Abstract
Polyketide synthases (PKSs) are crucial multidomain enzymes in diverse natural product biosynthesis. Parrots use a type I PKS to produce a unique pigment called psittacofulvin in their feathers. In domesticated budgerigars and lovebirds, the same amino acid substitution (R644W) within malonyl/acetyltransferase (MAT) domain of this enzyme has been shown to cause the blue phenotype with no psittacofulvin pigmentation, proposing a strong evolutionary constraint on the mechanism. Here, we identified seven previously unreported variants in PKS associated with defective psittacofulvin production in four diverse species, including three nonsense mutations. Intriguingly, three of the remaining nonsynonymous substitutions reside within the ketoacyl synthase (KS) domain, whereas one at MAT domain. The heterologous expression of these PKS variants in yeast confirmed complete or partial loss of psittacofulvin production. These findings establish PKS as a functionally conserved key-enzyme determining psittacofulvin-based hues among diverse parrots, highlighting multiple conserved domains essential for the PKS function.
Subject terms: Zoology, Genetics, Mutation
Mutations in polyketide synthase (PKS) disrupt psittacofulvin pigment production in parrots, altering feather hues. Seven unique variants in multiple conserved domains reveal a shared mechanism across four species and underscore PKS’s multidomain role in parrot coloration.
Introduction
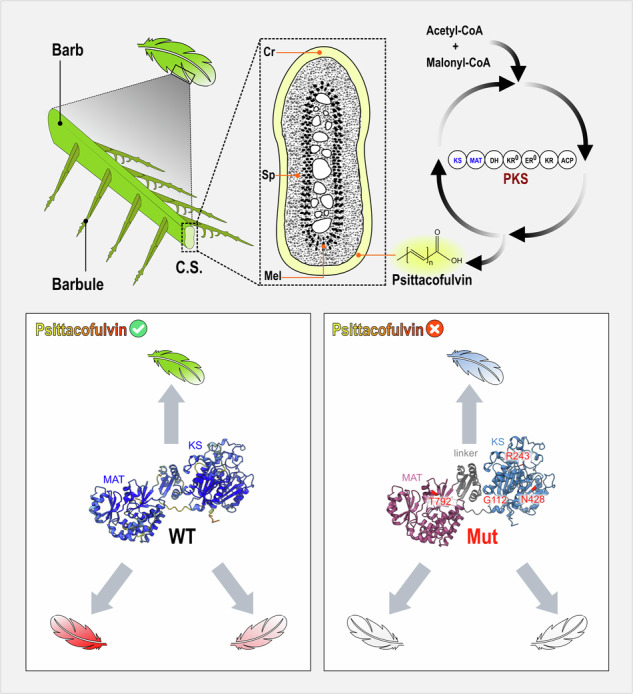
Plumage coloration in birds is a marvel of nature, showcasing a stunning palette ranging from vibrant hues to subtle tones. The colors, derived from pigments or structural properties in feathers, serve various purposes1. The pigment-based color diversity, particularly the bright yellows to reds observed in many species, is attributed to carotenoid pigments obtained from their diet2. But, in the case of parrots (order Psittaciformes), a distinct approach to achieving such vibrant hues has been recognized for over a century3. Despite assimilating dietary carotenoids into their bloodstream, parrots do not deposit them in feathers. Instead, they employ a unique class of polyene pigments called psittacofulvins4. Although the purpose of this unique phenomenon is not clear, psittacofulvins have likely played a role in the evolutionary success of parrots5. Various adaptive functions have been proposed, such as mate choice6 and protection of feathers from bacterial degradation7. Still, during a long period of time, little was known about the biosynthetic origin or the genetic basis of this pigment until a pioneering study was conducted by Cooke and his coworkers in the last decade8.
Cooke et al. shed light on the biochemistry and evolution of psittacofulvins offering valuable comparisons to carotenoid-based coloration. In captive budgerigars (Melopsittacus undulatus), a type I polyketide synthase (MuPKS) enzyme is responsible for polyene chain elongation resulting in psittacofulvin production. Since the initial discovery, PKS enzymes have been found in many animal genomes9, but only a few have undergone biochemical characterization10. Being the first characterized vertebrate candidate, MuPKS belongs to the class of iterative PKSs that catalyzes multiple chain elongation cycles using the same set of enzymatic domains to produce a diverse range of natural products known as polyketides9. Wild-type budgerigars of both sexes have yellow feathers containing yellow psittacofulvins and green feathers with a combination of yellow psittacofulvins and blue color, generated by feather microstructures8. A specific amino acid substitution (R644W) in the MuPKS gene of budgerigars disrupts the psittacofulvin synthesis pathway, resulting in a blue phenotype. Thus, as budgerigars with the blue phenotype lack yellow psittacofulvins production, feathers that are yellow in the wild-type are white in blue birds, and feathers that are green in the wild-type are blue. Interestingly, this blue phenotype is extraordinarily common among artificially selected captive-bred populations across the order Psittaciformes, inherited similarly as an autosomal recessive allele like budgerigars. This is primarily due to selective breeding, which plays a crucial role in preserving the blue phenotype across generations in various parrot species. Recently, the same amino acid mutation in PKS as found in budgerigars has been reported in two species of lovebirds: Fischer’s lovebird (Agapornis fischeri) and yellow-collared lovebird (Agapornis personatus) exhibiting the blue phenotype11. The presence of identical gene mutations in both lovebirds and budgerigars has been suggested as evidence of significant evolutionary constraint on psittacofulvin-based coloration11, that contributes to a range of hues from yellow to red gradient including pink and orange seen in various parrot species12. The latest study in the dusky lory (Pseudeos fuscata) identified the enzyme ALDH3A2 as a key player in balancing these yellow and red pigments, suggesting the involvement of additional factors beyond PKS in determining the final psittacofulvin hue13. However, given the widespread presence of the blue phenotype across various parrot species, a comprehensive study is needed to explore the conservation of genetic control over psittacofulvin coloration throughout the order Psittaciformes. We therefore decided to investigate whether PKS homologs have similar roles in blue variants across different members of Psittaciformes.
To test this, we chose four popular pet species from the order Psittaciformes14, showing different psittacofulvin-based hues in their feathers: Rose-ringed parakeet (Psittacula krameri), Alexandrine parakeet (Psittacula eupatria), and Moluccan Eclectus (Eclectus roratus) from the family psittaculidae, and Galah cockatoo (Eolophus roseicapilla) from the family cacatuidae. Our study revealed seven previously uncharacterized de novo variants in PKS including three nonsense variants and four nonsynonymous substitutions. Three of the substitutions reside within the important ketoacyl synthase (KS) domain, while only one was found to be in malonyl/acetyltransferase (MAT) domain. All the substitutions displayed complete or partial loss of pigmentation upon heterologous expression in yeast, establishing their significant role causing blue phenotype. This is the first evidence of variations in the ketoacyl synthase (KS) domain that severely disrupt PKS function, similar to effects previously observed in the MAT domain of budgerigars and lovebirds8,11. These findings highlight the significance of multiple domains in shaping the final pigment phenotype in parrot plumage. Besides, as polyketides provide a wide range of beneficial properties, polyketide synthases are essential to produce a variety of bioactive compounds with significant pharmaceutical, agricultural, and ecological importance15. Studying these enzymes and the polyketides they produce provides insights into eukaryotic biology and offers opportunities for biotechnological innovation. Our current study not only presents significant insights into PKS evolution across taxa but also helps to expand the knowledge underlying secondary metabolism and ecological interactions through collaborating functional domains.
