ABSTRACT
Background
Changes in functional genetic polymorphisms may increase or decrease the risk of cancer in patients. Nowadays, the association between polymorphisms in the interleukin‐8 (IL‐8) gene and the susceptibility of cancer risk have been investigated in many studies, however, above relationships remain unclear.
Aim
The current study aims to comprehensively evaluate the association between IL‐8 gene six polymorphisms and the whole cancer risk, especially −251 polymorphism and gastric cancer.
Methods and Results
Six polymorphisms (−251, −353, +678, +1633, +2767, +781) were collected. The expression of serum IL‐8 was calculated by ELISA assay. First, 104 case–control studies were conducted. Second, this research has made significant discoveries regarding the −251, −353 and +781 polymorphisms and the potential associations with cancer risk. Finally, the serum IL‐8 levels in gastric cancer patients with AA/TT genotypes were significantly higher than those with the same genotypes of healthy controls and TT genotypes in gastric cancer patients.
Conclusion
Overall, the investigation has revealed that IL‐8 gene polymorphisms significantly influence vulnerability to cancer development, especially for gastric cancer.
Keywords: biomarker, cancer susceptibility, gastric cancer, IL‐8, polymorphism
1. Introduction
Cancer, a broad range of diseases, can originate in nearly any tissue or organ within the human body. Cancer cells have the capability to disseminate to distant organs, thereby establishing secondary tumor sites [1, 2]. The subsequent phenomenon is referred to as metastasis, which significantly contributes to mortality in cancer patients. A neoplasm, also referred to as a malignant tumor, is a prevalent term used to describe the pathological condition known as cancer. In 2020, a global estimation revealed that almost 19.3 million new cancer instances were diagnosed, with an undesirable mortality rate of 10.0 million cancer patients [2]. Among the diverse array of cancer types, prostate, lung, stomach, colorectal, and liver cancer exhibit the highest prevalence in males. Conversely, breast, thyroid, colorectal, cervical, and lung cancer are the predominant neoplastic conditions commonly encountered in females [3].
According to current scientific literature, a significant proportion, ranging from 30% to 50%, of mortality resulting from malignant neoplastic diseases can be prevented by altering or avoiding pivotal risk factors. Furthermore, the implementation of established prevention strategies that are firmly grounded in empirical evidence may also involve in the reduction of cancer‐related fatalities. Reducing the cancer burden can be achieved by implementing strategies for early cancer detection and effectively managing individuals who develop cancer.
The early detection of cancer plays a dynamic role in signifying the efficacy of treatment interventions, thereby increasing the possibility of survival while minimizing morbidity and the financial burden associated with treatment [4]. Two strategies facilitate early detection: timely detection of symptomatic cancer is crucial in identifying malignancies at their initial stage. Conversely, screening attempts to detect individuals exhibiting particular cancer indications or precancerous conditions without any symptomatic manifestation and promptly refer them for further diagnosis and therapeutic intervention [5]. In addition, Genome‐Wide Association Studies (GWAS) have recognized many loci allied with cancer risk. These loci contain a multitude of Single Nucleotide Polymorphisms (SNPs) that exert regulatory control in gene expression. Consequently, these SNPs can influence an individual's genetic susceptibility to cancer via various mechanisms [6].
In recent decades, GWAS have recognized numerous loci associated with increased risk, encompassing many SNPs [7]. Several SNPs associated with cancer have been identified as having a causal relationship, while in some instances, the functional mechanisms responsible for the association between these SNPs and cancer risk have been elucidated [8, 9]. To date, multiple GWAS have been undertaken over the previous decade to investigate various types of malignancies, including but not restricted to breast, lung, prostate, colorectal, and others [6, 10, 11, 12].
The involvement of inflammation in cancer progression is multifaceted, encompassing various mechanisms such as immune suppression, tissue remodeling, DNA damage, and stimulation of cell proliferation. Chronic inflammation has suppressed the immune response, thereby impeding the identification and subsequent elimination of tumor cells [13]. The inhibitory effect of cytokines secreted by inflammatory cells on the functionality of immune cells facilitates the proliferation and dissemination of cancer cells [14]. IL‐8, also called CXCL8, is a cytokine intricately associated with the inflammatory response. It influences various cellular mechanisms, encompassing the convergence of cancer plasticity, angiogenesis, and immune suppression [15]. Several studies have documented that IL‐8 exhibits increased expression levels in certain tumor cell types, and the upregulation of IL‐8 has been associated with the processes of invasion and metastasis [16]. Cancer susceptibility has been extensively documented in six polymorphisms (−251, −353, +678, +1633, +2767, +781) within the IL‐8 gene.
Despite the existence of multiple meta‐analyses, the current sample size remains insufficient. Therefore, re‐analyzing the association between IL‐8 gene six polymorphisms and the risk of susceptibility for cancer is imperative [17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136]. Besides, we will evaluate the relationship between IL‐8 expression and gastric cancer based on the TCGA data and our own clinic information.
2. Materials and Methods
2.1. Bioinformatics Analysis
The differential expression of IL‐8 between various tumor types and adjacent para‐cancerous tissue was examined via data obtained from the Gene Expression Profiling Interactive Analysis (GEPIA) website. The data of overall survival and disease‐free survival concerning the expression of IL‐8 in each tumor were obtained from the above website. The present study investigates the clinical characteristics associated with the expression of IL‐8 and its association with gastric cancer, utilizing data obtained from TCGA database.
2.2. Data Eligibility and Credentials of Relevant Studies
Extensive literature was searched in Google Scholar, PubMed, Embase, Web of Science, and Chinese databases. The most recent search was conducted on June 23, 2023. The search strategy used keywords such as “Interleukin‐8,” “IL‐8,” “CXCL8,” “polymorphism,” “variant,” “cancer,” “carcinoma,” and “tumor.” A comprehensive search yielded 973 articles, from which 104 distinct articles met the predefined inclusion criteria. Each type of cancer is diagnosed by clinical pathologists through HE staining or immunohistochemistry. It should be noted that some of these specimens were obtained through puncture, while others were obtained through surgical resection. There are no requirements for the size of tumor tissue or the location of the lesion, as long as which is sufficient for the pathological diagnosis. All cancer patients and their control healthy population were sampled from peripheral blood and tested for SNPs in IL‐8 gene using different detection methods.
2.3. Study Criteria
The present analysis incorporated studies that fulfill the following criteria of inclusion: (a) association between cancer susceptibility and just more types of six IL‐8 polymorphisms (−251, −353, +678, +1633, +2767, +781); (b) study design about case–control groups; and (c) adequate availability of each genotype data for both cases and controls or alternatively for certain genetic models; (d) Each type of cancer patients and healthy control population must be informed of the purpose, methods, significance, and risks of the study. Moreover, it is required to fill out a detailed questionnaire, mainly including age, gender, BMI, smoking history, alcohol consumption history, family history of cancer, cancer staging, and so on. The investigation period depends on each type of cancer and should not exceed 1 year at most. Finally, it is necessary to sign the informed consent form for the enrolled population. Also the subsequent exclusion criteria were implemented: First, no control population was included in the analysis, which may have affected the interpretation of the results. Second, the genotype frequency data was unavailable, which could have provided valuable insights into the genetic composition of the study population. Finally, duplicated publications should be identified and deleted.
2.4. Data Extraction for Meta‐Analysis
The study encompassed the collection of several key variables, including the name of authors, publication year, country of origin, ethnicity of the participants, specific type of cancer under investigation, the number of cases and controls, source of the control group, assessment of Hardy–Weinberg Equilibrium (HWE) in the control group, and the employed for genotyping.
2.5. Data Analysis
The present study measured odds ratios (OR) accompanied by 95% confidence intervals (CI) to evaluate the link between IL‐8 six polymorphisms and cancer risk. This was determined by comparing the genotype frequencies in both groups (cases and controls). The statistical importance of the summary OR was assessed via Z‐test [137]. The heterogeneity assumption was determined using a chi‐square‐based Q‐test between the studies: a p value greater than 0.05 was obtained and the random effects model was employed; however, the fixed effects model was selected [138, 139]. We employed various statistical analyses, including allelic contrast, homozygote comparison, dominant genetic model, heterozygote comparison, and recessive genetic model. The evaluation of HWE was executed in the control group via the Pearson chi‐square test. In order to examine the potential publication bias, Begg's and Egger's tests were conducted [140]. All statistical analyses for this meta‐analysis were conducted via Stata software (Version 11.0; StataCorp LP, College Station, TX). Finally, the quality of studies in meta‐analysis was assessing by Newcastle‐Ottawa Scale method [141].
2.6. Information of Participants
In this study, 90 patients were newly diagnosed with gastric cancer from February 2018 to July 2022. These patients were recruited from the Affiliated Hospital of Jiangnan University, and were selected based on clinical signs, tumor location, and tumor grade and stage according to WHO criteria. The histological confirmation of gastric cancer diagnosis was executed by pathologists affiliated with the Department of Pathology at the Affiliated Hospital of Jiangnan University. An age‐matched healthy control group (n = 90) was also recruited during the same time period undergoing routine physical examinations in the outpatient. The Each study participant was required to provide a peripheral blood sample of 2 mL. The ethical approval was obtained from the Institutional Review Board (IRB) of the Affiliated Hospital of Jiangnan University. Each participant's written informed consent was also obtained before the sample collection.
2.7. Genotyping and Enzyme‐Linked Immunosorbent Assay (ELISA)
For the present study, −251 polymorphism genotypes were assessed with a TaqMan assay using the approach documented by Castro et al. [142]. The levels of IL‐8 in serum were quantified via an ELISA kit (Abcam Co. ltd.). For specific operational procedures and data processing, please refer to previous reference [12].
3. Results
3.1. Study Selection via Meta‐Analysis
A comprehensive search of various databases yielded 973 articles. Following a deep evaluation, 104 distinct publications were deemed suitable for inclusion in the present study (Figure 1). The comprehensive details regarding the incorporated studies have been presented in Table 1. The IL‐8 expression was remarkably elevated in tumor tissues in contrast to normal tissues in multiple types of cancer (Figure 2A). This observation is supported by the data presented in Figure 2B–E.
FIGURE 1.
Flowchart depicting the systematic search strategy employed for the identification of studies investigating IL‐8 gene polymorphisms and their potential association with overall cancer risk.
TABLE 1.
