentiviral supernatants in the presence of 8 μg mL−1 of polybrene (Sigma) for 16 h. Twenty-six hours post-infection, stable pools of cells were selected with 1.2 μg mL−1 puromycin (Invitrogen, San Diego, USA) for MCF-7 cells and 1.4 μg mL−1 puromycin for ZR-75-1 cells for 3 days before being used for subsequent experiments.
RNA isolation and qPCR
Total RNAs were isolated from cells using RNA-easy isolation reagent. cDNA was synthesized from 1 μg of purified RNA using HiScript II Q RT SuperMix for qPCR (+gDNA wiper) kit, and qPCR was performed using ChamQ Universal qPCR Master Mix kit according to the manufacturer’s instructions. All these kits were purchased from Vazyme (Jiangsu, China). Primer sequences for qPCR are:
SLC7A11-Forward: 5′-TCTCCAAAGGAGGTTACCTGC-3′;
SLC7A11-Reverse: 5′-AGACTCCCCTCAGTAAAGTGAC-3′.
GAPDH-Forward: 5′-GCAAATTCCATGGCACCGT-3′;
GAPDH-Reverse: 5′-TCGCCCCACTTGATTTTGG-3′.
Measurement of lipid peroxidation
To assess lipid peroxidation, lipid reactive oxygen species (Lipid ROS) were detected by flow cytometry. Cells were stained with 5 μM BODIPY C11 (Thermo Fisher, Cat#D3861) for 30 min after the indicated treatments. After staining, labeled cells were washed, trypsinized, resuspended in PBS, and then subjected to flow cytometry analysis. The gating strategy for flow cytometry in analysis of lipid ROS is shown in Supplementary Fig. 10.
Measurement of malondialdehyde (MDA) level
MDA content was measured by the malondialdehyde (MDA) lipid peroxidation microplate assay (M496, DOJINDO) according to the manufacturers’ instructions. MDA levels were normalized to protein concentration.
Measurements of GSSG/GSH
The levels of GSSH and GSH were analyzed using the GSSG/GSH Quantification Kit II (G263, DOJINDO). GSH and GSSG levels were measured by lysing cells in GSH assay buffer according to the manufacturer’s instruction. The GSH and GSSG concentrations were calculated according to the standard curve.
Measurement of iron level
Total cellular iron level was measured using the colorimetric Total Iron Content Colorimetric Assay Kit (E1042, PPLYGEN). To measure total labile iron (Fe2+), cell supernatants were prepared according to the manufacturer’s instructions. The data were normalized to protein content.
Cystine uptake assay
Cystine uptake by cells was measured using the Cystine Uptake Assay Kit (UP05, DOJINDO, Japan). Cells were cultured in 96-well black plates before being subjected to cystine uptake analysis according to the manufacturer’s instructions. Black 96-well plate was used to prevent light from entering adjacent wells. Microplate readers were used to measure fluorescence.
Immunoprecipitation and immunoblotting
For immunoprecipitation, around 1 × 107 cells were washed twice in cold PBS and lysed in IP buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 20 mM beta-glycerol) plus 10 mM NaF, 2 mM Na3VO4, and protease inhibitor cocktail (Bimake, Houston, USA). Whole-cell lysates were clarified by centrifugation and incubated with the appropriate primary antibodies overnight at 4 °C. Antibody-bound proteins were precipitated with protein A agarose (REPLIGEN, Boston, USA), washed five times with lysis buffer and eluted in 1 × SDS sample loading buffer. Eluted proteins were separated by SDS PAGE, transferred to PVDF membranes (Millipore), immunoblotted with the appropriate primary antibodies, and horseradish peroxidase-conjugated secondary antibodies (Jackson Immunoresearch, USA), and detected by Enhanced Chemiluminescence. USP35 rabbit polyclonal antibodies for immunoprecipitation and immunoblot were generated against the His-tagged USP35 C-terminus (aa 856–1018) by HUABIO (Hangzhou, Zhejiang, China) and affinity purified using the His-tagged USP35 C-terminus as the affinity reagent. Negative control IgG was purchased from Invitrogen (USA).