Results
Phenotypic characterization of the blue phenotypes belonging to diverse parrot species
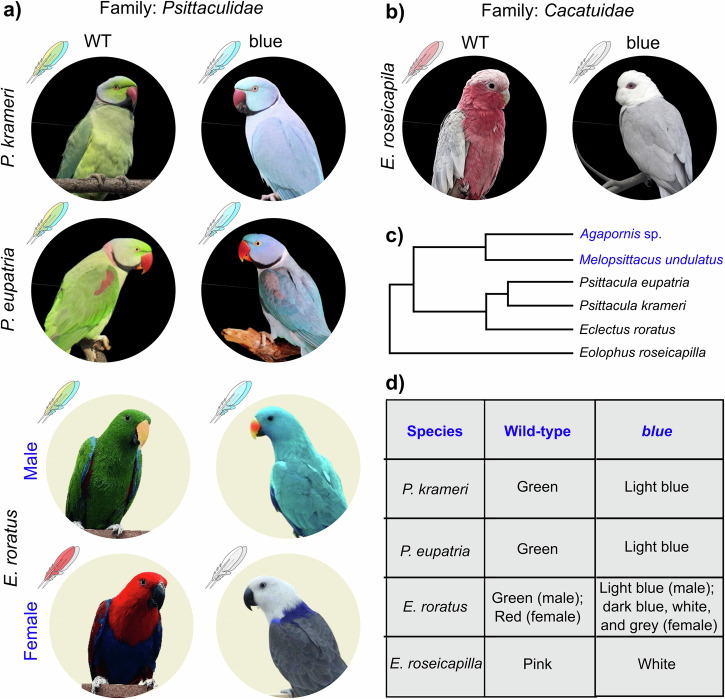
Psittacofulvin-based hues range from yellow to red and the absence of any of these colors is designated as the blue phenotype among breeders. In the following segment, we have characterized variations in blue phenotypes among diverse parrot species. In the blue phenotype of predominantly green parrots, such as P. krameri and P. eupatria, the wild-type green becomes light blue due to the absence of yellow pigment (Fig. 1a). Whereas sexually dichromatic E. roratus, previously shown to use yellow and red psittacofulvins to color their feathers16, displays the blue phenotype differently (Fig. 1a). In E. roratus, males are mostly bright green, and females are predominantly bright red. In this case, blue males display a uniform light blue color across the entire body while females carrying the blue phenotype appear mostly white throughout the body with dark blue on their back and gray on the wings (Fig. 1a). Both sexes of E. roseicapilla have a pale silver to gray back, a pale gray rump, a pink face, and a light pink mobile crest. However, E. roseicapilla individuals carrying the blue phenotype display a white body lacking the pink pigment with unchanged gray wings (Fig. 1b). Key features of overall plumage coloration in targeted species are summarized in Table S1.
Fig. 1. Phenotypic characterization of diverse parrots.
a Color morphs analyzed from family psittaculidae b Color morphs analyzed from family cacatuidae (wild-type birds are at the left and blue phenotypes are at right) (c) Phylogenetic relation between previously analyzed species (marked in blue) and species analyzed in the current study (d) Key changes in psittacofulvin-based hues in different species. [Pictures were taken by Shatadru Ghosh Roy or available in the public domain].
The blue phenotype lacks psittacofulvin pigmentation in the barb cortex
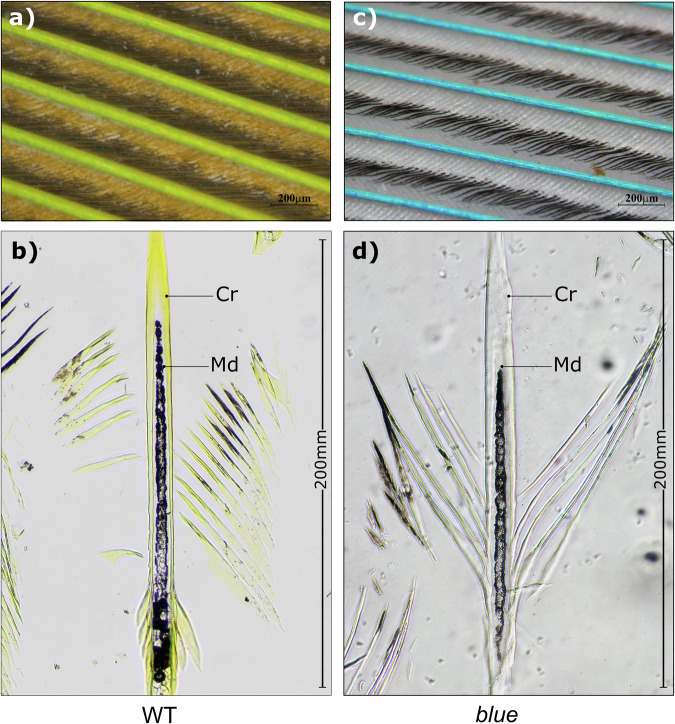
In budgerigar feathers, yellow psittacofulvin pigment has been shown to occur in the barbules and the outer cortex of the ramus17. We started by examining the status of psittacofulvin pigmentation in a typical parrot feather. As a standard representative, we examined contour feathers of P. krameri. Under a light microscope, a typical green contour feather from wild type green P. krameri shows green barbs and yellow barbules with black tips. The black tips are likely explained by melanin pigmentation (Fig. 2a). In contrast, light microscopic observation of contour feathers from P. krameri exhibiting the blue phenotype displayed blue barbs and white barbules with black tip (Fig. 2c). To characterize the microstructure responsible for the blue feather, we next examined longitudinal sections of contour feathers from the same green and P. krameri (blue) samples. We found that the outer cortex of green barbs is filled with yellow pigment while the blue barbs lack yellow pigment. The medullary region showed black melanin pigment surrounding central air vacuoles in both samples (Fig. 2).
Fig. 2. Histological sections of remex barb from P. krameri under a light microscope.
a Green barbs and yellow to black barbules of contour feathers from wild type P. krameri b Light microscope images of longitudinal section of the contour barb from wild type P. krameri. c blue barbs and white to black barbules of contour feathers from P. krameri (blue) d Light microscope images of longitudinal section of the contour barb from P. krameri (blue). [Cr- Cortex; Md- Medulla. Scale bars for images are 200 µm.].
Coding SNPs in conserved residues of PKS completely segregate with the blue phenotype
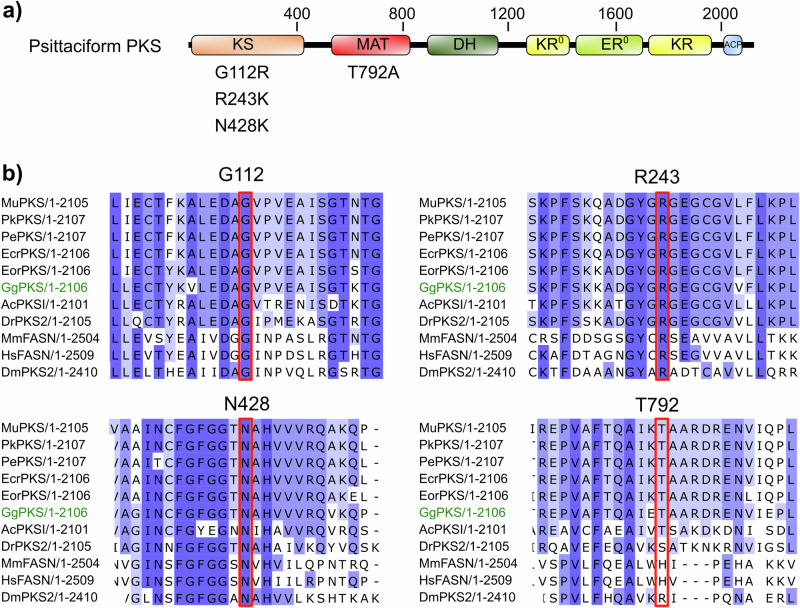
The enzyme MuPKS has been shown to be required for the synthesis of psittacofulvins in budgerigars by an iterative mechanism, homologous to that of vertebrate fatty-acid synthase. This raises the question whether other parrot species exhibiting similar psittacofulvin-based plumage coloration follow a similar biochemical pathway8,16. To test whether PKS is functionally conserved across the order Psittaciformes and if mutations in this gene could also be responsible for blue phenotype in other pet parrots, we sought to characterize the function of PKS in several closely related parrots (Fig. 1c). For species with genomes available but with no annotation we identified PKS locus using nucleotide BLAST with six exons of MuPKS (LOC101880715) coding sequence as a query8. For species without an available genome assembly, we sequenced all PKS exons using intronic primers (Tables S2, S3) designed based on the closest related species. Multiple sequence alignment of deduced amino acids for all the sample homologs showed more than 90% of sequence identity among each other including MuPKS (Table S4). Additionally, motif prediction of the deduced homologs confirmed the presence of all conserved domains that characterize the PKS protein reported to date (Fig. 3a). Consequently, to detect non-synonymous variants in protein-coding regions of different PKS homologs, we Sanger-sequenced all six exons of the PKS gene, including the splice sites and adjacent intronic parts, from wild-type and blue mutants of each species and compared them by pairwise alignment.