Characteristics of included studies about polymorphisms in IL‐8 gene polymorphisms and cancer risk.
| Author | Year | Country | Ethnicity | Cancer type | Case | Control | SOC | HWE | Genotype |
|---|---|---|---|---|---|---|---|---|---|
| −251 | |||||||||
| Ahirwar [21] | 2010 | India | Asian | Bladder cancer | 205 | 270 | PB | 0.005 | AS‐PCR |
| Smith [100] | 2004 | UK | Caucasian | Breast cancer | 119 | 235 | PB | 0.131 | ARMS‐PCR |
| Zhang [127] | 2017 | China | Asian | Breast cancer | 442 | 447 | HB | 0.948 | PCR‐RFLP |
| Kamali‐Sarvestani [63] | 2007 | Iran | Asian | Breast cancer | 257 | 233 | HB | 0.26 | AS‐PCR |
| Snoussi [102] | 2010 | Tunisia | African | Breast cancer | 409 | 301 | PB | 0.173 | AS‐PCR |
| Wang [116] | 2022 | China | Asian | Breast cancer | 1232 | 1232 | HB | 0.231 | PCR‐RFLP |
| Wang [119] | 2014 | China | Asian | Breast cancer | 474 | 501 | HB | 0.005 | PCR‐RFLP |
| Althubyani [24] | 2020 | Eygypt | African | Colorectal cancer | 70 | 70 | HB | 0.932 | TaqMan |
| Burada [33] | 2013 | Romania | Caucasian | Colorectal cancer | 144 | 233 | HB | 0.291 | TaqMan |
| Ankathil [25] | 2019 | Malaysia | Asian | Colorectal cancer | 280 | 280 | HB | < 0.001 | PCR‐RFLP |
| Walczak [113] | 2012 | Poland | Caucasian | Colorectal cancer | 191 | 205 | PB | 0.001 | PCR‐RFLP |
| Theodoropoulos [107] | 2006 | Greece | Caucasian | Colorectal cancer | 222 | 196 | HB | 0.327 | PCR‐RFLP |
| Landi [74] | 2003 | Spain | Caucasian | Colorectal cancer | 352 | 308 | HB | 0.047 | TaqMan |
| Basavaraju [27] | 2015 | USA | Caucasian | Colorectal cancer | 388 | 491 | PB | 0.711 | TaqMan |
| Küry [73] | 2008 | France | Caucasian | Colorectal cancer | 923 | 1121 | HB | 0.033 | TaqMan |
| Tsilidis [108] | 2009 | USA | Caucasian | Colorectal cancer | 205 | 362 | PB | 0.058 | TaqMan |
| Mustapha [88] | 2012 | Malaysia | Asian | Colorectal cancer | 255 | 264 | HB | < 0.001 | AS‐PCR |
| Gunter [58] | 2006 | Italy | Caucasian | Colorectal cancer | 205 | 191 | HB | 0.84 | TaqMan |
| Wilkening [120] | 2008 | Sweden | Caucasian | Colorectal cancer | 300 | 580 | HB | 0.476 | TaqMan |
| Vogel [112] | 2007 | Denmark | Caucasian | Colorectal cancer | 355 | 753 | PB | 0.627 | PCR‐CE‐SSCP |
| Malespín‐Bendana [83] | 2021 | Costa Rica | Mixed | Gastric cancer | 46 | 81 | HB | 0.907 | PCR‐RFLP |
| Kamali‐Sarvestani [64] | 2006 | Iran | Asian | Gastric cancer | 19 | 153 | HB | 0.797 | AS‐PCR |
| Qadri [91] | 2014 | India | Asian | Gastric cancer | 130 | 200 | HB | 0.066 | PCR‐RFLP |
| Kamangar [65] | 2006 | USA | Caucasian | Gastric cancer | 112 | 207 | PB | 0.055 | TaqMan |
| Felipe [51] | 2012 | Brasil | Mixed | Gastric cancer | 104 | 196 | HB | 0.065 | PCR‐RFLP |
| Garza‐Gonzalez [55] | 2007 | USA | Mixed | Gastric cancer | 78 | 207 | HB | 0.492 | PCR‐RFLP |
| Ye [124] | 2009 | Korea | Asian | Gastric cancer | 153 | 206 | PB | 0.72 | PCR‐RFLP |
| Shirai [133] | 2006 | Japan | Asian | Gastric cancer | 181 | 468 | HB | 0.343 | PCR‐RFLP |
| Song [103] | 2009 | China | Asian | Gastric cancer | 125 | 140 | HB | 0.72 | PCR‐RFLP |
| Burada [32] | 2012 | Craiova | Caucasian | Gastric cancer | 105 | 242 | HB | 0.386 | AS‐PCR |
| Vinagre [111] | 2011 | Brasil | Mixed | Gastric cancer | 102 | 103 | HB | 0.15 | PCR‐RFLP |
| Wang [117] | 2016 | China | Asian | Gastric cancer | 132 | 296 | HB | 0.946 | PCR‐RFLP |
| Zeng [20] | 2005 | China | Asian | Gastric cancer | 104 | 94 | HB | 0.212 | PCR‐RDB |
| Zeng [20] | 2005 | China | Asian | Gastric cancer | 102 | 102 | HB | 0.042 | PCR‐RDB |
| Chang [129] | 2017 | Korea | Asian | Gastric cancer | 283 | 176 | HB | 0.136 | PCR‐RFLP |
| Chang [129] | 2017 | Korea | Asian | Gastric cancer | 283 | 284 | HB | 0.082 | PCR‐RFLP |
| Bo [30] | 2010 | China | Asian | Gastric cancer | 208 | 190 | HB | 0.389 | PCR‐RFLP |
| Taguchi [105] | 2005 | Japan | Asian | Gastric cancer | 396 | 252 | HB | 0.994 | PCR‐RFLP |
| Oliveira [47] | 2015 | Brazil | Mixed | Gastric cancer | 240 | 207 | HB | 0.488 | PCR‐RFLP |
| Kumar [72] | 2015 | India | Asian | Gastric cancer | 200 | 182 | PB | 0.801 | AS‐PCR |
| Kang [66] | 2009 | Korea | Asian | Gastric cancer | 334 | 322 | PB | 0.226 | PCR‐RFLP |
| Canedo [39] | 2008 | France | Caucasian | Gastric cancer | 333 | 693 | PB | 0.459 | TaqMan |
| Lu [82] | 2005 | China | Asian | Gastric cancer | 250 | 300 | PB | 0.516 | PCR‐DHPLC |
| Lee [76] | 2005 | China | Asian | Gastric cancer | 461 | 303 | HB | 0.184 | PCR‐RFLP |
| Savage [96] | 2006 | USA | Caucasian | Gastric cancer | 287 | 428 | PB | 0.391 | TaqMan |
| Zhang [128] | 2010 | China | Asian | Gastric cancer | 519 | 504 | PB | 0.754 | PCR‐RFLP |
| Li [79] | 2010 | China | Asian | Gastric cancer | 101 | 137 | HB | 0.579 | PCR‐DHPLC |
| Ohyauchi [89] | 2005 | Japan | Asian | Gastric cancer | 212 | 244 | HB | 0.847 | DS |
| Ko [69] | 2009 | Korea | Asian | Gastric cancer | 81 | 589 | PB | < 0.001 | Snapshot |
| Crusius [45] | 2008 | France | Caucasian | Gastric cancer | 236 | 1139 | PB | 0.705 | Real‐Time PCR |
| Leung [78] | 2006 | China | Asian | Gastric cancer | 83 | 179 | HB | 0.638 | TaqMan |
| Szoke [104] | 2008 | Hungary | Caucasian | Gastric cancer | 35 | 168 | HB | 0.165 | ARMS‐PCR |
| Ramis [94] | 2017 | Brazil | Mixed | Gastric cancer | 9 | 38 | PB | 0.691 | PCR‐RFLP |
| Fu [53] | 2016 | China | Asian | Glioma | 127 | 284 | HB | 0.251 | PCR‐RFLP |
| Liu [81] | 2015 | China | Asian | Glioma | 300 | 300 | HB | 0.772 | PCR‐RFLP |
| Chien [44] | 2011 | China | Asian | Hepatocellular carcinoma | 131 | 340 | HB | 0.445 | PCR‐RFLP |
| Wang [114] | 2014 | China | Asian | Hepatocellular carcinoma | 205 | 208 | HB | 0.266 | PCR‐RFLP/PCR‐SSP |
| Liao [17] | 2011 | China | Asian | Hepatocellular carcinoma | 390 | 150 | HB | 0.104 | PCR‐RFLP |
| Elsamanoudy [132] | 2015 | Egypt | African | Hepatocellular carcinoma | 112 | 105 | HB | 0.551 | PCR‐RFLP |
| Qin [92] | 2012 | China | Asian | Hepatocellular carcinoma | 150 | 150 | HB | 0.104 | PCR‐RFLP |
| Lu [19] | 2015 | China | Asian | Hepatocellular carcinoma | 454 | 446 | HB | 0.115 | PCR‐RFLP |
| Rafrafi [93] | 2013 | Tunisia | African | Lung cancer | 170 | 225 | PB | 0.181 | PCR‐RFLP |
| Yamamoto [122] | 2017 | Japan | Asian | Lung cancer | 462 | 379 | HB | 0.939 | TaqMan |
| Campa [38] | 2004 | Norway | Caucasian | Lung cancer | 239 | 210 | PB | 0.317 | TaqMan |
| Kaanane [62] | 2022 | Morocco | African | Lung cancer | 150 | 150 | PB | 0.169 | TaqMan |
| Bhat [29] | 2013 | India | Asian | Lung cancer | 190 | 200 | HB | < 0.001 | PCR‐RFLP |
| Campa [37] | 2005 | Germany | Caucasian | Lung cancer | 2144 | 2116 | PB | 0.203 | TaqMan |
| Vogel [134] | 2008 | Denmark | Caucasian | Lung cancer | 403 | 744 | PB | 0.672 | PCR‐RFLP |
| Li [130] | 2015 | China | Asian | Lung cancer | 132 | 150 | HB | 0.894 | PCR‐HRM |
| Tai [18] | 2007 | China | Asian | Nasopharyngeal carcinoma | 105 | 109 | HB | 0.886 | PCR‐RFLP |
| Huang [61] | 2018 | China | Asian | Nasopharyngeal carcinoma | 176 | 352 | HB | 0.109 | PCR‐RFLP |
| Nasr [28] | 2007 | Tunisia | African | Nasopharyngeal carcinoma | 160 | 169 | PB | 0.349 | PCR‐SSP |
| Wei [131] | 2007 | China | Asian | Nasopharyngeal carcinoma | 280 | 290 | PB | 0.164 | PCR‐RFLP |
| Matos [46] | 2019 | Brazil | Mixed | Oral cancer | 66 | 130 | HB | 0.493 | PCR |
| Vairaktaris [110] | 2007 | Germany | Caucasian | Oral cancer | 158 | 156 | HB | < 0.001 | PCR‐RFLP |
| Qin [92] | 2012 | China | Asian | Oral cancer | 150 | 150 | HB | 0.104 | PCR‐RFLP |
| Liu [82] | 2012 | China | Asian | Oral cancer | 270 | 350 | HB | 0.454 | PCR‐RFLP |
| Singh [99] | 2016 | India | Asian | Oral cancer | 300 | 300 | HB | < 0.001 | PCR‐RFLP |
| Campa [36] | 2017 | Germany | Caucasian | Oral cancer | 153 | 725 | HB | 0.524 | TaqMan |
| Shimizu [98] | 2008 | Japan | Asian | Oral cancer | 69 | 91 | HB | 0.296 | PCR‐FLP |
| Kietthubthew [67] | 2010 | Thailand | Asian | Oral cancer | 63 | 99 | PB | 0.813 | TaqMan |
| Moreno‐Guerrero [87] | 2021 | Mexico | Mixed | Neuroblastoma | 27 | 38 | HB | 0.152 | PCR‐RFLP |