For immunoblotting, rabbit polyclonal antibodies against BRD4 (Cat#ab289893, 1:1000). were purchased from Abcam (UK). Rabbit antibodies against SLC7A11 (Cat#12691, 1:1000), phospho-RIP (Ser166) (Cat#44590, 1:1000), phospho-MLKL (Ser345) (Cat#37333, 1:1000), and mouse monoclonal antibodies against α-tubulin (Cat#3873, 1:5000) and β-actin (Cat#3700, 1:5000) were from Cell Signaling Technology (USA). Rabbit antibody against HA (Cat#0039, 1:1000) was purchased from Beyotime (Jiangsu, China). Mouse antibody against PARP1 (Cat#66520, 1:1000) was purchased from Proteintech (Wuhan, China).
Immunohistochemistry (IHC) and hematoxylin-eosin (H&E)
Formalin-fixed and paraffin-embedded (FFPE) ER+ breast tumor samples were from the Pathology Department at The First Affiliated Hospital of Wenzhou Medical University of Wenzhou Medical University. FFPE breast tumor sections (4 μm thickness) were subjected to immunohistochemistry (IHC) as described16 with some modifications. Antigen retrieval was performed in 10 mM citrate buffer, pH 6.0 using pressure cooker (at 125 °C for 5 min). Rabbit anti-USP35 polyclonal antibodies (ab128592, Abcam) were used at 1:500 dilution. Rabbit anti-BRD4 polyclonal antibodies (cat#28486, Proteintech) were used at 1:1000 dilution. Rabbit anti-4-HNE polyclonal antibodies (ab46545, Abcam) were used at 1:100 dilution. IHC staining was scored by two pathologists blindly in the following way. The staining intensity was scored at four levels (0, 1, 2, and 3). The percentage of positive staining cells was scored at 6 levels (0% = 0, <1% = 1, 1–10% = 2, 11–33% = 3, 34–66% = 4, 67–100% = 5). IHC score was calculated as the sum of the score for staining intensity and the score for percentage of positive stained cells blindly by two individuals. For H&E staining, tissue sections were immersed in hematoxylin for 1 min, washed with water for 5 min, and stained with eosin for 20 s. The slides were mounted after dehydration.
Deubiquitination assay
pBABE-Vector or pBABE-USP35WT, USP35C450A plasmids were cotransfected with vectors expressing Flag-BRD4 and HA-ubiquitin into 293T17 cells. After forty-eight hours, cells were treated with MG132 (10 μM) for 8 h before being lysed in immunoprecipitation buffer. Clarified cell lysates were incubated with anti-Flag beads at 4 °C overnight. The anti-Flag beads were washed with lysis buffer for five times, and then eluted with 1X sample loading buffer for immunoblotting analysis.
Cell viability assay
MCF-7 cells (4000 per well) and ZR-75-1 cells (8000 per well) were seeded in 96-well plates, and allowed to adhere overnight. Cells were treated with different inhibitors, including 5 μΜ Ferrostatin-1, 10 μΜ Z-VAD-FMK, and 2 μΜ Necrostatin-1, and ferroptosis inducers including 10 μM Erastin and 5 μM RSL3 (Selleck) the following day. In accordance with the manufacturer’s instructions, the viability of the cells was assessed 48 h later using the CCK-8 Kit (Dojindo). The absorbance at 450 nm was represented as the relative cell viability.
Colony formation assay
Breast cancer cells MCF-7 (8000 per well) and ZR-75-1 (12,000 per well) were cultured in 24-well tissue culture plates and treated with DMSO or 5 μM ( + )-JQ-1 (BRD4 inhibitor) (Selleck) for 3 d. After one week of culturing, cells were fixed with 10% neutral formalin and stained with 0.5% crystal violet solution. The stained dye was eluted with 10% acetic acid and finally measured at 540 nm using a Varioskan flash microplate reader.
TCGA and CPTAC data access and analysis
The breast cancer dataset was downloaded from TCGA34. The correlation between USP35 mRNA and ferroptosis in breast cancer patients from TCGA was analyzed using the Assistant for Clinical Bioinformatics (https://www.aclbi.com/static/index.html#/). The correlation between USP35 and BRD4 protein level in breast cancer patients from CPTAC database was analyzed.