Fig. 3. Non-synonymous substitutions found in Sanger sequencing.
a Position of affected residues on the domain structure of a PKS, homologous to mammalian fatty-acid synthase. b Conservation of affected amino acid residues found in different PKS homologs. The shades of blue in the alignment represent the percentage identity of the amino acid residues. Affected residue is marked with red box. [Mu: M. undulatus; Pk: P. krameri; Pe: P. eupatria; Ecr: E. roratus; Eor: E. roseicapilla; Gg: G. gallus; Ac: A. carolinensis; Dr: D. rerio; Mm: M. musculus; Hs: H. sapiens; Dm: D. melanogaster].
Out of the 9 blue specimens of P. krameri tested in our study, we found a single nonsense mutation (c.2005G>T) in 4 specimens, resided at exon six of PKS homolog, altering the glutamate 668 into a premature stop codon (SNP frequency: 44.44%). One missense mutation (c.334 G > A) at exon three was also detected in 5 blue specimens, changing glycine 112 to arginine (SNP frequency: 55.56%). Among 11 blue specimens of P. eupatria, 6 individuals showed a nonsense mutation (c.2647 A > T) at exon six changing the lysine 883 into a premature stop codon (SNP frequency: 54.55%), while 5 individuals showed a missense mutation (c.728 G > A) at exon five, changing the arginine 243 to lysine (SNP frequency: 45.45%). However, all 5 blue specimens of E. roratus we tested exhibited a single nonsense mutation (c.5869 C > T) at exon six of PKS homolog, changing glutamine 1957 into premature stop codon (SNP frequency: 100%). Interestingly, PKS coding sequences of all 5 blue individuals of E. roseicapilla tested in this study, showed two homozygous substitutions at exon 6 simultaneously: one (c.1284 T > G) modifying the asparagine 428 into a lysine and another (c.2374 A > G) changing the amino acid threonine 792 to alanine, having SNP frequency of 100% for both. A list of the detected mutations across different species has been summarized in Table 1, along with the number of specimens genotyped in finer detail in Table S5–S8. However, its needs to be mentioned that two allelic variations found in the species P. krameri and P. eupatria was never found in same individuals as trans-heterozygous. They were always segregated independently in homozygous manner. However, two variants found in E. roseicapilla (N428K and T792A) were always segregated together in homozygous manner.
Table 1.
Allelic variation found in Sanger sequencing of PKS homologs
| Species | Allelic variation found in PKS homologs | ||
|---|---|---|---|
| P. krameri | WT (n = 7) | G334A (n = 5) | G2005T (n = 4) |
| P. eupatria | WT (n = 25) | G728A (n = 5) | A2647T (n = 6) |
| E. roratus | WT (n = 3) | C5869T (n = 5) | |
| E. roseicapilla | WT (n = 4) | T1284G; A2374G (n = 5) | |
Homozygous variants found in blue specimens are marked in blue columns. Sample numbers are shown in parentheses. blue variants of same species in two different columns represent independent segregation, whereas blue variants of E. roseicapilla in the same column represent co-segregation.
A multiple sequence alignment of diverse species, including major vertebrate classes and invertebrates, showed that three out of four affected amino acids (G112, R243, and N428) are universally conserved in other parrot members analyzed in this study along with chicken (Gallus gallus), anole lizard (Anolis carolinensis), zebra fish (Danio rerio), house mouse (Mus musculus), human (Homo sapiens) and fruit fly (Drosophila melanogaster) (Fig. 3b). T792 showed conservation in the order Psittaciformes, chicken, and anole lizard but not in zebra fish, house mouse, human and fruit fly. Type 1 polyketide synthases have been shown to contain multiple domains with independent catalytic activities18. Domain structure analysis showed that three of the non-synonymous polymorphisms (G112R, R243K and N428K) found in this study reside on the ketoacyl synthase domain, whereas T792A maps to the MAT domain (Fig. 3a).
Missense variants disrupt feather pigment synthesis in yeast
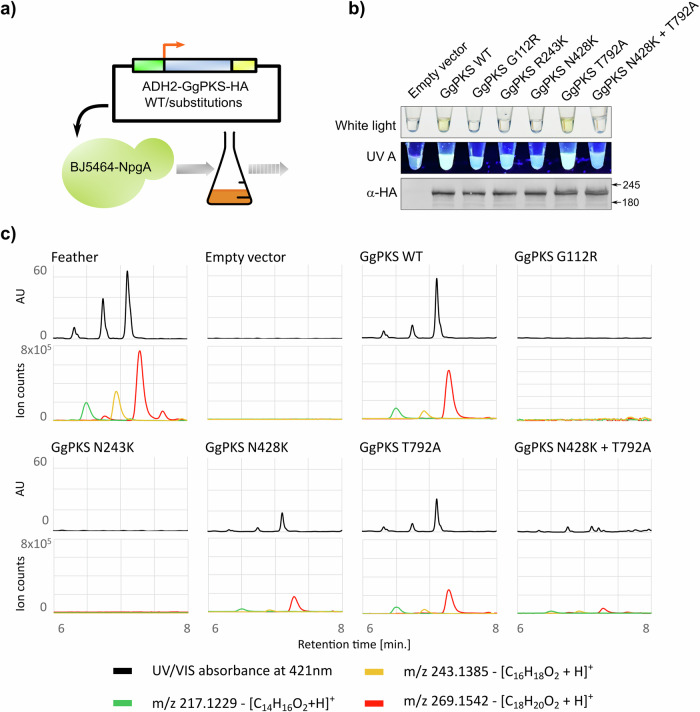
To investigate the functional impact of non-synonymous substitutions found in this study, we used strain BJ5464-NpgA of baker’s yeast (Saccharomyces cerevisiae) for psittacofulvin production upon transfection with the avian polyketide synthase (PKS). This strain contains a promiscuous phosphopantetheinyl transferase, NpgA that converts various polyketide synthase apoenzymes to holoenzymes19. We started with expressing PkPKS (OR452361) and PePKS (OR452362) under inducible promoter but organic extracts from yeast expressing these homologs showed no visible yellow pigmentation. Previously, it has been shown that organic extracts from yeast cells expressing chicken PKS (GgPKS; LOC420486) exhibited a yellow pigment, sharing similar pigment components with yeast expressing MuPKS as detected by LC-MS analysis8,13. Moreover, yeast strains expressing chicken PKS yielded even higher polyketide concentrations compared to yeast strains expressing PKS from budgerigar8,13. As, multiple sequence alignment showed that GgPKS share around 80% of sequence identity with all PKS homologs tested in this study (Table S4) and the identified missense variants are conserved in GgPKS, we decided to validate the affected residues by using heterologous expression of GgPKS in the BJ5464-NpgA strain. The expression of GgPKS in yeast was confirmed through western blot analysis (Figs. 4b and S1). We found that extracts from yeast expressing the GgPKS wild-type (WT) allele displayed a yellow pigment, contrasting with clear extracts from yeast expressing an empty control vector (Fig. 4a, b). This difference confirmed GgPKS’s ability to synthesize the yellow pigment. Additionally, extracts from yeast expressing the wild-type allele exhibited notable fluorescence under UV-A illumination as observed in the yellow feathers of certain parrot species20 as well as in octadecaoctaenal purified from red parrot feathers12. Next, we expressed the same GgPKS construct with the candidate substitutions created by site-directed mutagenesis. Extracts from three independent yeast strains respectively expressing GgPKS with substitutions G112R (found in blue P. krameri), R243K (found in blue P. eupatria), or N428K (found in blue E. roseicapilla) remained clear (Fig. 4b), while extracts from a yeast strain expressing GgPKS with another non-synonymous substitution found in E. roseicapilla T792A, showed a yellow coloration like the WT extract (Fig. 4b). As all blue specimens of E. roseicapilla, exhibited two homozygous substitutions (N428K and T792A) simultaneously, we decided to check pigment production by a GgPKS with N428K and T792A together. In accordance with N428K data, co-expression of N428K and T792A resulted in clear extracts (Fig. 4b).