| Kilic [68] | 2016 | Turkey | Caucasian | Thyroid cancer | 101 | 109 | HB | 0.586 | PCR |
| Wu [121] | 2013 | China | Asian | Urothelial carcinoma | 300 | 594 | HB | 0.075 | PCR‐RFLP |
| Kuyl [135] | 2004 | Netherlands | Caucasian | Kaposi's sarcoma | 84 | 153 | HB | 0.382 | PCR‐RFLP |
| Chen [43] | 2016 | China | Asian | Osteosarcoma | 190 | 190 | HB | < 0.001 | PCR‐RFLP |
| Howell [59] | 2003 | UK | Caucasian | Melanoma | 142 | 233 | HB | 0.16 | ARMS–PCR |
| Cacev [35] | 2008 | Croatia | Caucasian | Colon cancer | 160 | 160 | PB | 0.346 | PCR‐RFLP |
| Koensgen [70] | 2014 | Germany | Caucasian | Ovarian cancer | 267 | 426 | HB | 0.026 | PCR‐RFLP |
| Franz [52] | 2017 | Brazil | Mixed | Prostate cancer | 175 | 185 | HB | 0.127 | PCR‐SSP |
| Chen [42] | 2016 | China | Asian | Prostate cancer | 439 | 524 | HB | 0.129 | PCR‐RFLP |
| Taheri [106] | 2019 | Iran | Asian | Prostate cancer | 355 | 200 | HB | 0.689 | ARMS‐PCR |
| Yang [123] | 2006 | USA | Caucasian | Prostate cancer | 520 | 418 | PB | 0.168 | ht‐SNP |
| McCarron [136] | 2002 | UK | Caucasian | Prostate cancer | 238 | 235 | HB | 0.131 | PCR |
| Michaud [85] | 2006 | USA | Caucasian | Prostate cancer | 484 | 613 | PB | 0.777 | PCR |
| Leibovici [77] | 2005 | USA | Caucasian | Bladder cancer | 463 | 440 | HB | NA | TaqMan |
| Zamora‐Ros [125] | 2015 | Spain | Caucasian | Colorectal cancer | 344 | 303 | HB | NA | TaqMan |
| Oliveira [48] | 2013 | Brazil | Mixed | Gastric cancer | 200 | 240 | HB | NA | PCR‐RFLP |
| Pan [90] | 2014 | China | Asian | Gastric cancer | 308 | 308 | PB | NA | MALDI‐TOF MS |
| Boonyanugomol [31] | 2019 | Korea | Asian | Gastric cancer | 10 | 72 | HB | NA | PCR‐RFLP |
| Zhang [126] | 2010 | USA | Caucasian | Prostate cancer | 162 | 173 | PB | NA | MOLD‐TOF‐MS |
| −353 | |||||||||
| Wei [131] | 2007 | China | Asian | Nasopharyngeal carcinoma | 280 | 290 | PB | 0.406 | PCR‐RFLP |
| Wang [114] | 2014 | China | Asian | Hepatocellular carcinoma | 205 | 208 | HB | 0.474 | PCR‐RFLP/PCR‐SSP |
| Zhang [127] | 2017 | China | Asian | Breast cancer | 442 | 447 | HB | < 0.001 | PCR‐RFLP |
| +678 | |||||||||
| Wei [131] | 2007 | China | Asian | Nasopharyngeal carcinoma | 280 | 290 | PB | 0.064 | PCR‐RFLP |
| Ahirwar [21] | 2010 | India | Asian | Bladder cancer | 205 | 270 | PB | < 0.001 | AS‐PCR |
| Wang [114] | 2014 | China | Asian | Hepatocellular carcinoma | 205 | 208 | HB | 0.161 | AS‐PCR |
| +1633 | |||||||||
| Chien [44] | 2011 | China | Asian | Hepatocellular carcinoma | 131 | 340 | HB | 0.562 | PCR‐RFLP |
| Liu [82] | 2012 | China | Asian | Oral cancer | 270 | 350 | HB | 0.569 | PCR‐RFLP |
| Koensgen [70] | 2014 | Germany | Caucasian | Ovarian cancer | 246 | 62 | HB | 0.865 | PCR‐RFLP |
| Huang [61] | 2018 | China | Asian | Nasopharyngeal carcinoma | 176 | 352 | HB | 0.109 | PCR‐RFLP |
| +2767 | |||||||||
| Chien [44] | 2011 | China | Asian | Hepatocellular carcinoma | 131 | 340 | HB | 0.392 | PCR‐RFLP |
| Liu [82] | 2012 | China | Asian | Oral cancer | 270 | 350 | HB | 0.029 | PCR‐RFLP |
| Koensgen [70] | 2014 | Germany | Caucasian | Ovarian cancer | 268 | 426 | HB | 0.029 | PCR‐RFLP |
| Hsieh [60] | 2007 | China | Asian | Leiomyoma | 162 | 156 | HB | 0.078 | PCR‐RFLP |
| Huang [61] | 2018 | China | Asian | Nasopharyngeal carcinoma | 176 | 352 | HB | 0.012 | PCR‐RFLP |
| −781 | |||||||||
| Liu [81] | 2015 | China | Asian | Glioma | 300 | 300 | HB | 0.049 | PCR‐RFLP |
| Kamangar [65] | 2006 | USA | Caucasian | Gastric cancer | 111 | 208 | PB | 0.158 | TaqMan |
| Bo [30] | 2010 | China | Asian | Gastric cancer | 208 | 190 | HB | 0.225 | PCR‐RFLP |
| Chien [44] | 2011 | China | Asian | Hepatocellular carcinoma | 131 | 340 | HB | 0.776 | PCR‐RFLP |
| Liu [82] | 2012 | China | Asian | Oral cancer | 270 | 350 | HB | 0.781 | PCR‐RFLP |
| Qin [92] | 2012 | China | Asian | Oral cancer | 150 | 150 | HB | 0.041 | PCR‐RFLP |
| Rafrafi [93] | 2013 | Tunisia | African | Lung cancer | 170 | 225 | PB | 0.329 | PCR‐RFLP |
| Wang [114] | 2014 | China | Asian | Hepatocellular carcinoma | 205 | 208 | HB | 0.549 | PCR‐RFLP/PCR‐SSP |
| Koensgen [70] | 2014 | Germany | Caucasian | Ovarian cancer | 267 | 426 | HB | 0.1 | PCR‐RFLP |
| Chen [43] | 2016 | China | Asian | Osteosarcoma | 190 | 190 | HB | 0.116 | PCR‐RFLP |
| Taheri [106] | 2019 | Iran | Asian | Prostate cancer | 355 | 200 | HB | 0.639 | ARMS‐PCR |
| Kaanane [62] | 2022 | Morocco | African | Lung cancer | 150 | 150 | PB | 0.307 | TaqMan |
| Alkanli [23] | 2023 | Turkey | Caucasian | Bladder cancer | 88 | 89 | HB | 0.608 | PCR‐RFLP |
| Moreno‐Guerrero [87] | 2021 | Mexico | Mixed | Neuroblastoma | 27 | 38 | HB | 0.313 | PCR‐RFLP |
| Song [103] | 2009 | China | Asian | Gastric cancer | 125 | 140 | HB | 0.48 | PCR‐RFLP |
| Fu [53] | 2016 | China | Asian | Glioma | 127 | 284 | HB | 0.788 | PCR‐RFLP |
| Zhang [127] | 2017 | China | Asian | Breast cancer | 442 | 447 | HB | 0.327 | PCR‐RFLP |
| Huang [61] | 2018 | China | Asian | Nasopharyngeal carcinoma | 176 | 352 | HB | 0.671 | PCR‐RFLP |
| Ghazy [56] | 2021 | Saudi Arabia | Asian | Prostate cancer | 40 | 40 | HB | 0.673 | real‐time PCR |
| Liao [17] | 2011 | China | Asian | Hepatocellular carcinoma | 150 | 150 | HB | 0.041 | PCR‐RFLP |
| Elsamanoudy [132] | 2015 | Egypt | African | Hepatocellular carcinoma | 112 | 105 | HB | 0.178 | PCR‐RFLP |
| Qin [92] | 2012 | China | Asian | Hepatocellular carcinoma | 150 | 150 | HB | 0.041 | PCR‐RFLP |
| Lu [19] | 2015 | China | Asian | Hepatocellular carcinoma | 454 | 446 | HB | 0.062 | PCR‐RFLP |
Abbreviations: ARMS: amplification refractory mutation system; AS: allele specific primer; CE‐SSCP: capillary electrophoresis‐single strand conformation polymorphism; DHPLC: denaturing high performance liquid chromatography; HB: hospital‐based; HRM: high resolution melt; HWE: Hardy–Weinberg equilibrium of control group; MALDI‐TOF MS: matrix‐assisted laser desorption/ionization time of flight mass spectrometry; PB: population‐based; PCR‐RFLP: polymerase chain reaction followed by restriction fragment length polymorphism; SOC; source of control; SSP: sequence specific primer.
FIGURE 2.
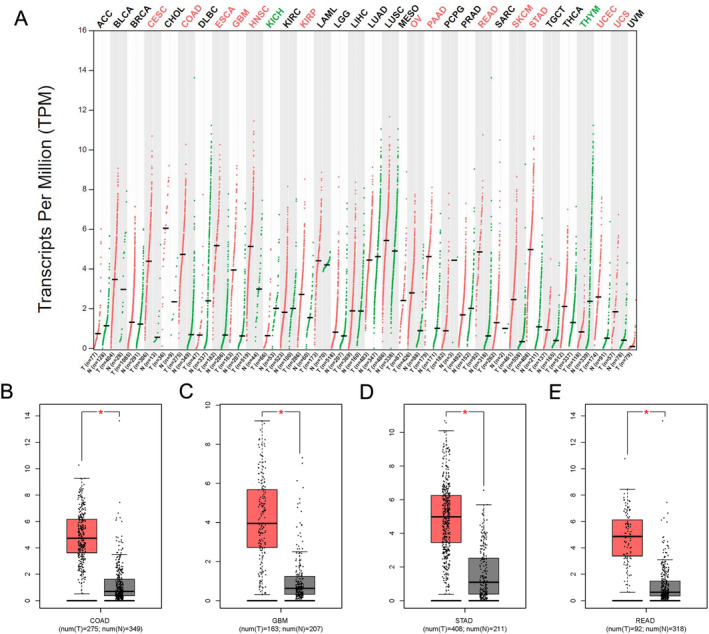
Bioinformatics examinations of IL‐8 gene. (A) The expression profile of IL‐8 gene in all tumor samples and paired normal tissues. (B) IL‐8 gene expression in colon adenocarcinoma. *p < 0.05. (C) IL‐8 gene expression in glioblastoma multiforme. (D) IL‐8 gene expression in stomach adenocarcinoma. (E) IL‐8 gene expression in rectum adenocarcinoma. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B‐cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
3.2. Relationship Between the Expression of IL‐8 and Gastric Cancer From TCGA Data
First, the expression of IL‐8 was remarkably elevated in tumor tissue compared to normal tissue (p < 0.001) (Figure 3A). Second, clinicopathological factors of gastric cancer were analyzed, age more than 65 had higher expression of IL‐8 (p < 0.05) (Figure 3B), however, no positive result was observed in subgroup including gender, grade and TNM stage (Table 2) (Figure 3C–F). Furthermore, prognostic factors for recurrence‐free survival were calculated using univariate and multivariate analyses. Age, grade and stage were three significant prognostic factors (p < 0.05) (Table 3) (Figure 3G,H).
FIGURE 3.
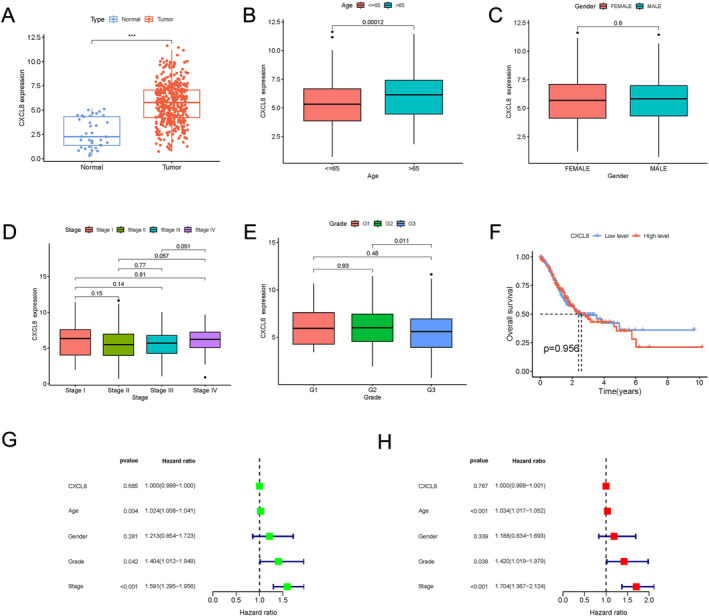
TCGA database information shows IL‐8 expression in gastric cancer. (A) The expression of the IL‐8 gene in samples of gastric carcinoma and paired normal tissues. (B) The expression of the IL‐8 gene in gastric cancer samples from individuals less than 65 years and older than 65 years. (C) The IL‐8 gene expression difference between female and male gastric cancer samples. (D) The expression of IL‐8 at various gastric cancer stages. (E) The expression of IL‐8 among different grades of gastric cancer. (F) The overall survival between the level of IL‐8 gene expression. Analyses of prognostic factors for progression‐free survival in univariate (G) and multivariate (H).