ZR-75-1 orthotopic xenograft model
To construct the orthotopic ER+ breast cancer mouse model, ZR-75-1 cells (4 × 106 cells) with vector and USP35 overexpression mixed 1:1 with Matrigel (Corning, cat#354234) were injected into the fourth mammary fat pads of female nude mice (Vital River Laboratories Animal Technology, Beijing, China). A 60-day release estrogen pellet (1.5 mg per pellet, Innovative Research of America) was implanted into mice 2 days before inoculations of ZR-75-1 cells. After the sizes of xenograft tumors reached around 100 mm3, mice were randomized into two groups (five mice per group), and were treated with vehicle or (+)-JQ-1 (40 mg kg−1) by intraperitoneal injection every other day. Tumor volumes were measured every 2 days and calculated according to the formula, volume = length × width2/2. Mice were euthanized at the end of the experiment, and xenografted tumors were dissected, weighed, and photographed. The animal study was approved by the Institutional Animal Care and Use Committee of Wenzhou Medical University.
Statistical analysis and reproducibility
In this study, statistical significance was calculated using the GraphPad Prism Software (Version 7). Statistical differences between two groups were analyzed by either nonparametric (Mann-Whitney test) or parametric Student’s t test, unless otherwise indicated, unpaired t-test was performed to evaluate statistical significance. Statistical differences among three groups were analyzed by One-way analysis of variance (ANOVA). Two-way ANOVA was used to compare differences among multiple comparisons. P < 0.05 was considered statistically significant. *, **, *** represented the difference within the group. #, ##, ### represented the difference between groups. All data presented were from experiments repeated at least three times with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Materials
Acknowledgements
We would like to thank Dr. Wenyu Wang (the Sixth Affiliated Hospital of Sun Yat-sen University, Guangzhou, China) for providing us the Flag-BRD4-pcDNA4.0 plasmid. We used the Biorender site (https://www.biorender.com) for figure drawing. This study was supported in part by grants from the Natural Science Foundation of Zhejiang Province, China (LQ23H160012, LZ23H160001, LTGY23H080005), National Natural Science Foundation of China (82302925, 82272702), Wenzhou Science and Technology Bureau of China (ZY2023021), and Medical and Health Science and Technology Plan Project of Zhejiang Province, China (2021KY1083), the Wenzhou Medical University Startup Funds, and the Key Discipline of Zhejiang Province in Medical Technology (First Class, Category A).
Author contributions
J.C., T.W., T.Z., Z.J., R.Y., J.Y., J.W., Q.C. performed experiments; J.C., Z.J., G.W., L.H., H.L., R.L., M.L., H.G. analyzed and interpreted the data; J.C., R.L., M.L., H.G. conceived and supervised the study; J.C., H.G. drafted and wrote the manuscript. All authors read and approved the final version of the manuscript for publication.
Peer review
Peer review information
Communications Biology thanks Paul Kennedy, Emma Saulters and Julie S Di Martino for their contribution to the peer review of this work. Primary Handling Editors: Georgios Giamas and Mengtan Xing. A peer review file is available.
Data availability
All data generated or analysed during this study are included in this published article. The uncropped western blot images for all of the western blot figures in the manuscript are also included in the Supplementary information. The numeral source data for all graphs in the manuscript are provided in Supplementary Data 1. Materials generated in this study will be freely available to any researcher upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethics statements
All human breast tumor samples were obtained from patients with informed consent. The current study (protocol# 2019006) was approved by the Institutional Review Board for human study of Wenzhou Medical University and conducted according to the principles expressed in the Helsinki Declaration. The animal study (protocol# wydw2023-0484) was approved by the Institutional Animal Care and Use Committee of Wenzhou Medical University and conducted in compliance with relevant local guidelines.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiawei Cao, Email: jiaweicao@wmu.edu.cn.
Rixu Lin, Email: linrixu316@163.com.
Min Liu, Email: rahosjoint@163.com.
Haihua Gu, Email: haihuagu@wmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-025-07513-1.