Fig. 4. Biochemical validation of candidate missense variants.
a Graphical representation of methanol extraction from yeast strain BJ5464-NpgA expressing HA-tagged GgPKS Wild-type or with the substitutions found in blue phenotypes. b The extracts, illuminated with white light or UV A and corresponding GgPKS expression in total soluble protein extracts from the same yeast cultures by western blot with α-HA antibody. c the results of the analysis using UHPLC-HRAM QTOF showed that the compounds produced in the feathers of the parrot P. krameri, which elute from the Phenyl-Hexyl column at 6.3, 6.7, and 7.1 min and have characteristic exact molecular masses of 217.1229 m/z, 243.1385 m/z, and 269.1542 m/z—corresponding to carboxy-psittacofulvins with chain lengths of 14, 16, and 18 carbons, respectively—are also produced in yeast cultures expressing wild-type GgPKS. These compounds are not found in extracts from yeast lacking GgPKS (empty vector) or with GgPKS containing G112R or R243K substitutions. A partial production of these compounds is seen in GgPKS with N428K, T792A substitutions and their combinations. Wild-type GgPKS sample shows the highest amount of psittacofulvins, followed by lower peaks in the T792A sample, with the N428K sample yielding the least. When N428K and T792A mutations are combined, the production of psittacofulvins is even lower than when these two mutations are expressed individually.
To investigate the effect of G112R, R243K, N428K, T792A, and N428K combined with T792A mutations on psittacofulvin expression in finer detail, we subjected the yeast extracts (and green feather of P. krameri as control) to a sensitive analysis using ultra-high performance liquid chromatography with photodiode array detector coupled to high-resolution accurate-mass mass spectrometry (UHPLC-PDA-HRAM MS). The results confirmed the presence of the same psittacofulvins as parrot feathers in GgPKS WT and their absence in the empty vector control (Fig. 4c). Subsequent analysis by HRAM-MS confirmed the identities of the three compounds as C14-carboxyl psittacofulvin (217.1229 m/z), C16-carboxyl psittacofulvin (243.1385 m/z) and C18-carboxyl psittacofulvin (269.1542 m/z) which correspond to compounds found predominantly in green feathers of parrots13 and extracts of yeast expressing wild-type MuPKS8. In the extracts from yeast expressing GgPKS with substitutions G112R and R243K we detected no or negligible level of known psittacofulvins respectively (Fig. 4c), that clearly implies that G112 and R243 are vital residues for the biosynthetic activity of GgPKS. Interestingly, in the extracts from yeast expressing GgPKS with substitution N428K, T792A, and their combination, some compounds were detected, (Fig. 4c) characteristic by similar absorbance, and the same molecular masses as the psittacofulvins in GgPKS WT. However, in similarly scaled chromatograms and extracted ion chromatograms, it was evident that the GgPKS WT sample produced the highest amount of psittacofulvins, followed by a lower yield in the T792A sample, with the N428K sample yielding the least. Our estimation also suggests that if the N428K and T792A mutations are combined, the production of psittacofulvins is even lower than when these two mutations are expressed separately (Fig. 4c). The raw files of PDA and MS data for feather pigment, pigment extracted from yeast expressing GgPKS WT and mutants have been provided in Zenodo (https://zenodo.org/records/14184888).
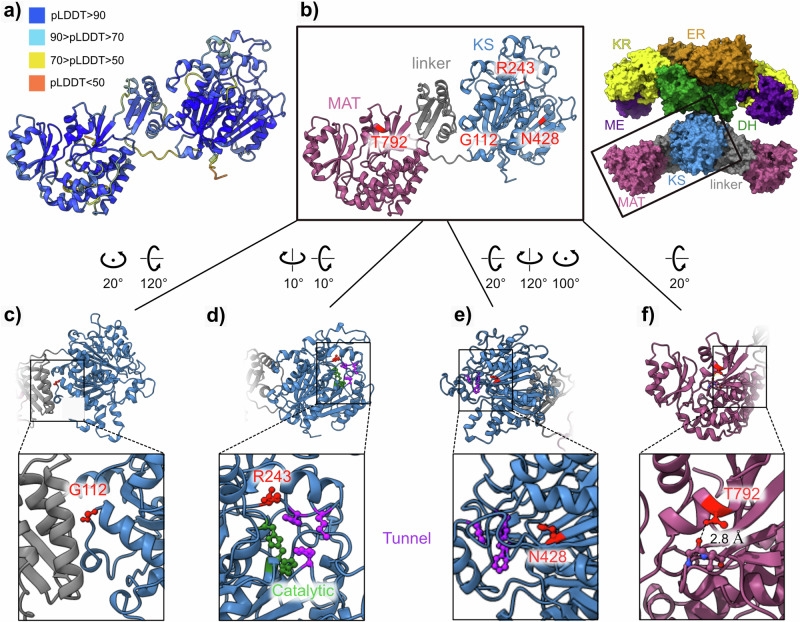
Structural prediction of causal SNPs
To obtain a structural understanding of the SNPs causal effects on enzyme function, we predicted the structure of the KS, linker, and MAT domains of P. krameri PKS using Alphafold21 and mapped onto it the positions of the four SNPs (Fig. 5a, b). Taking advantage of the sequence conservation among fatty acid synthases, we were able to correspond residues with a key function in porcine fatty acid synthase, for which a high-resolution crystal structure was determined22, with those of P. krameri PKS (Figs. 5b, 3a). Three SNPs reside at the conserved ketoacyl synthase (KS) domain, and one resides at the conserved MAT domain (Fig. 5b). The KS domain within the mammalian fatty acid synthases (mFAS) has been shown to catalyze the condensation reaction crucial for fatty acid biosynthesis. G112 in the P. krameri (blue) PKS is conserved among vertebrate fatty acid synthases (Fig. 3b) and resides at a loop at the interface between the KS domain and a linker domain, presumably providing flexibility to create a turn in the polypeptide chain (Fig. 5c). Replacing this conserved glycine residue with a much larger residue, i.e., arginine, may disrupt the local structure of the loop and the packing of the KS domain and the linker domain due to steric hindrance. R243 in the P. eupatria (blue) PKS is conserved among vertebrate fatty acid synthases (Fig. 3b) and is positioned near the three residues that correspond to those that create the entrance of the substrate binding tunnel in mFAS22 (Fig. 5d), suggesting that this arginine residue participates in substrate recognition or positioning relative to the active site. The SNP identified a change of an arginine to a lysine, a difference that would not be expected to have a major effect due to the similarity between these amino acids. Nevertheless, if this arginine is indeed involved in substrate binding, even such a small change in the properties of the residue could lead to decreased substrate binding and consequently to the dramatic effect seen in pigment synthesis. N428 in E. roseicapilla (blue) is conserved also among fatty acid synthases across taxa (Fig. 3b) and is placed in a β sheet near one of the substrate-binding tunnel residues and is buried under an α helix (Fig. 5e). Replacement of this asparagine to a longer residue, i.e., a lysine, may create a steric clash with the nearby helix, presumably leading to local unfolding and possibly to disruption of the substrate binding tunnel. Finally, T792 is in the conserved MAT domain. In the predicted structure, the OH group in the sidechain of this residue forms a hydrogen bond with the backbone of an adjacent loop, suggesting it is important for domain folding and stabilization (Fig. 5f). An alanine at this position would not be able to form such a bond, and thus, replacing T792 with an alanine would presumably deteriorate domain folding and/or stability and may affect enzyme activity.