TABLE 2.
Correlation between IL‐8 expression and clinicopathological factors in gastric cancer from TCGA database.
| Covariates | Group | Total | IL‐8 expression | chi | p | |
|---|---|---|---|---|---|---|
| Low | High | |||||
| Age | ≤ 65 | 184 (45.21%) | 110 (53.66%) | 74 (36.63%) | 11.228 | 8.00E‐04 |
| > 65 | 223 (54.79%) | 95 (46.34%) | 128 (63.37%) | |||
| Gender | Female | 145 (35.19%) | 74 (35.92%) | 71 (34.47%) | 0.0426 | 0.8365 |
| Male | 267 (64.81%) | 132 (64.08%) | 135 (65.53%) | |||
| G1 | 12 (2.98%) | 6 (2.99%) | 6 (2.97%) | 2.0173 | 0.3647 | |
| G2 | 148 (36.72%) | 67 (33.33%) | 81 (40.1%) | |||
| G3 | 243 (60.3%) | 128 (63.68%) | 115 (56.93%) | |||
| Grade | I | 58 (14.95%) | 25 (12.76%) | 33 (17.19%) | 5.0362 | 0.1692 |
| II | 122 (31.44%) | 68 (34.69%) | 54 (28.12%) | |||
| III | 169 (43.56%) | 88 (44.9%) | 81 (42.19%) | |||
| IV | 39 (10.05%) | 15 (7.65%) | 24 (12.5%) | |||
| T | T1 | 22 (5.45%) | 11 (5.37%) | 11 (5.53%) | 4.0643 | 0.2546 |
| T2 | 88 (21.78%) | 38 (18.54%) | 50 (25.13%) | |||
| T3 | 181 (44.8%) | 101 (49.27%) | 80 (40.2%) | |||
| T4 | 113 (27.97%) | 55 (26.83%) | 58 (29.15%) | |||
| M | M0 | 365 (93.35%) | 186 (94.42%) | 179 (92.27%) | 0.4218 | 0.516 |
| M1 | 26 (6.65%) | 11 (5.58%) | 15 (7.73%) | |||
| N | N0 | 124 (31.55%) | 62 (31.31%) | 62 (31.79%) | 1.0709 | 0.7841 |
| N1 | 109 (27.74%) | 51 (25.76%) | 58 (29.74%) | |||
| N2 | 78 (19.85%) | 41 (20.71%) | 37 (18.97%) | |||
| N3 | 82 (20.87%) | 44 (22.22%) | 38 (19.49%) | |||
TABLE 3.
Prognostic factors for recurrence‐free survival in univariate and multivariate analyses.
| Covariates | Univariate analysis | p | Multivariate | p |
|---|---|---|---|---|
| HR (95%CI) | HR (95%CI) | |||
| CXCL8 | 0.999 (0.999–1) | 0.684 | 0.999 (0.999–1) | 0.766 |
| Age | 1.024 (1.007–1.041) | 0.003 | 1.034 (1.016–1.052) | 0 |
| Gender | 1.212 (0.854–1.722) | 0.28 | 1.188 (0.834–1.692) | 0.339 |
| Grade | 1.404 (1.011–1.949) | 0.0423 | 1.419 (1.018–1.978) | 0.038 |
| Stage | 1.591 (1.294–1.956) | 1.03E‐05 | 1.703 (1.366–2.123) | 2.16E‐06 |
3.3. Meta‐Analysis
The analysis revealed a significant increase in the correlation among the −251 polymorphism and cancer risk (such as: A‐allele vs. T‐allele, OR = 1.078, 95%CI = 1.020–1.140, p = 0.008, Table 4). Furthermore, a higher prevalence of associations was observed among Asian in relation to the −251 polymorphism (such as: AA+AT vs. TT: OR = 1.168, 95%CI = 1.047–1.303, p < 0.005, Figure 4A, Table 4). Substantial relationships in four distinct types of cancer were observed (such as gastric cancer: such as AT vs. TT, OR = 1.292, 95%CI = 1.117–1.494, p = 0.001, Figure 4B, Table 4). There was an elevated link between −353 polymorphism and cancer risk, such as A‐allele versus T‐allele, OR = 1.255, 95%CI = 1.079–1.459, p = 0.003 (Figure 4C, Table 4). For −781 polymorphism, a single potential link was noted in the ethnicity subgroup: Caucasian, TT versus TC + CC, OR = 1.472, 95%CI = 1.078–2.009, p = 0.015 (Figure 4D, Table 4).
TABLE 4.
Stratified analyses of IL‐8 genes common polymorphisms on cancer risk.
| Variables | No | Case/Controls | M‐allele versus W‐allele OR (95%CI) P h P | MM versus WW OR (95%CI) P h P | MW versus WW OR (95%CI) P h P | MM + MW versus WW OR (95%CI) P h P | MM versus MW + WW OR (95%CI) P h P |
|---|---|---|---|---|---|---|---|
| IL‐8 −251 | |||||||
| Total | 101 | 25 750/31995 | 1.078 (1.020–1.140)0.000 0.008 | 1.171 (1.052–1.303)0.000 0.004 | 1.119 (1.033–1.211)0.000 0.006 | 1.111 (1.032–1.196)0.000 0.005 | 1.100 (1.004–1.206)0.000 0.041 |
| Ethnicity | |||||||
| Asian | 52 | 13 058/14784 | 1.118 (1.022–1.222)0.000 0.014 | 1.295 (1.088–1.541)0.000 0.004 | 1.167 (1.037–1.314)0.000 0.010 | 1.168 (1.047–1.303)0.000 0.005 | 1.211 (1.031–1.423)0.000 0.020 |
| Caucasian | 33 | 10 574/14766 | 1.030 (0.969–1.095)0.000 0.342 | 1.043 (0.930–1.171)0.004 0.469 | 1.016 (0.907–1.139)0.000 0.780 | 1.023 (0.923–1.134)0.000 0.664 | 1.026 (0.948–1.111)0.125 0.524 |
| African | 6 | 1071/1020 | 0.979 (0.660–1.453)0.000 0.917 | 0.947 (0.458–1.960)0.000 0.884 | 1.013 (0.657–1.563)0.004 0.952 | 0.981 (0.578–1.663)0.000 0.942 | 0.962 (0.577–1.606)0.000 0.883 |
| Mixed | 10 | 1047/1425 | 1.076 (0.876–1.321)0.023 0.483 | 1.093 (0.737–1.619)0.047 0.659 | 1.402 (1.041–1.889)0.086 0.026 | 1.205 (0.907–1.602)0.019 0.198 | 0.874 (0.646–1.183)0.122 0.384 |
| Cancer type | |||||||
| Gastric cancer | 36 | 6562/9650 | 1.115 (0.988–1.257)0.000 0.077 | 1.248 (1.004–1.553)0.000 0.046 | 1.292 (1.117–1.494)0.000 0.001 | 1.200 (1.054–1.368)0.000 0.006 | 1.135 (0.921–1.398)0.000 0.234 |
| Hepatocellular carcinoma | 6 | 1442/1399 | 1.023 (0.844–1.241)0.014 0.814 | 0.963 (0.690–1.344)0.096 0.825 | 1.209 (0.793–1.842)0.000 0.377 | 1.149 (0.779–1.693)0.000 0.484 | 0.828 (0.647–1.060)0.255 0.134 |
| Prostate cancer | 7 | 2373/2348 | 0.958 (0.862–1.064)0.200 0.422 | 0.936 (0.753–1.163)0.168 0.549 | 0.929 (0.770–1.119)0.121 0.436 | 0.927 (0.782–1.099)0.143 0.383 | 0.936 (0.783–1.120)0.151 0.471 |
| Oral cancer | 8 | 1229/2001 | 1.075 (0.882–1.311)0.003 0.473 | 1.206 (0.743–1.960)0.001 0.448 | 0.863 (0.674–1.104)0.055 0.240 | 0.965 (0.803–1.160)0.265 0.703 | 1.369 (0.777–2.414)0.000 0.277 |
| Lung cancer | 8 | 3890/4174 | 0.888 (0.757–1.042)0.000 0.147 | 0.830 (0.617–1.115)0.001 0.215 | 0.926 (0.794–1.080)0.174 0.326 | 0.889 (0.728–1.086)0.010 0.251 | 0.861 (0.685–1.083)0.003 0.201 |
| Glioma | 2 | 427/584 | 1.278 (1.067–1.532)0.406 0.008 | 1.660 (1.162–2.371)0.315 0.005 | 0.982 (0.729–1.322)0.386 0.904 | 1.171 (0.889–1.544)0.817 0.262 | 1.581 (0.929–2.691)0.085 0.092 |
| Bladder cancer | 2 | 668/710 | 1.590 (1.227–2.061)0.000 0.000 | 2.196 (1.377–3.504)0.000 0.001 | 0.977 (0.635–1.503)0.000 0.917 | 1.264 (0.989–1.616)0.563 0.061 | 2.221 (1.463–3.372)0.000 0.000 |
| Breast cancer | 6 | 2933/2949 | 1.158 (0.885–1.514)0.000 0.285 | 1.406 (0.829–2.386)0.000 0.206 | 1.233 (0.831–1.829)0.000 0.299 | 1.283 (0.845–1.948)0.000 0.242 | 1.199 (0.868–1.655)0.000 0.271 |
| Colorectal cancer | 14 | 4234/5357 | 1.121 (0.999–1.257)0.000 0.053 | 1.320 (1.016–1.715)0.000 0.038 | 1.101 (0.876–1.385)0.000 0.409 | 1.107 (0.898–1.364)0.000 0.342 | 1.230 (1.018–1.486)0.001 0.032 |
| Nasopharyngeal carcinoma | 4 | 721/920 | 1.079 (0.717–1.623)0.000 0.715 | 1.131 (0.535–2.392)0.000 0.746 | 1.116 (0.703–1.772)0.005 0.640 | 1.124 (0.658–1.920)0.000 0.670 | 1.075 (0.646–1.789)0.018 0.780 |
| Other cancer | 8 | 1271/1903 | 1.089 (0.905–1.310)0.011 0.367 | 1.130 (0.816–1.565)0.087 0.461 | 1.096 (0.875–1.372)0.102 0.424 | 1.122 (0.879–1.433)0.026 0.356 | 1.066 (0.830–1.370)0.225 0.617 |
| Source of control | |||||||
| HB | 70 | 16 121/19095 | 1.116 (1.048–1.188)0.000 0.971 | 1.244 (1.090–1.420)0.000 0.001 | 1.149 (1.040–1.269)0.000 0.006 | 1.157 (1.054–1.269)0.000 0.002 | 1.154 (1.032–1.291)0.000 0.012 |