References
- 1.Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Sun, Y. S. et al. Risk factors and preventions of breast cancer. Int. J. Biol. Sci.13, 1387–1397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stopeck, A. T. et al. The role of targeted therapy and biomarkers in breast cancer treatment. Clin. Exp. Metastasis29, 807–819 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Nakao, M., Fujiwara, S. & Iwase, H. Cancer navigation strategy for endocrine therapy-resistant breast tumors. Trends Cancer4, 404–407 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Stockwell, B. R. Ferroptosis turns 10: emerging mechanisms, physiological functions, and therapeutic applications. Cell185, 2401–2421 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banjac, A. et al. The cystine/cysteine cycle: a redox cycle regulating susceptibility versus resistance to cell death. Oncogene27, 1618–1628 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Friedmann Angeli, J. P. et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol.16, 1180–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang, W. S. et al. Regulation of ferroptotic cancer cell death by GPX4. Cell156, 317–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verma, N. et al. Synthetic lethal combination targeting BET uncovered intrinsic susceptibility of TNBC to ferroptosis. Sci. Adv.610.1126/sciadv.aba8968 (2020). [DOI] [PMC free article] [PubMed]
- 10.Stathis, A. & Bertoni, F. BET proteins as targets for anticancer treatment. Cancer Discov.8, 24–36 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Sui, S. et al. Ferritinophagy is required for the induction of ferroptosis by the bromodomain protein BRD4 inhibitor (+)-JQ1 in cancer cells. Cell Death Dis.10, 331 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clague, M. J. et al. Deubiquitylases from genes to organism. Physiol. Rev.93, 1289–1315 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Liu, Y. et al. USP14 regulates cell cycle progression through deubiquitinating CDK1 in breast cancer. Acta Biochim. Biophys. Sin.54, 1610–1618 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang, J. et al. Deubiquitinase USP35 restrains STING-mediated interferon signaling in ovarian cancer. Cell Death Differ.28, 139–155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, S. et al. The deubiquitylating enzyme USP35 restricts regulated cell death to promote survival of renal clear cell carcinoma. Cell Death Differ.30, 1757–1770 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao, J. et al. USP35, regulated by estrogen and AKT, promotes breast tumorigenesis by stabilizing and enhancing transcriptional activity of estrogen receptor alpha. Cell Death Dis.12, 619 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mompean, M. et al. The structure of the necrosome RIPK1-RIPK3 core, a human hetero-amyloid signaling complex. Cell173, 1244–1253.e1210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon, S. J. et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife3, e02523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi, J., Zhu, J., Wu, J., Thompson, C. B. & Jiang, X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc. Natl Acad. Sci. USA117, 31189–31197 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang, D. et al. Ferroptosis surveillance independent of GPX4 and differentially regulated by sex hormones. Cell186, 2748–2764.e2722 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang, D. & Kroemer, G. Ferroptosis. Curr. Biol.30, R1292–R1297 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Cao, J. Y. et al. A genome-wide haploid genetic screen identifies regulators of glutathione abundance and ferroptosis sensitivity. Cell Rep.26, 1544–1556.e1548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jardim, B. V. et al. Glutathione and glutathione peroxidase expression in breast cancer: an immunohistochemical and molecular study. Oncol. Rep.30, 1119–1128 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Tang, Z. et al. Deubiquitinase USP35 modulates ferroptosis in lung cancer via targeting ferroportin. Clin. Transl. Med11, e390 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delmore, J. E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell146, 904–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagarajan, S. et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep.8, 460–469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi, J. et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell25, 210–225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature468, 1067–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, T., Jiang, L., Tavana, O. & Gu, W. The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res.79, 1913–1924 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, W. et al. RBMS1 regulates lung cancer ferroptosis through translational control of SLC7A11. J. Clin. Invest.13110.1172/JCI152067 (2021). [DOI] [PMC free article] [PubMed]
- 31.Yan, B. et al. The ubiquitin-specific protease 5 mediated deubiquitination of LSH links metabolic regulation of ferroptosis to hepatocellular carcinoma progression. MedComm (2020)4, e337 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, W. et al. Stromal induction of BRD4 phosphorylation results in chromatin remodeling and BET inhibitor resistance in colorectal cancer. Nat. Commun.12, 4441 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang, Z. et al. Gab2 promotes cancer stem cell like properties and metastatic growth of ovarian cancer via downregulation of miR-200c. Exp. Cell Res.382, 111462 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article. The uncropped western blot images for all of the western blot figures in the manuscript are also included in the Supplementary information. The numeral source data for all graphs in the manuscript are provided in Supplementary Data 1. Materials generated in this study will be freely available to any researcher upon reasonable request.