Fig. 5. Structural analysis of causal SNPs.
a Predicted structure of the KS, linker, and MAT domains of the P. krameri (blue) PKS generated using AlphaFold (Jumper et al., 21) and colored according to per-residue confidence (pLDDT) scale (see legend). Note the high confidence in prediction (indicated in blue). b Left - the prediction shown in (a) colored by domain, showing the positions of the four residues identified as SNPs (indicated in red). Right - the crystal structure of porcine fatty-acid synthase (PDB: 2VZ8) (Maier et al. 22) with each domain differently colored is shown for context. Note that the structure is a homodimer containing two polypeptide chains. The boxed region includes the relevant domains for which the P. krameri structure was predicted. KS - β-ketoacyl synthase domain, MAT - malonyl/acetyltransferase domain, DH – dehydrase domain, ME - pseudo-methyltransferase domain, KR - β-ketoacyl reductase domain, and ER - enoyl reductase domain. c Closer view of G112 (in red), showing it is positioned in a loop, at the interface of a linker domain (gray) and the KS domain (blue). d Closer view of R243 (in red), showing it is positioned near the catalytic residues (colored green) and the residues that form the entrance to the substrate binding tunnel (colored purple). e Closer view of N428, showing its proximity to the residues that form the entrance of the substrate binding tunnel (colored purple) and that it is buried under an α helix. f Closer view of T792, showing a predicted hydrogen bond (indicated with a dashed line) between the sidechain OH group and the main chain of the adjacent loop.
Discussion
Psittacofulvin-based hues in parrots span a vibrant spectrum of colors from yellow to red. Under strong artificial selection by breeders, the naturally occurring color diversity of parrots was further expanded amongst captive populations of several species. The blue phenotype is one of the most common variations among them. Parrots create green feathers by combining yellow psittacofulvins around the barb cortex with blue structural colors produced by melanin pigments and feather nanostructure23. The absence of this cortical pigment psittacofulvin, characterizes the blue phenotype. Studies on blue mutants from family Psittaculidae (budgerigar and love birds) have already confirmed this physical basis of color variation8,11. Here, we analysed blue parrots belonging to three new members of this family and one from the family Cacatuidae from the order Psittaciformes. Notably, where the psittacofulvin-based hue deviates from its major yellow color the scenario changes a bit. For instance, loss of cortical psittacofulvin in red feather of female E. roratus or pink feather of both sexes of E. roseicapilla, results in white, blue, or gray feather. These variations are a result of altering constructive interference within medullary ultrastructure, depending on two interplaying components: melanosomes and spongy structure23. Moreover, our observation suggests that despite independent evolution of parrot morphotype apart from psittacula parrots24, loss of pink psittacofulvin in E. roseicapilla (blue) follows a similar mechanism to that found in the parrots displaying yellow psittacofulvins. This illustrates the shared genetic and biochemical origins of evolutionary adaptations of diverse plumage across various environmental habitats.
To understand the complete spectrum of psittacofulvin-based coloration, we focused on the genetic control of diverse psittacofulvin-based hues. Earlier research proposed that psittacofulvin synthesis takes place within the axial plate through the function of the multidomain enzyme PKS8,11. We identified PKS homologs in four different parrot members and our alignment data showed that polyketide synthase belonging to birds with yellow, red, or pink psittacofulvin hue share significant sequence similarities at the nucleotide as well as the amino acid level. The presence of all conserved structural domains suggests functional resemblance between these homologs. To test this hypothesis, we Sanger sequenced PKS homologs of these four parrot members carrying the blue phenotype. We uncovered seven unique de novo variations (Table 1) in four diverse species carrying the blue phenotype, three of which are nonsense mutations. Given the fixed segregation pattern of nonsense mutations observed in blue specimens of P. krameri, P. eupatria and E. roratus, we concluded these mutations to be causal in each respective species. However, we proceeded with functional validation of the missense variants. Intriguingly, three of these non-synonymous substitutions were located within the conserved ketoacyl synthase (KS) domain while one is found at MAT domain. Upon heterologous expression of GgPKS carrying candidate causal variants (G112R, R243K, and N428K) at the ketoacyl synthase domain, respective yeast extracts showed no pigmentation under white light and UV-A illumination, whereas one of the two variations found in E. roseicapilla (T792A) showed no changes in pigment production. As N428K and T792A were always found to segregate simultaneously, we checked their mutual expression and found clear extract under white light and UV-A illumination. Nevertheless, we went for more sensitive analysis using UHPLC-PDA-HRAM MS. This advanced chromatographic analysis confirmed the disruptive effect of G112R, and R243K mutations as null allele. These missense substitutions found in blue samples of P. krameri and P. eupatria are highly conserved across taxa and capable of functional disruption as per domain structure analysis. Thus, we inferred them as effective as the nonsense variants found in respective species in causing the blue phenotype. Fascinatingly, demonstrating the limitation of fluorescence-based observation, the advanced UHPLC-PDA-HRAM MS analysis revealed crucial details about the nature of the two missense mutations, N428K and T792A, found in E. roseicapilla. Conservation status and domain structure analysis suggested that variant N428K is capable of disrupting PKS activity. Our UHPLC-PDA-HRAM data also showed a significant reduction in psittacofulvin production in the yeast extract expressing N428K. This evidence proposed that variant N428K acts as a strong loss-of-function allele. On the other hand, T792A likely acts as a hypomorphic allele. Although it is partially conserved and located in important MAT domain, T792A disrupts PKS activity to a lesser extent. It produces more psittacofulvin than N428K, but still less than the wild type. Together, our results suggest that while independent expression of either variant (N428K or T792A) partially disrupts psittacofulvin production, the combined effect of both variants leads to a more severe reduction than either one alone (Fig. 4c). However, it is important to note that our HPLC analysis is currently more qualitative, and validating these findings will require more advanced quantitative experiments. In conclusion, despite their independent functional impacts, lower yield of combined expression of N428K and T792A suggests that the severity of these two variants depends on their segregation pattern. Therefore, we propose that N428K and T792A are capable of partial reduction of psittacofulvin production, while a combinatorial effect of these two variants determines the final blue phenotype in E. roseicapilla.
Despite having a shared ancestry, Parrots’ evolutionary divergence has been shaped by their varied tropical habitats. Although, the role of polyketide synthase (PKS) has been established as essential for psittacofulvin biosynthesis in domesticated mutants lacking psittacofulvin-based pigmentation8,11; whether this role is conserved across the entire order remains unclear. A recent discovery pointed a shared chemical basis for yellow-to-red psittacofulvin coloration across all parrot superfamilies, spanning 50–80 million years of evolution13. Our study further confirms that PKS drives a common biochemical pathway for yellow-to-red psittacofulvin-based coloration across diverse parrot lineages. Also, the evidence of multiple causal variants in the same conserved domains implies a significant role of this domain in determining shared traits across order Psittaciformes. High similarity between PKS homologs hints that the key evolutionary changes leading to varied psittacofulvin pigmentation across parrots were not variants in the ancestral PKS coding sequence, but rather variations affecting carbon chain length or oxidation state in later steps. Therefore, enzymes like ALDH3A2, which shifts red to yellow psittacofulvins by changing the oxidation status, highlight the role of trans-acting enzymes in final color determination13. We report multiple causal mutations within different functional domains of PKS, that expands the latest research suggesting psittacofulvin-based coloration as an evolutionarily constrained mechanism. It creates new research opportunities to explore how similar phenotypes evolve independently in different species. We know that years of selective breeding have resulted in the emergence of similar color phenotypes across diverse parrot species. Understanding this selection dynamic is also critical. Two of our sample species, P. krameri (E668* and G112R) and P. eupatria (K883* and R243K), exhibit multiple potential causative SNPs, each independently capable of producing the blue phenotype. When multiple causative SNPs appear in a species, haplotype reconstruction helps determine if they exist on the same haplotype, indicating a single selective sweep, or on different haplotypes, suggesting independent sweeps. However, our use of Sanger sequencing limits our ability to resolve haplotypes with the precision that next-generation sequencing could provide. Future studies with whole-genome sequencing could enable accurate haplotype reconstruction, clarifying whether the SNPs represent one or multiple adaptive events.