| PB | 31 | 9629/12900 | 0.998 (0.894–1.114)0.000 0.001 | 1.039 (0.863–1.251)0.000 0.685 | 1.059 (0.925–1.212)0.000 0.405 | 1.024 (0.907–1.157)0.000 0.698 | 1.002 (0.851–1.180)0.000 0.983 |
| IL‐8 −353 | 3 | 927/945 | 1.255 (1.079–1.459)0.449 0.003 | 1.463 (1.068–2.004)0.653 0.018 | 1.269 (0.980–1.643)0.732 0.070 | 1.339 (1.052–1.705)0.524 0.018 | 1.297 (1.031–1.632)0.784 0.026 |
| IL‐8 + 678 | 3 | 690/768 | 1.020 (0.866–1.201)0.970 0.816 | 1.166 (0.837–1.623)0.814 0.364 | 0.881 (0.696–1.115)0.390 0.291 | 0.952 (0.770–1.178)0.785 0.652 | 1.203 (0.875–1.655)0.702 0.256 |
| IL‐8 + 1633 | 4 | 823/1104 | 0.968 (0.804–1.166)0.166 0.733 | 0.937 (0.651–1.349)0.197 0.727 | 0.978 (0.789–1.211)0.569 0.837 | 0.963 (0.769–1.206)0.304 0.740 | 0.935 (0.719–1.215)0.372 0.614 |
| IL‐8 + 2767 | 5 | 1007/1624 | 0.930 (0.799–1.082)0.149 0.347 | 0.875 (0.626–1.224)0.094 0.435 | 0.924 (0.774–1.102)0.577 0.380 | 0.913 (0.774–1.078)0.532 0.283 | 0.905 (0.638–1.283)0.034 0.575 |
| IL‐8 + 781 | |||||||
| Total | 23 | 4398/5178 | 0.942 (0.836–1.062)0.000 0.329 | 0.904 (0.694–1.178)0.000 0.454 | 0.966 (0.846–1.104)0.003 0.617 | 0.953 (0.824–1.102)0.000 0.518 | 0.917 (0.733–1.146)0.000 0.446 |
| Ethnicity | |||||||
| Asian | 16 | 3473/3937 | 0.948 (0.854–1.054)0.005 0.323 | 0.884 (0.692–1.131)0.003 0.327 | 0.969 (0.841–1.117)0.021 0.667 | 0.956 (0.833–1.098)0.013 0.523 | 0.893 (0.715–1.116)0.004 0.321 |
| Caucasian | 3 | 466/723 | 1.189 (0.856–1.653)0.043 0.302 | 1.528 (0.773–3.020)0.063 0.223 | 1.225 (0.761–1.973)0.067 0.404 | 1.276 (0.746–2.181)0.024 0.374 | 1.472 (1.078–2.009)0.366 0.015 |
| African | 3 | 432/480 | 0.826 (0.406–1.681)0.000 0.598 | 0.809 (0.199–3.297)0.000 0.767 | 0.746 (0.461–1.205)0.070 0.231 | 0.750 (0.370–1.521)0.001 0.426 | 0.932 (0.282–3.077)0.001 0.907 |
| Mixed | 1 | 27/38 | 0.424 (0.208–0.866)0.000 0.018 | 0.164 (0.033–0.810)0.000 0.027 | 0.933 (0.271–3.209)0.000 0.913 | 0.536 (0.167–1.718)0.000 0.294 | 0.172 (0.044–0.671)0.000 0.011 |
| Cancer type | |||||||
| Gastric cancer | 3 | 444/538 | 1.119 (0.931–1.346)0.456 0.232 | 1.267 (0.864–1.859)0.675 0.226 | 1.118 (0.845–1.479)0.411 0.434 | 1.148 (0.883–1.494)0.368 0.303 | 1.171 (0.826–1.659)0.859 0.375 |
| Hepatocellular carcinoma | 6 | 1202/1399 | 0.778 (0.595–1.019)0.000 0.068 | 0.602 (0.357–1.017)0.004 0.058 | 0.826 (0.575–1.187)0.001 0.301 | 0.763 (0.529–1.100)0.000 0.147 | 0.659 (0.415–1.045)0.011 0.076 |
| Prostate cancer | 2 | 395/240 | 0.613 (0.183–2.056)0.001 0.428 | 0.352 (0.026–4.717)0.003 0.431 | 0.535 (0.089–3.207)0.031 0.494 | 0.429 (0.048–3.804)0.006 0.447 | 0.622 (0.183–2.114)0.018 0.447 |
| Oral cancer | 2 | 420/500 | 0.877 (0.724–1.063)0.470 0.181 | 0.866 (0.548–1.368)0.289 0.536 | 0.765 (0.579–1.012)0.956 0.061 | 0.783 (0.600–1.021)0.792 0.070 | 1.014 (0.627–1.639)0.247 0.955 |
| Lung cancer | 2 | 320/375 | 1.181 (0.819–1.704)0.127 0.373 | 1.617 (0.650–4.022)0.083 0.302 | 0.939 (0.674–1.306)0.791 0.707 | 1.065 (0.785–1.443)0.363 0.687 | 1.650 (0.694–3.924)0.089 0.257 |
| Glioma | 2 | 427/584 | 1.117 (0.883–1.414)0.235 0.355 | 1.343 (0.888–2.031)0.366 0.163 | 0.990 (0.728–1.346)0.270 0.949 | 1.069 (0.780–1.463)0.228 0.680 | 1.346 (0.906–2.002)0.543 0.142 |
| Other cancer | 6 | 1190/1542 | 0.976 (0.747–1.274)0.000 0.856 | 0.858 (0.426–1.727)0.000 0.667 | 0.980 (0.868–1.107)0.096 0.358 | 1.080 (0.790–1.477)0.009 0.629 | 0.801 (0.451–1.422)0.000 0.448 |
| Source of control | |||||||
| HB | 20 | 3967/4595 | 0.920 (0.806–1.050)0.000 0.215 | 0.849 (0.635–1.135)0.000 0.268 | 0.971 (0.834–1.130)0.001 0.701 | 0.942 (0.798–1.110)0.000 0.474 | 0.863 (0.678–1.099)0.000 0.232 |
| PB | 3 | 431/583 | 1.087 (0.846–1.397)0.191 0.514 | 1.336 (0.735–2.428)0.147 0.343 | 0.911 (0.693–1.198)0.919 0.505 | 1.002 (0.776–1.294)0.512 0.987 | 1.383 (0.799–2.393)0.170 0.247 |
Abbreviations: HB: hospital‐based; P: Z‐test for the statistical significance of the OR; PB: population‐based; P h: value of Q‐test for heterogeneity test; SOC; source of control.
FIGURE 4.
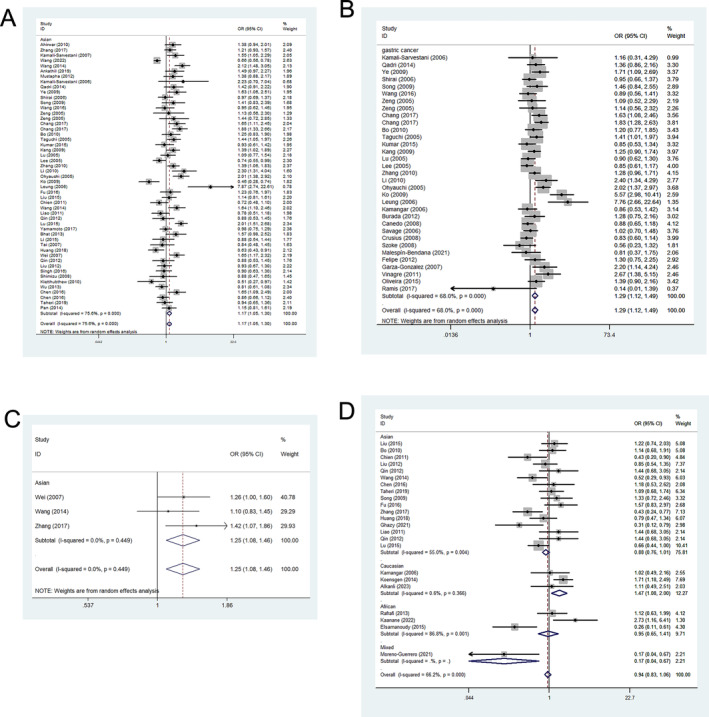
Forest plots corresponding to cancer‐related risk between the IL‐8 polymorphisms. The squares and horizontal lines respectively correspond to the study‐specific ORs and 95% CIs, with square area being indicative of weight (the inverse of the variance). Diamonds additionally reflect the summary OR and 95% CI. (A) Relationship between −251 polymorphism and cancer risk in Asians based on the dominant genetic model. (B) Relationship between −251 polymorphism and gastric cancer based on heterozygote comparison. (C) Relationship between −353 polymorphism and cancer risk based on allelic contrast. (D) Relationship between +781 polymorphism and cancer risk in Caucasians based on the recessive genetic model.
3.4. IL‐8 Expression in the Serum of Gastric Cancer Patients
In this study, 180 serum samples (90 patients were newly diagnosed with gastric cancer and 90 individuals were from age‐matched healthy control group) were collected. These samples were specifically selected to represent various genotypes of the IL‐8 −251 variant via ELISA. Specifically, this study exhibited that the serum IL‐8 levels in gastric cancer patients with AA/TT genotypes were significantly higher than those with TT genotypes and also higher than those with same AA/TT genotypes from healthy controls (p < 0.01, as illustrated in Figure 5).
FIGURE 5.
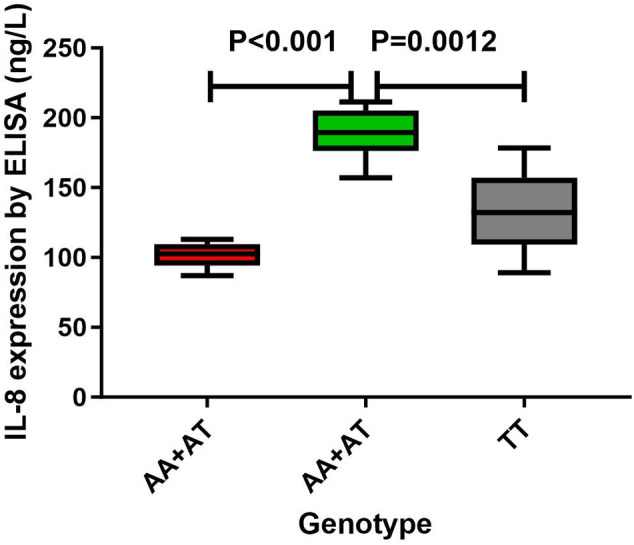
Serum analysis of IL‐8 levels in −251 genotype of gastric cancer using mean values (horizontal lines, mean values). Serum IL‐8 levels in gastric cancer patients carrying AA/TT genotypes were remarkably higher than that carrying TT genotypes (p < 0.01). Serum IL‐8 concentrations were also significantly higher in gastric cancer patients with the AA/TT genotypes as compared to healthy controls with the same genotypes (p < 0.01).
4. Discussion
Globally, cancer remains a predominant cause of both mortality and morbidity, resulting in approximately 9 million deaths annually [143]. IL‐8 is a prominent pro‐inflammatory mediator that has been extensively studied as a potential risk factor in the pathogenesis and progression of cancer. In addition, a number of SNPs within the IL‐8 gene, situated in its promoter region, have been implicated in the modulation of IL‐8 expression levels. For example, the A allele of the −251 SNP has been found to be associated with increased protein expression compared to the T allele. Therefore, it is hypothesized that the presence of SNPs in the IL‐8 gene may indirectly influence the expression of IL‐8, potentially influencing the development and progression of tumors [144].