Type I polyketide synthases are typically large, multifunctional enzymes organized in modules, each containing various domains needed for polyketide synthesis18. The domain structure of budgerigar polyketide synthase (MuPKS) is homologous to mammalian fatty-acid synthase and type I bacterial polyketide synthases8. These domains work collaboratively to synthesize polyketides. The β-ketoacyl synthase (KS) domain initiates and extends the polyketide chain through condensation reactions, while the malonyl/acetyltransferase (MAT) domain selects and loads the precursor units onto the acyl carrier protein (ACP). The ACP domain acts as a carrier, shuttling the growing polyketide chain between various enzymatic domains. The β-ketoacyl reductase (KR), dehydrase (DH), and enoyl reductase (ER) domains modify the polyketide structure, introducing ketone reductions, dehydrations, and double bond reductions, respectively18. Cooke et al. demonstrated in budgerigars that variation within the malonyl-CoA:ACP transacylase (MAT) domain of PKS can reduce the enzyme activity and halt yellow psittacofulvin production8. Structural analysis has revealed that the three causal variants (G112R, R243K and N428K) found in this study reside within the KS domain. This domain adds two-carbon units to the growing polyene chain by condensing malonyl-CoA with the existing acyl chain, contributing to the iterative elongation process22. Alterations in polyene chain length have been suggested to be associated with variations in pigment saturation25,26. Hence, change in any of the detected conserved residues can affect product chain length, thereby directly influencing the pigment intensity. However, our biochemical assay revealed that replacement of asparagine 428 to a longer residue lysine is not capable enough to disrupt the PKS function completely, possibly needs additional contribution from another hypomorphic variant T792A, resided at MAT domain. Our analysis of the affected domains and their associated residues represents a promising avenue for modulating psittacofulvin expression, offering insights into the mechanisms underlying pigment production in parrots. It also facilitates advancements in both research and practical applications within the field of engineered PKSs involved in the biosynthesis of a variety of compounds with important biological activities, including antibiotic, antifungal, anticancer, and immunosuppressive properties.
Material and methods
Feather samples and light microscopy
Contour feathers of wild type and blue phenotype of P. krameri were obtained from local breeders and pet shops. Feathers were washed in distilled water, dried overnight at 60 °C and prepared for light microscopy. Images were taken using Leica M165 FC microscope.
Feather histology
Parts of contour feathers were washed in PBS, fixed in 4% paraformaldehyde overnight and washed in 50–100% ethanol followed by xylene. After dehydration, the tissues were incubated in 65 °C hot paraffin baths in vacuum oven for 1–2 h each. Then, the samples were embedded in paraffin mold and sectioned at 8μm. Longitudinal sections were observed and imaged under Nikon light microscope ECLIPSE Ci-L.
Sequence alignment
Homologs of PKS gene were identified using MuPKS (LOC101880715) coding sequence as a query8. Identity was calculated via the Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi). For pairwise sequence alignment EMBOSS Needle (https://www.ebi.ac.uk/Tools/psa/emboss needle/) was used. To identify the conserved sequences, a domain search was done using Jalview version 2.11.2.0 (Tcoffee alignment). Clustal Omega (https://www.ebi.ac.uk/jdispatcher/msa/clustalo) was used for Multiple Sequence Alignment. Percent Identity Matrix was created by Clustal2.1.
Phylogenetic mapping
Phylogenetic tree was generated using Timetree of Life27 (https://timetree.org/) with the help of published molecular phylogeny of the parrots (Psittaciformes)28,29.
Blood sample collections
Whole blood from a clipped toe was collected with necessary pedigree data and dried on an appropriate filter paper from healthy birds owned by breeders. The collection was done by the Sde-tzvi lab (https://dnalab.co.il/) as part of their sex identification services. The mutants examined in this study are shown in Table 1 and Table S5–S8.
DNA extraction from whole blood
Total DNA was extracted from whole blood using an alkaline extraction method. Part of the paper with dried blood was soaked in 45 μL 0.2 M NaOH solution, followed by incubation at 75 °C for 5 min. 1 μl from this solution was used as template for PCR amplification mixture of 30ul.
Genotyping
The coding sequences of PKS homologs were amplified from 50 ng of genomic DNA per reaction using intronic primers (refer to Tables S2, S3) having final concentration of 0.6 mM through PCR amplification by2x PCRBIO HS Taq Mix Red. Primers were designed within intronic regions to produce amplicons approximately 500–700 bp in size. The PCR was run in Bio-Rad T100 Thermal Cycler according to the manufacturer’s instructions. The concentrations of the PCR products were measured using a spectrophotometer (DeNovix DS-11 FX) at OD260 and verified for integrity by running them on a 1% agarose gel in TBE. The amplified products were then sequenced using the forward primers listed in Table S2, S3 via Sanger sequencing. Sanger reactions are performed using the eBgDy™ Terminator 3.1 Cycle Sequencing Kit (Epoch Life Science) per kit recommendations on a Biometra Trio (analytik jena). Reactions are cleaned using HighPrep DTR - DX beads (MAGBIO) and sequenced on a 3500xL Genetic Analyzer (Applied Biosystems). The genotypes of each bird were determined by manually examining the sequencing chromatograms in Chromas software version 2.6.6).
GgPKS expression constructs
For cloning HA-tagged GgPKS into the pCY5N vector, total cDNA was first prepared from total RNA extracted using TRIzol reagent from Gallus gallus embryos by qScript cDNA synthesis kit. The coding sequence of GgPKS was cloned using Gibson assembly protocol into pCY5N vector under an ADH2 promoter instead of the original ADH1 promoter with HA tag at C-terminal. Nucleotide changes found in different species were introduced into pCY5N-ADH2-GgPKS-HA by site directed mutagenesis (listed in Table S9).
Yeast transformation and GgPKS expression
The yeast strain BJ5464-NpgA was transformed with an expression plasmid carrying an inducible chicken polyketide synthase (PKS; LOC420486 [Gallus gallus] construct (ADH2 promoter, PKS CDS, CYC1 terminator) and the URA3 auxotrophic marker, or with the plasmid lacking the PKS construct as control. We opted for the use of an expression construct carrying the PKS sequence from chicken, since in a previous study, yeast strains expressing chicken PKS yielded higher polyketide concentrations compared to yeast strains expressing PKS from budgerigar8. To initiate the transformation process, the yeast strain BJ5464-NpgA was grown in 5 ml of YPD medium until reaching the mid-log phase. Subsequently, 1 ml of the culture was collected in a 1.5 ml tube through centrifugation at 800 × g for 5 min. After discarding the supernatant, the cell pellet was resuspended in 1 ml of water and centrifuged once more. The supernatant was again removed, and the cell pellet was resuspended in 1 ml of 100 mM lithium acetate, followed by a brief centrifugation. After discarding the supernatant, the pellet underwent a sequential addition of the following components, with vortexing between each addition: 240 μL of 50% (w/v) polyethylene glycol, 37 μL of 1 M lithium acetate, and 10 μL of purified plasmid. The yeast mixture was then incubated at room temperature for 30 min and subsequently heat-shocked at 42 °C for 30 min. Transformants were selected on synthetic dextrose (SD-Ura) plates. For protein expression and pigment synthesis, overnight cultures (5 ml) were cultured in SD-Ura at 30 °C and transferred to 200 ml YPD with 1% glucose in 1 L flasks, with an initial OD600 of 0.02. The cultures were allowed to grow for 72 h at 30 °C. The cells were then harvested through centrifugation and stored at −80 °C.
Yeast western blot
About 100 mg (wet weight) of a frozen yeast pellet was reconstituted in 150 ml of zymolyase buffer (composed of 50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 1 M sorbitol) containing 30 mM DTT, followed by a 15-min incubation at room temperature. The cells underwent centrifugation for 5 min at 1500 × g, and the supernatant was discarded. The cell pellet was then resuspended in 100 ml of zymolyase buffer with 1 mM DTT and approximately 1 mg/ml zymolyase, undergoing a 1-h incubation at 30 °C with mixing at 1000 RPM. Afterward, the spheroplasts were pelleted through centrifugation (5 min at 1500 × g), the supernatant was discarded, and the pellet was washed with 500 ml of zymolyase buffer. The spheroplasts underwent another round of centrifugation (5 min at 1500 × g), the supernatant was discarded, and the pellet was resuspended in 500 µl of cold PBS with 1x protease inhibitor cocktail (Sigma P8849). Subsequently, the cells were lysed by sonication (30 s at 3 W). The homogenate was clarified by centrifugation (10 min at 16,000 × g at 4 °C) and then resuspended in gel loading buffer (composed of 100 mM-Tris/HCl, pH 7.0, 10% glycerol, 4% SDS, 5% beta-mercaptoethanol, 0.02% bromophenol blue). Following a 5-min incubation in a boiling water bath, any remaining cell debris was eliminated by centrifugation for 2 min in a microfuge. Subsequently, 20 μL of the extract per gel slot was subjected to electrophoresis on a 9% polyacrylamide gel using standard PAGE-SDS techniques (Laemmli, 1970). The proteins were then transferred to a nitrocellulose membrane through a wet transfer. To detect GgPKS-HA, a western blot was performed. The membrane was initially blocked for 1 h at room temperature with 5% nonfat dry milk in TBS with 0.1% Tween-20 (TBST). After a brief rinse in TBST, the membrane was incubated overnight at 4 °C with an anti-HA rat monoclonal antibody (Sigma 11867423001) for 1 h, diluted at 1:1000 in 0.5% nonfat dry milk in PBST. Following three washes (5 min each) with TBST, the membrane was incubated for 1 h at room temperature with goat anti-rat IgG, Alexa Fluor™ 680 (Thermo A-21096) secondary antibody, diluted at 1:5000 in 5% nonfat dry milk in TBST. The membrane underwent three additional washes (5 min each) and was developed using the iBright™ CL1500 Imaging System.
Pigment extraction
The feather pigment extraction followed a similar procedure as the study of Cooke et al. on psittacofulvin pigments in parrots8. Green flight feathers from P. krameri were cleaned, rinsed in ethanol, hexane, and dried. About 5 mg of feather barbs were trimmed, weighed, and incubated for 1 h at 95 °C in 1 ml of 2% HCl in pyridine. After cooling, the solvent was evaporated under nitrogen, and the residue was dissolved in 0.5 ml of 2% acetic acid in ethyl acetate. The organic phase was washed with water, centrifuged, and dried. Pigments were redissolved in methanol and analysed by LC-MS.
For the extraction of pigments shown in photographs (Fig. 4b), around 500 mg of yeast cells (wet weight) underwent homogenization in a 2 ml tube with about 0.5 g of glass beads in 0.5 ml methanol. The resulting homogenate underwent centrifugation for 5 min at 16,000 × g to clear it. The supernatant was subsequently moved to a fresh tube and concentrated under a stream of N2 until 50 μL remained, and then transferred to 0.5 ml qubit tubes for photography purposes.
HPLC
The psittacofulvin pigments were extracted from freeze-dried biomass according to slightly modified method8. Briefly, approximately 2 × 40 mg of each yeast strain was soaked with 1 mL of cold methanol (MeOH) in 2 mL screw cap tubes for 10 min at 4 °C. After sonication for 1 min, the samples were vortexed and then homogenized by bead mill homogenizer (PPRECELLYS® Evolution, Bertin Technologies, France) using 0.5 ml of 1.0 mm diameter zirconia/silica beads (BioSpec Products, Inc., Bartlesville, OK, USA) 4 × 20 s at speed of 5800 rpm with ice cooling. After centrifugation (24,400 × g, 10 min, 4 °C), the 800 μL of supernatant from each replicate was transferred into one 2 mL centrifuge tube and pooled sample was evaporated to dryness with a stream of N2 at 40 °C. Then, 1 mL of methyl tert-butyl ether (MTBE) and 1 ml of water, centrifuged (4755 × g, 10 min, 4 °C), and the organic phase was washed again with 1 ml of 2% acetic acid. A volume of 800 μL of the supernatant was pipetted into 1.5 mL centrifuge tube and dried by stream of N2 at 40 °C. The sample was then resuspended in 80 μL of MeOH, centrifuged (24,400 × g, 10 min, 4 °C), and 70 μL was transferred to a conical 150 μL insert for UPLC-PDA-MS analysis.
The extracts were analysed by ultra-high performance liquid chromatography with photodiode array (PDA) detector coupled to high-resolution accurate-mass mass spectrometry (UHPLC-PDA-HRAM MS). The analytical system consisted of an Ultimate 3000 ultra-high-performance chromatograph with PDA detector module (Thermo Fischer Scientific, Waltham, MA, USA) and Q-TOF high-accuracy mass spectrometer (HRAM) Impact II (Bruker Daltonik, Bremen, Germany). Separation was performed using Kinetex Phenyl-Hexyl column (1.7 µm, 100 × 2.1 mm, Phenomenex, Torrance, CA, USA) with 0.2% formic acid (solvent A) and MeOH (solvent B) as mobile phase. Gradient elution started at 5% B (0–0.5 min), then increased to 50% B in 2.2 min and then to 100% B in 8 min, where it was held until 15 min, then the gradient was returned to the initial conditions (5% of B) in 15.5 min and the system was equilibrated until the end of the analysis in 20 min. The constant flow rate was 300 mL/min and column temperature was set at 40 °C. Injection volume was 5 mL. Absorbance spectra were measured in a range of 250–750 nm at an acquisition rate of 5 Hz. Chromatograms were monitored at four discrete wavelengths (444, 472, 421, and 350 nm). APCI ionization in positive mode was used for full-scan MS analysis with a resolution of 60,000 and sampling frequency of 1 Hz. The data were collected and pre-processed with oTof Control 4.0, HyStar 3.2 and DataAnalysis 4.3 software (all Bruker Daltonik, Bremen, Germany). The absorbance profiles were compared with published absorbance spectra of psittacofulvins12 and the identity of the compounds was determined by MS and compared to previously reported compounds in yeast extracts8.
Structural analysis
The PROSITE webtool (http://prosite.expasy.org/) was used to predict the motifs in the protein sequence of the deduced homologs. Structure prediction was performed using AlphaFold2 (Jumper et al.21). The crystal structure of the porcine fatty-acid synthase (PDB: 2VZ8) was retrieved from the PDB database (https://www.rcsb.org/) and used for structure comparison and analyses. Figures were prepared using ChimeraX30.
Statistics and reproducibility
For all experiments, sample sizes and replicates are specified. Feather microscopy, histology, pigment extraction, and LC-MS were performed in triplicate, with each replicate representing independent biological samples. Yeast transformation experiments were repeated three times with independent transformations. Computational analyses, including sequence alignment, phylogenetic mapping and structural predictions, used standardized workflows to ensure reproducibility. HPLC analyses were verified with consistent results across replicates. Detailed protocols are provided above to facilitate reproducibility.