In previous studies, a number of meta‐analyses have been conducted this association, however, the conclusion remains not clear and definite. For instance, Farbod et al. discovered that the IL‐8 −251 T/A polymorphism exhibited a significant association with susceptibility to breast cancer [145]. Additional, Chen et al. proposed that −251 polymorphism of the IL‐8 exhibited a significant association with susceptibility to prostate cancer. Moreover, Wang et al. suggested that this specific polymorphism could potentially act as a genetic biomarker for the identification of gastric cancer in Asian individuals [146]. On the other hand, Rezaei, Antikchi, and Gao et al. reported negative results regarding several types of cancer [147, 148, 149]. Therefore, it is necessary to make an up‐dated analysis. In our current investigation, a comprehensive meta‐analysis was executed to elucidate the potential correlation between six IL‐8 polymorphisms and the susceptibility to various types of cancer, which had two advantages: on one hand, current study included the most largest samples than previous meta‐analysis, on the other hand, serum IL‐8 expression was added, and was analyzed the relationship between different genotypes and IL‐8 expression, which was the novel exploration. Finally, 104 case–control studies were incorporated into the analysis. The findings exhibited a statistically substantial correlation between the IL‐8 −251 polymorphism and susceptibility to various types of cancer. A stratified analysis examined the connection between the −251 polymorphism and various cancer types. The findings revealed that the −251 polymorphism was identified as a risk factor for gastric, glioma, bladder, and colorectal cancer. Specifically, individuals carrying the A‐allele were more susceptible to developing these cancer types. However, no substantial correlation was detected between the −251 polymorphism and the incidence of hepatocellular carcinoma, prostate cancer, or oral cancer. The differential impact of a shared gene polymorphism across various cancer types can be attributed to several factors. First, the etiology of various cancer exhibits considerable heterogeneity. Second, it has been observed that the identical polymorphism of a specific gene exhibits distinct functions in the initiation and progression of diverse tumor types. Third, it is noteworthy that the target sites of gene polymorphism exhibit variations across different tumor types. Fourth, the multifaceted functionality of the same gene polymorphism site acted throughout distinct stages of disease progression. The subgroup analysis based on ethnicity indicates a significant association between the −251 polymorphism and an elevated risk of cancer, specifically in Asians. However, this association was not observed in Caucasians, Africans, or Mixed populations. Furthermore, variant genotypes at −353 were significantly correlated with an elevated susceptibility to cancer. Finally, individuals carrying the +781 A allele may exhibit an increased susceptibility to cancer risk within the Caucasian population.
The incidence of gene polymorphisms exhibits significant variation across diverse ethnic populations, thereby signifying a crucial property of these genetic variations due to following two primary factors: genetic disparities and environmental variations. Ethnic groups inherently possess distinct genetic and environmental backgrounds contributing to the observed differences. Additionally, diverse populations often exhibit dissimilar patterns of linkage disequilibrium, further contributing to the observed variations. Polymorphism has the potential to exhibit close linkage with distinct nearby causal variants across diverse populations.
In this study, a significant discovery was indicated that gastric cancer patients with AA/AT genotypes exhibited elevating levels of IL‐8 expression than healthy controls, suggesting this may offer a valuable biomarker for the early detection for gastric cancer. These results imply that such individuals may be more susceptible to gastric cancer development. Consequently, it is crucial to closely monitor these individuals and implement timely interventions, preventive measures, and treatment strategies upon definitive diagnosis.
IL‐8 gene polymorphisms (−251, +353, +781) were related to cancer susceptibility, especially −251 site and gastric cancer, suggesting these polymorphisms may offer value as biomarkers suitable for use when early detecting cancer. Besides, above significant polymorphisms of IL‐8 may have some potential clinical applications: such as some related inhibitors. Future studies may be able to apply these results to guide diagnostic and therapeutic approaches to abrogate cancer‐related risk.
Several limitations should be taken into consideration when interpreting the findings of the meta‐analysis. First, the modulation of cancer risk is influenced by the interactions among genes, gene–environment, and polymorphisms within the same gene. Therefore, future research endeavors should incorporate these factors to comprehensively understand cancer susceptibility. Second, it is imperative to incorporate various covariates such as sex, age, family history, environmental factors, cancer stage, and lifestyle into the analysis. Third, it should be noted that the control group consisted of individuals who did not strictly meet the criteria for being classified as healthy controls. Fourth, the investigation encompassed limited case–control studies regarding the polymorphisms (+678, +1633, +2767). Future research efforts should prioritize the examination of the above four polymorphisms. Fifth, case–control studies of small numbers of subjects, seeking to identify low‐penetrance susceptibility genes, may be confounded by interstudy variability and lack of reproducibility, so further work is required about larger patients combined with age‐ and sex‐matched controls to explore the trends and resolve apparent conflicts with other studies. Sixth, current analysis was the lack of haplotype reconstruction. Because, haplotypes are considered more powerful to detect susceptibility alleles than individual polymorphisms. Seventh, in some case–controls studies, the use of hospital controls is not ideal. The use of hospital controls probably has minimal effect on the allele frequencies, which may increase the potential bias. In final, further investigation is warranted to elucidate the underlying mechanisms, utilizing the available epigenetic data, related to the influence of distinct genotypes on tumor proliferation and invasion processes. In spite of these limitations, this meta‐analysis also possessed two notable advantages. First, in order to enhance the statistical power of the analysis, a substantial cohort of cases and controls was aggregated from multiple research investigations. Second, the inclusion of case–control studies in the present meta‐analysis was deemed satisfactory according to the predetermined selection criteria.
In conclusion, the present study evaluated the involvement of three polymorphisms (−251, +353, +781) of the IL‐8 gene in cancer risk, especially for gastric cancer. Hence, it is imperative to conduct additional meticulously planned and extensive investigations, specifically focusing on the interplay between genes and both genetic and environmental factors. Future investigations in this field are anticipated to yield enhanced and comprehensive insights into the correlation between genetic polymorphisms of the IL‐8 gene and susceptibility to cancer development.
Author Contributions
Bin Xun: writing main manuscript; preparing figures. Yidan Yan: data curation (equal); formal analysis (equal). Bin Xun: validation (equal); visualization (equal). Yidan Yan: validation (equal); visualization (equal). Bin Xun: supervision (equal); writing – review and editing (equal).
Ethics Statement
Approval of the research protocol by an Institutional Reviewer Board.
Conflicts of Interest
The authors have stated explicitly that there are no conflicts of interest in connection with this article.
Acknowledgements
The authors have nothing to report.
Funding: The authors received no specific funding for this work.
Data Availability Statement
All data and material in this study were available. In addition, the present work has not been published and not in consideration elsewhere. Also, the current work has been published as preprint https://www.researchsquare.com/article/rs‐3348999/v1.
References
- 1. Bray F. A.‐O., Laversanne M., Weiderpass E., et al., “The ever‐increasing importance of cancer as a leading cause of premature death worldwide,” Cancer 127, no. 16 (2021): 3029–3030. [DOI] [PubMed] [Google Scholar]
- 2. Sung H. A.‐O., Ferlay J., Siegel R. L., et al., “Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries,” CA: A Cancer Journal for Clinicians 71, no. 3 (2021): 209–249. [DOI] [PubMed] [Google Scholar]
- 3. Global Burden of Disease 2019 Cancer Collaboration , Kocarnik J. M., Compton K., et al., “Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability‐Adjusted Life Years for 29 Cancer Groups From 2010 to 2019: A Systematic Analysis for the Global Burden of Disease Study 2019,” JAMA Oncology 8, no. 3 (2022): 420–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Santucci C., Carioli G., Bertuccio P., et al., “Progress in cancer mortality, incidence, and survival: a global overview,” European Journal of Cancer Prevention 29, no. 5 (2020): 367–381. [DOI] [PubMed] [Google Scholar]
- 5. Parascandola M. A.‐O., Neta G., Salloum R. G., et al., “Role of Local Evidence in Transferring Evidence‐Based Interventions to Low‐ and Middle‐Income Country Settings: Application to Global Cancer Prevention and Control,” JCO Global Oncology 8 (2022): e2200054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sud A., Kinnersley B., and Houlston R. S., “Genome‐wide association studies of cancer: current insights and future perspectives,” Nature Reviews Cancer 17, no. 11 (2017): 692–704. [DOI] [PubMed] [Google Scholar]
- 7. Yang W., Zhang T., Song X., Dong G., Xu L., and Jiang F., “SNP‐Target Genes Interaction Perturbing the Cancer Risk in the Post‐GWAS,” Cancers 14, no. 22 (2022): 5636, 10.3390/cancers14225636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edwards S. L., Beesley J., French J. D., et al., “Beyond GWASs: illuminating the dark road from association to function,” 1537–6605. [DOI] [PMC free article] [PubMed]
- 9. Freedman M. L., Monteiro A. N. A., Gayther S. A., et al., “Principles for the post‐GWAS functional characterization of cancer risk loci,” 1546–1718. [DOI] [PMC free article] [PubMed]
- 10. Lu Z., Fan L., Zhang F., et al., “HSPA12A was identified as a key driver in colorectal cancer GWAS loci 10q26.12 and modulated by an enhancer‐promoter interaction,” 1432–1738. [DOI] [PubMed]
- 11. Mathias C., Marin A. M., Kohler A. F., et al., “LncRNA‐SNPs in a Brazilian Breast Cancer Cohort: A Case‐Control Study,” 14: 2073–4425, 10.3390/genes14050971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shao H. B., Ren K., Gao S.‐L., et al., “Human methionine synthase A2756G polymorphism increases susceptibility to prostate cancer,” 1945–4589. [DOI] [PMC free article] [PubMed]