Reporting Summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We are grateful to all the anonymous breeders for providing the blood and feather samples for allowing us to use the samples sent to Sde-tzvi lab (https://dnalab.co.il/). We would also like to thank Center for Advance Genomics of Ilse Katz Institute at Ben-Gurion University. This work was supported by the Israel Science Foundation (ISF) (grant 941/21to U.A.), Portuguese Foundation for Science and Technology (FCT, https://www.fct.pt) research contracts to M.C. (CEECINST/00014/2018/CP1512/CT0002) and P.M.A. 2020.01494.CEECIND, by a research contract to S.A. from the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement No. 101000504).
Author contributions
Conceptualization: U.A. Methodology: S.G.R., J.B., P.M., A.B., M.A., and U.A. Investigation: S.G.R., J.B., P.M., A.B., and I.G.H. Visualization: S.G.R., I.G.H., J.B., and P.M. Supervision: U.A. Writing—original draft: S.G.R. Writing—review & editing: U.A., M.C., R.A., P.M.A., S.A., I.G.H., J.B., and P.M.
Peer review
Peer review information
Communications Biology thanks Simon Yung Wa Sin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: David Favero. A peer review file is available.
Data availability
Plasmids are available on request. DNA-seq data are deposited at GenBank repository with accession numbers OR452361 (P. krameri), OR452362 (P. eupatria), OR452363 (E. roratus) and OR452364 (E. roseicapilla). Uncropped and unedited blot images of Fig. 4B are provided in Supplementary Fig. 1. The raw PDA and MS data for feather pigments, as well as pigments extracted from yeast expressing GgPKS WT and mutants, are available on Zenodo (https://zenodo.org/records/14184888).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-025-07537-7.
References
- 1.Prum, R. O., Torres, R., Kovach, C., Williamson, S. & Goodman, S. M. Coherent light scattering by nanostructured collagen arrays in the caruncles of the malagasy asities (Eurylaimidae: Aves). J. Exp. Biol.202, 3507–3522 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Prum, R. O. & Brush, A. H. The evolutionary origin and diversification of feathers. Q. Rev. Biol. 77, 261–295 (2002). [DOI] [PubMed]
- 3.Krukenberg, C. F. W. Die federfarbstoffe der psittaciden. Vergleichend-physiologische Studien Reihe2, 29–36 (1882). [Google Scholar]
- 4.Stradi, R., Pini, E. & Celentano, G. The chemical structure of the pigments in Ara macao plumage. Compar. Biochem. Physiol. B Biochem. Mol. Biol.130, 57–63 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Tinbergen, J., Wilts, B. D. & Stavenga, D. G. Spectral tuning of amazon parrot feather coloration by psittacofulvin pigments and spongy structures. J. Exp. Biol.216, 4358–4364 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Masello, J. F. & Quillfeldt, P. Body size, body condition and ornamental feathers of Burrowing Parrots: Variation between years and sexes, assortative mating and influences on breeding success. Emu103, 149–161 (2003).
- 7.Burtt, E. H., Schroeder, M. R., Smith, L. A., Sroka, J. E. & McGraw, K. J. Colourful parrot feathers resist bacterial degradation. Biol. Lett.7, 214–216 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooke, T. F. et al. Genetic mapping and biochemical basis of yellow feather pigmentation in budgerigars. Cell171, 427–439.e21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres, J. P. & Schmidt, E. W. The biosynthetic diversity of the animal world. J. Biol. Chem. 294, 17684 –17692 (2019). [DOI] [PMC free article] [PubMed]
- 10.Li, F. et al. Sea Urchin Polyketide Synthase SpPks1 Produces the Naphthalene Precursor to Echinoderm Pigments. J. Am. Chem. Soc.144, 9363–9371 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke, F. et al. Convergent evolution of parrot plumage coloration. PNAS Nexus10.1093/pnasnexus/pgae107 (2024). [DOI] [PMC free article] [PubMed]
- 12.Adamec, F. et al. Spectroscopic investigation of a brightly colored psittacofulvin pigment from parrot feathers. Chem. Phys. Lett.648, 195–199 (2016).
- 13.Arbore, R. et al. A molecular mechanism for bright color variation in parrots. Science386, eadp7710 (2024). [DOI] [PMC free article] [PubMed]
- 14.Chan, D. T. C., Poon, E. S. K., Wong, A. T. C. & Sin, S. Y. W. Global trade in parrots – Influential factors of trade and implications for conservation. Glob. Ecol. Conserv.30, e01784 (2021).
- 15.Ross, C., Opel, V., Scherlach, K. & Hertweck, C. Biosynthesis of antifungal and antibacterial polyketides by Burkholderia gladioli in coculture with Rhizopus microsporus. Mycoses57, 48–55 (2014). [DOI] [PubMed] [Google Scholar]
- 16.McGraw, K. J. & Nogare, M. C. Carotenoid pigments and the selectivity of psittacofulvin-based coloration systems in parrots. Compare. Biochem. Physiol. B Biochem. Mol. Biol.138, 229–233 (2004). [DOI] [PubMed] [Google Scholar]
- 17.D’Alba, L., Kieffer, L. & Shawkey, M. D. Relative contributions of pigments and biophotonic nanostructures to natural color production: a case study in budgerigar (Melopsittacus undulatus) feathers. J. Exp. Biol.215, 1272–1277 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Leibundgut, M., Maier, T., Jenni, S. & Ban, N. The multienzyme architecture of eukaryotic fatty acid synthases. Curr. Opin. Struct. Biol.18, 714-725 (2008). [DOI] [PubMed]
- 19.Ma, S. M. et al. Complete reconstitution of a highly reducing iterative polyketide synthase. Science326, 589–592 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hausmann, F., Arnold, K. E., Marshall, N. J. & Owens, I. P. F. Ultraviolet signals in birds are special. Proc. R. Soc. B Biol. Sci.270, 61–67 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maier, T., Leibundgut, M. & Ban, N. The Crystal Structure of a Mammalian Fatty Acid Synthase. https://www.science.org (2008). [DOI] [PubMed]
- 23.Ghosh Roy, S. et al. Mutations in SLC45A2 lead to loss of melanin in parrot feathers. G3: Genes, Genomes, Genetics10.1093/g3journal/jkad254 (2023). [DOI] [PMC free article] [PubMed]
- 24.Bonin, J. & Homberger, D. Possible convergent evolution of yellow psittacofulvin colors in the feathers of cockatoos (Cacatuidae) and parrots (Psittacidae). FASEB J.26, 722.23 (2012).
- 25.De Oliveira Neves, A. C., Galván, I. & Van Den Abeele, D. Impairment of mixed melanin-based pigmentation in parrots. J. Exp. Biol.223, jeb225912 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Price-Waldman, R. & Stoddard, M. C. Avian coloration genetics: recent advances and emerging questions. J. Heredity112, 395–416 (2021). [DOI] [PubMed]
- 27.Kumar, S. et al. TimeTree 5: an expanded resource for species divergence times. Mol. Biol. Evol.39, msac174 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright, T. F. et al. A multilocus molecular phylogeny of the parrots (Psittaciformes): support for a gondwanan origin during the cretaceous. Mol. Biol. Evol.25, 2141–2156 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, B. T. et al. Phylogenomic analysis of the parrots of the world distinguishes artifactual from biological sources of gene tree discordance. Syst. Biol.72, 228–241 (2023). [DOI] [PubMed] [Google Scholar]
- 30.Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci.27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmids are available on request. DNA-seq data are deposited at GenBank repository with accession numbers OR452361 (P. krameri), OR452362 (P. eupatria), OR452363 (E. roratus) and OR452364 (E. roseicapilla). Uncropped and unedited blot images of Fig. 4B are provided in Supplementary Fig. 1. The raw PDA and MS data for feather pigments, as well as pigments extracted from yeast expressing GgPKS WT and mutants, are available on Zenodo (https://zenodo.org/records/14184888).