- 13. Landskron G., De la Fuente M., Thuwajit P., et al., “Chronic inflammation and cytokines in the tumor microenvironment,” 2314–7156. [DOI] [PMC free article] [PubMed]
- 14. Greten F. R. and Grivennikov S. I., “Inflammation and Cancer: Triggers, Mechanisms, and Consequences,” 1097–4180. [DOI] [PMC free article] [PubMed]
- 15. Fousek K., Horn L. A., and Palena C., Interleukin‐8: A chemokine at the intersection of cancer plasticity, angiogenesis, and immune suppression 1879‐016X. [DOI] [PMC free article] [PubMed]
- 16. Waugh D. J. and Wilson C., “The interleukin‐8 pathway in cancer,” 6735–6741. [DOI] [PubMed]
- 17. Liao S. C., Interleukin‐8 Gene Polymorphism and Genetic Susceptibility to Hepatitis, Cirrhosis and Hepatocellular Carcinoma in HBV Background (Guangxi, China: Guangxi Medical University, 2011). [Google Scholar]
- 18. Qin S. H., et al., “IL‐8 and IFN in Nasopharyngeal Carcinoma Patients‐γ Research on Genetic Polymorphism,” Journal of Sichuan University 05 (2007): 862–865. [PubMed] [Google Scholar]
- 19. Lu X. H., et al., “Association Between IL‐8 Gene −251T/A and +781C/T Polymorphisms and Genetic Susceptibility to Liver Cancer in Nantong Area,” Journal of Interventional Radiology 24, no. 4 (2015): 314–319. [Google Scholar]
- 20. Zeng Z. R., et al., “Relationship Between Interleukin 8 −251 Gene Polymorphism and Gastric Cancer in High and Low Incidence Areas of China,” Journal of Sun Yat Sen University 05 (2005): 537–540. [Google Scholar]
- 21. Ahirwar D. K., Mandhani A., and Mittal R. D., “IL‐8 −251 T > A Polymorphism Is Associated With Bladder Cancer Susceptibility and Outcome After BCG Immunotherapy in a Northern Indian Cohort,” Archives of Medical Research 41, no. 2 (2010): 97–103. [DOI] [PubMed] [Google Scholar]
- 22. Aleagha O. E., et al., “Evaluation of Interleukin 8 Polymorphisms (−251T/A and +781C/T) in Pat Ients With Hepatocellular Carcinoma: A Meta‐Analysis,” Clinical and Experimental Hepatology 7, no. 3 (2020): 278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alkanli N., Ay A., and Cevik G., “Investigation of Roles of IL‐8 (+ 781 C/T) and MMP‐2 (−735 C/T) Gene Variations in Early Diagnosis of Bladder Cancer and Progression,” Molecular Biology Reports 50, no. 1 (2023): 443–451. [DOI] [PubMed] [Google Scholar]
- 24. Althubyani S. A., et al., “A Preliminary Study of Cytokine Gene Polymorphism Effects on Saudi Patients With Colorectal Cancer,” Saudi Medical Journal 41, no. 12 (2020): 1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ankathil R., Mustapha M. A., Abdul Aziz A. A., et al., “Contribution of Genetic Polymorphisms of Inflammation Response Genes on Sporadic Colorectal Cancer Predisposition Risk in Malaysian Patients—A Case Control Study,” Asian Pacific Journal of Cancer Prevention: APJCP 20, no. 6 (2019): 1621–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Antikchi M. H., et al., “Cumulative Evidence for Association Between IL‐8 −251T > A and IL‐18‐60 7C > A Polymorphisms and Colorectal Cancer Susceptibility: A Systematic Review and Meta‐Analysis,” Journal of Gastrointestinal Cancer 52, no. 1 (2007): 31–40. [DOI] [PubMed] [Google Scholar]
- 27. Basavaraju U., Shebl F. M., Palmer A. J., et al., “Cytokine Gene Polymorphisms, Cytokine Levels and the Risk of Colorectal Neoplasia in a Screened Population of Northeast Scotland,” European Journal of Cancer Prevention: The Official Journal of the European Cancer Prevention Organisation (ECP) 24, no. 4 (2015): 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ben Nasr H., et al., “Association of IL‐8 (−251)T/A Polymorphism With Susceptibility to and Aggressiveness of Nasopharyngeal Carcinoma,” Human Immunology 68, no. 9 (2019): 761–769. [DOI] [PubMed] [Google Scholar]
- 29. Bhat I. A., Pandith A. A., Bhat B. A., et al., “Lack of Association of a Common Polymorphism in the 3′ ‐UTR of Interleukin 8 With Non Small Cell Lung Cancer in Kashmir,” Asian Pacific Journal of Cancer Prevention: APJCP 14, no. 7 (2013): 4403–4408. [DOI] [PubMed] [Google Scholar]
- 30. Bo S., Dianliang Z., Hongmei Z., Xinxiang W., Yanbing Z., and Xiaobo L., “Association of Interleukin‐8 Gene Polymorphism With Cachexia From Patients With Gastric Cancer,” Journal of Interferon & Cytokine Research: The Official Journal of the International Society for Interferon and Cytokine Research 30, no. 1 (2010): 9–14. [DOI] [PubMed] [Google Scholar]
- 31. Boonyanugomol W., et al., “Genetic Polymorphisms of CXCL8 (−251) are Associated With the Suscepti Bility of Helicobacter pylori Infection Increased the Risk of Inflamma Tion and Gastric Cancer in Thai Gastroduodenal Patients,” Iranian Journal of Allergy, Asthma, and Immunology 18, no. 4 (2019): 393–401. [DOI] [PubMed] [Google Scholar]
- 32. Burada F., Angelescu C., Mitrut P., et al., “Interleukin‐4 Receptor −3223C→T Polymorphism Is Associated With Increased Gastric Adenocarcinoma Risk,” Canadian Journal of Gastroenterology = Journal Canadien de Gastroenterologie 26, no. 8 (2012): 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burada F., Dumitrescu T., Nicoli R., Ciurea M., Rogoveanu I., and Ioana M., “Cytokine Promoter Polymorphisms and Risk of Colorectal Cancer,” Clinical Laboratory 59, no. 7–8 (2013): 773–779. [DOI] [PubMed] [Google Scholar]
- 34. Cacev T., et al., “Influence of Interleukin‐8 and Interleukin‐10 on Sporadic Colon Cancer Development and Progression,” Carcinogenesis 29, no. 8 (2008): 1572–1580. [DOI] [PubMed] [Google Scholar]
- 35. Cacev T., Radosevic S., Krizanac S., and Kapitanovic S., “Influence of Interleukin‐8 and Interleukin‐10 on Sporadic Colon Cancer Development and Progression,” Carcinogenesis 29, no. 8 (2008): 1572–1580. [DOI] [PubMed] [Google Scholar]
- 36. Campa D., et al., “Association of Common Polymorphisms in Inflammatory Genes With Risk of Developing Cancers of the Upper Aerodigestive Tract,” Cancer Causes & Control: CCC 18, no. 4 (2007): 449–455. [DOI] [PubMed] [Google Scholar]
- 37. Campa D., et al., “Lack of Association Between −251 T>A Polymorphism of IL8 and Lung Canc Er Risk,” Cancer Epidemiology, Biomarkers & Prevention: A Publication of the Am Erican Association for Cancer Research, Cosponsored by the American So Ciety of Preventive Oncology 14, no. 10 (2005): 2457–2458. [DOI] [PubMed] [Google Scholar]
- 38. Campa D., et al., “Association of a Common Polymorphism in the Cyclooxygenase 2 Gene With Risk of Non‐small Cell Lung Cancer,” Carcinogenesis 25, no. 2 (2004): 229–235. [DOI] [PubMed] [Google Scholar]
- 39. Canedo P., Castanheira‐Vale A. J., Lunet N., et al., “The Interleukin‐8 −251*T/*A Polymorphism Is Not Associated With Risk for Gastric Carcinoma Development in a Portuguese Population,” European Journal of Cancer Prevention: The Official Journal of the European Cancer Prevention Organisation (ECP) 17, no. 1 (2008): 28–32. [DOI] [PubMed] [Google Scholar]
- 40. Castro F. A., et al., “Inflammatory Gene Variants and the Risk of Biliary Tract Cancers and s Tones: A Population‐Based Study in China,” BMC Cancer 12 (2014): 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen C.‐H., et al., “Association Between Interleukin‐8 rs4073 Polymorphism and Prostate Can Cer: A Meta‐Analysis,” Journal of the Formosan Medical Association = Taiwan Yi Zhi 119, no. 7 (2019): 1201–1210. [DOI] [PubMed] [Google Scholar]
- 42. Chen J., Ying X. M., Huang X. M., Huang P., and Yan S. C., “Association Between Polymorphisms in Selected Inflammatory Response Genes and the Risk of Prostate Cancer,” Oncotargets and Therapy 9 (2016): 223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen Y., Yang Y., Liu S., Zhu S., Jiang H., and Ding J., “Association Between Interleukin 8 −251 A/T and +781 C/T Polymorphisms and Osteosarcoma Risk in Chinese Population: A Case–Control Study,” Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine 37, no. 5 (2016): 6191–6196. [DOI] [PubMed] [Google Scholar]
- 44. Chien M.‐H., Yeh C. B., Li Y. C., et al., “Relationship of Interleukin‐8 Gene Polymorphisms With Hepatocellular Carcinoma Susceptibility and Pathological Development,” Journal of Surgical Oncology 104, no. 7 (2011): 798–803. [DOI] [PubMed] [Google Scholar]
- 45. Crusius J. B. A., et al., “Cytokine Gene Polymorphisms and the Risk of Adenocarcinoma of the Stom Ach in the European Prospective Investigation Into Cancer and Nutrition (EPIC‐EURGAST),” Annals of Oncology: Official Journal of the European Society for Medical Oncology 19, no. 11 (2008): 1894–1902. [DOI] [PubMed] [Google Scholar]
- 46. de Matos F. R., Santos E. M., Santos H. B. P., et al., “Association of Polymorphisms in IL‐8, MMP‐1 and MMP‐13 With the Risk and Prognosis of Oral and Oropharyngeal Squamous Cell Carcinoma,” Archives of Oral Biology 108 (2019): 104547. [DOI] [PubMed] [Google Scholar]
- 47. de Oliveira J. G., et al., “Influence of Functional Polymorphisms in TNF‐α, IL‐8, and IL‐10 Cytokine Genes on mRNA Expression Levels and Risk of Gastric Cancer,” Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine 36, no. 12 (2015): 9159–9170. [DOI] [PubMed] [Google Scholar]
- 48. de Oliveira J. G., Rossi A. F. T., Nizato D. M., Miyasaki K., and Silva A. E., “Profiles of Gene Polymorphisms in Cytokines and Toll‐Like Receptors With Higher Risk for Gastric Cancer,” Digestive Diseases and Sciences 58, no. 4 (2013): 978–988. [DOI] [PubMed] [Google Scholar]
- 49. Ding K., et al., “Interleukin Polymorphisms and Protein Levels Associated With Lung Canc Er Susceptibility and Phenotypes,” Expert Review of Clinical Immunology 17, no. 9 (2021): 1029–1040. [DOI] [PubMed] [Google Scholar]
- 50. Farbod M., et al., “Association of IL‐8 −251T≫A and IL‐18 ‐607C≫A Polymorphisms With Susceptibility to Breast Cancer—A Meta‐Analysis,” Klinicka Onkologie: Casopis Ceske a Slovenske Onkologicke Spolecnosti 35, no. 3 (2022): 181–189. [DOI] [PubMed] [Google Scholar]
- 51. Felipe A. V., Silva T. D., Pimenta C. A., Kassab P., and Forones N. M., “Lnterleukin‐8 Gene Polymorphism and Susceptibility to Gastric Cancer in a Brazilian Population,” Biological Research 45, no. 4 (2012): 369–374. [DOI] [PubMed] [Google Scholar]
- 52. Franz J. M., Portela P., Salim P. H., et al., “CXCR2 +1208 CT Genotype May Predict Earlier Clinical Stage at Diagnosis in Patients With Prostate Cancer,” Cytokine 97 (2017): 193–200. [DOI] [PubMed] [Google Scholar]
- 53. Fu J. W., Wang K. W., and Qi S. T., “Role of IL‐8 Gene Polymorphisms in Glioma Development in a Chinese Population,” Genetics and Molecular Research: GMR 15, no. 3 (2016). [DOI] [PubMed] [Google Scholar]
- 54. Gao J., et al., “Certain Interleukin Polymorphisms Might Influence Predisposition to Lu Ng Cancer: A Meta‐Analysis of 35 Published Studies,” IUBMB Life 72, no. 5 (2020): 957–964. [DOI] [PubMed] [Google Scholar]
- 55. Garza‐Gonzalez E., et al., “Assessment of the Toll‐Like Receptor 4 Asp299Gly, Thr399Ile and Interl Eukin‐8 −251 Polymorphisms in the Risk for the Development of Distal g Astric Cancer,” BMC Cancer 7 (2012): 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ghazy A. A. and Alenzi M. J., “Relevance of Interleukins 6 and 8 Single Nucleotide Polymorphisms in Prostate Cancer: A Multicenter Study,” Prostate Cancer 2021 (2021): 3825525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grębowski R., Saluk J., Bijak M., Szemraj J., and Wigner P., “Variability, Expression, and Methylation of IL‐6 and IL‐8 Genes in Bladder Cancer Pathophysiology,” International Journal of Molecular Sciences 24, no. 7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gunter M. J., et al., “Inflammation‐Related Gene Polymorphisms and Colorectal Adenoma,” Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology 15, no. 6 (2006): 1126–1131. [DOI] [PubMed] [Google Scholar]
- 59. Howell W. M., et al., “Cytokine Gene Single Nucleotide Polymorphisms and Susceptibility to an d Prognosis in Cutaneous Malignant Melanoma,” European Journal of Immunogenetics: Official Journal of the British Society for Histocompatibility and Immunogenetics 30, no. 6 (2003): 409–414. [DOI] [PubMed] [Google Scholar]
- 60. Hsieh Y.‐Y., Chang C. C., Tsai C. H., Lin C. C., and Tsai F. J., “Interleukin (IL)‐12 Receptor beta1 Codon 378 G Homozygote and Allele, but Not IL‐1 (Beta‐511 Promoter, 3953 Exon 5, Receptor Antagonist), IL‐2 114, IL‐4‐590 Intron 3, IL‐8 3'‐UTR 2767, and IL‐18 105, Are Associated With Higher Susceptibility to Leiomyoma,” Fertility and Sterility 87, no. 4 (2007): 886–895. [DOI] [PubMed] [Google Scholar]
- 61. Huang C.‐Y., Chang W. S., Tsai C. W., et al., “The Contribution of Interleukin‐8 Genotypes and Expression to Nasopharyngeal Cancer Susceptibility in Taiwan,” Medicine 97, no. 36 (2018): e12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kaanane H., Senhaji N., Berradi H., et al., “The Influence of Interleukin‐6, Interleukin‐8, Interleukin‐10, Interleukin‐17, TNF‐A, MIF, STAT3 on Lung Cancer Risk in Moroccan Population,” Cytokine 151 (2022): 155806. [DOI] [PubMed] [Google Scholar]
- 63. Kamali‐Sarvestani E., Aliparasti M. R., and Atefi S., “Association of Interleukin‐8 (IL‐8 or CXCL8) −251T/A and CXCR2 +1208C/T Gene Polymorphisms With Breast Cancer,” Neoplasma 54, no. 6 (2007): 484–489. [PubMed] [Google Scholar]
- 64. Kamali‐Sarvestani E., Bazargani A., Masoudian M., Lankarani K., Taghavi A. R., and Saberifiroozi M., “Association of H Pylori cagA and vacA Genotypes and IL‐8 Gene Polymorphisms With Clinical Outcome of Infection in Iranian Patients With Gastrointestinal Diseases,” World Journal of Gastroenterology 12, no. 32 (2006): 5205–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kamangar F., et al., “Polymorphisms in Inflammation‐Related Genes and Risk of Gastric Cancer (Finland),” Cancer Causes & Control: CCC 17, no. 1 (2006): 117–125. [DOI] [PubMed] [Google Scholar]
- 66. Kang J. M., Kim N., Lee D. H., et al., “The Effects of Genetic Polymorphisms of IL‐6, IL‐8, and IL‐10 on Helicobacter pylori ‐Induced Gastroduodenal Diseases in Korea,” Journal of Clinical Gastroenterology 43, no. 5 (2009): 420–428. [DOI] [PubMed] [Google Scholar]
- 67. Kietthubthew S., et al., “Association of Polymorphisms in Proinflammatory Cytokine Genes With Th e Development of Oral Cancer in Southern Thailand,” International Journal of Hygiene and Environmental Health 213, no. 2 (2010): 146–152. [DOI] [PubMed] [Google Scholar]
- 68. Kilic I., et al., “Investigation of VEGF and IL‐8 Gene Polymorphisms in Patients With Differentiated Thyroid Cancer,” Clinical Laboratory 62, no. 12 (2016): 2319–2325. [DOI] [PubMed] [Google Scholar]
- 69. Ko K.‐P., et al., “Soybean Product Intake Modifies the Association Between Interleukin‐10 Genetic Polymorphisms and Gastric Cancer Risk,” Journal of Nutrition 139, no. 5 (2009): 1008–1012. [DOI] [PubMed] [Google Scholar]
- 70. Koensgen D., Bruennert D., Ungureanu S., et al., “Polymorphism of the IL‐8 Gene and the Risk of Ovarian Cancer,” Cytokine 71, no. 2 (2015): 334–338. [DOI] [PubMed] [Google Scholar]
- 71. Krajewski W., Karabon L., Partyka A., et al., “Polymorphisms of Genes Encoding Cytokines Predict the Risk of High‐Grade Bladder Cancer and Outcomes of BCG Immunotherapy,” Central‐European Journal of Immunology 45, no. 1 (2020): 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kumar S., Kumari N., Mittal R. D., Mohindra S., and Ghoshal U. C., “Association Between Pro‐ (IL‐8) and Anti‐Inflammatory (IL‐10) Cytokine Variants and Their Serum Levels and H. pylori ‐Related Gastric Carcinogenesis in Northern India,” Meta Gene 6 (2015): 6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Küry S., et al., “Low‐Penetrance Alleles Predisposing to Sporadic Colorectal Cancers: A French Case‐Controlled Genetic Association Study,” BMC Cancer 8 (2008): 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Landi S., et al., “Association of Common Polymorphisms in Inflammatory Genes Interleukin (IL)6, IL8, Tumor Necrosis Factor Alpha, NFKB1, and Peroxisome Prolife Rator‐Activated Receptor Gamma With Colorectal Cancer,” Cancer Research 63, no. 13 (2003): 3560–3566. [PubMed] [Google Scholar]
- 75. Lee K.‐M., et al., “Polymorphisms in Immunoregulatory Genes, Smoky Coal Exposure and Lung Cancer Risk in Xuan Wei, China,” Carcinogenesis 28, no. 7 (2007): 1437–1441. [DOI] [PubMed] [Google Scholar]
- 76. Lee W.‐P., et al., “The −251T Allele of the Interleukin‐8 Promoter Is Associated With Incr Eased Risk of Gastric Carcinoma Featuring Diffuse‐Type Histopathology in Chinese Population,” Clinical Cancer Research: An Official Journal of the American Associa Tion for Cancer Research 11, no. 18 (2005): 6431–6441. [DOI] [PubMed] [Google Scholar]
- 77. Leibovici D., et al., “Polymorphisms in Inflammation Genes and Bladder Cancer: From Initiatio n to Recurrence, Progression, and Survival,” Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology 23, no. 24 (2005): 5746–5756. [DOI] [PubMed] [Google Scholar]
- 78. Leung W. K., et al., “ H. pylori Genotypes and Cytokine Gene Polymorphisms Influence the Deve Lopment of Gastric Intestinal Metaplasia in a Chinese Population,” American Journal of Gastroenterology 101, no. 4 (2006): 714–720. [DOI] [PubMed] [Google Scholar]
- 79. Li Z.‐W., et al., “Inflammatory Cytokine Gene Polymorphisms Increase the Risk of Atrophic Gastritis and Intestinal Metaplasia,” World Journal of Gastroenterology 16, no. 14 (2010): 1788–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liu C. M., Yeh C. J., Yu C. C., et al., “Impact of Interleukin‐8 Gene Polymorphisms and Environmental Factors on Oral Cancer Susceptibility in Taiwan,” Oral Diseases 18, no. 3 (2012): 307–314. [DOI] [PubMed] [Google Scholar]
- 81. Liu H., et al., “Association Between Interleukin 8–251 T/A and +781 C/T Polymorphisms a Nd Glioma Risk,” Diagnostic Pathology 10 (2015): 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lu W., et al., “Genetic Polymorphisms of Interleukin (IL)‐1B, IL‐1RN, IL‐8, IL‐10 and Tumor Necrosis Factor {Alpha} and Risk of Gastric Cancer in a Chinese Population,” Carcinogenesis 26, no. 3 (2015): 631–636. [DOI] [PubMed] [Google Scholar]
- 83. Malespín‐Bendaña W., Machado J. C., Une C., Alpízar‐Alpízar W., Molina‐Castro S., and Ramírez‐Mayorga V., “The TNF‐A‐857*T Polymorphism Is Associated With Gastric Adenocarcinoma Risk in a Costa Rican Population,” American Journal of the Medical Sciences 362, no. 2 (2021): 182–187. [DOI] [PubMed] [Google Scholar]
- 84. Marcus M. W., et al., “Incorporating Epistasis Interaction of Genetic Susceptibility Single n Ucleotide Polymorphisms in a Lung Cancer Risk Prediction Model,” International Journal of Oncology 49, no. 1 (2016): 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Michaud D. S., et al., “Genetic Polymorphisms of Interleukin‐1B (IL‐1B), IL‐6, IL‐8, and IL‐10 and Risk of Prostate Cancer,” Cancer Research 66, no. 8 (2006): 4525–4530. [DOI] [PubMed] [Google Scholar]
- 86. Moghimi M., et al., “Association of Il‐8 −251T>A (RS4073) Polymorphism With Susceptibility,” Arquivos de Gastroenterologia 57, no. 1 (2020): 91–99. [DOI] [PubMed] [Google Scholar]
- 87. Moreno‐Guerrero S. S., et al., “Association of Genetic Polymorphisms and Serum Levels of IL‐6 and IL‐8 With the Prognosis in Children With Neuroblastoma,” Cancers 13, no. 3 (2021): 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mustapha M. A., et al., “Risk Modification of Colorectal Cancer Susceptibility by Interleukin‐8 −251T>A Polymorphism in Malaysians,” World Journal of Gastroenterology 18, no. 21 (2012): 2668–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ohyauchi M., et al., “The Polymorphism Interleukin 8–251 A/T Influences the Susceptibility of Helicobacter pylori Related Gastric Diseases in the Japanese Population,” Gut 54, no. 3 (2005): 330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Pan X.‐F., Wen Y., Loh M., et al., “Interleukin‐4 and ‐8 Gene Polymorphisms and Risk of Gastric Cancer in a Population in Southwestern China,” Asian Pacific Journal of Cancer Prevention: APJCP 15, no. 7 (2014): 2951–2957. [DOI] [PubMed] [Google Scholar]
- 91. Qadri Q., Rasool R., Afroze D., et al., “Study of TLR4 and IL‐8 Gene Polymorphisms in H.Pylori‐Induced Inflammation in Gastric Cancer in an Ethnic Kashmiri Population,” Immunological Investigations 43, no. 4 (2014): 324–336. [DOI] [PubMed] [Google Scholar]
- 92. Qin X., Deng Y., Liao X. C., et al., “The IL‐8 Gene Polymorphisms and the Risk of the Hepatitis B Virus/Infected Patients,” DNA and Cell Biology 31, no. 6 (2012): 1125–1130. [DOI] [PubMed] [Google Scholar]
- 93. Rafrafi A., et al., “Association of IL‐8 Gene Polymorphisms With Non Small Cell Lung Cancer in Tunisia: A Case Control Study,” Human Immunology 74, no. 10 (2013): 1368–1374. [DOI] [PubMed] [Google Scholar]
- 94. Ramis I. B., et al., “Polymorphisms of the IL‐6, IL‐8 and IL‐10 Genes and the Risk of Gastri c Pathology in Patients Infected With Helicobacter pylori ,” Journal of Microbiology, Immunology, and Infection 50, no. 2 (2017): 153–159. [DOI] [PubMed] [Google Scholar]
- 95. Rezaei F., et al., “Association between IL‐8 (−251T/A) and IL‐6 (−174G/C) Polymorphisms and Oral Cancer Susceptibility: A Systematic Review and Me ta‐Analysis. Medicina (Kaunas, Lithuania),” 57, no. 5 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Savage S. A., et al., “Interleukin‐8 Polymorphisms Are Not Associated With Gastric Cancer Ris k in a Polish Population,” Cancer Epidemiology, Biomarkers & Prevention: A Publication of the Am Erican Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology 15, no. 3 (2006): 589–591. [DOI] [PubMed] [Google Scholar]
- 97. Sha B. E., et al., “Safety and Immunogenicity of a Polyvalent Peptide C4‐V3 HIV Vaccine in Conjunction With IL‐12,” AIDS 18, no. 8 (2004): 1203–1206. [DOI] [PubMed] [Google Scholar]
- 98. Shimizu Y., et al., “A Single Nucleotide Polymorphism in the Matrix Metalloproteinase‐1 and Interleukin‐8 Gene Promoter Predicts Poor Prognosis in Tongue Cancer,” Auris, Nasus, Larynx 35, no. 3 (2008): 381–389. [DOI] [PubMed] [Google Scholar]
- 99. Singh P. K., Chandra G., Bogra J., et al., “Association of Genetic Polymorphism in the Interleukin‐8 Gene With Risk of Oral Cancer and Its Correlation With Pain,” Biochemical Genetics 54, no. 1 (2016): 95–106. [DOI] [PubMed] [Google Scholar]
- 100. Smith K. C., et al., “Cytokine Gene Polymorphisms and Breast Cancer Susceptibility and Progn Osis,” European Journal of Immunogenetics: Official Journal of the British Society for Histocompatibility and Immunogenetics 31, no. 4 (2004): 167–173. [DOI] [PubMed] [Google Scholar]
- 101. Snoussi K., et al., “Genetic Variation in IL‐8 Associated With Increased Risk and Poor Prog Nosis of Breast Carcinoma,” Human Immunology 67, no. 1–2 (2006): 13–21. [DOI] [PubMed] [Google Scholar]
- 102. Snoussi K., et al., “Combined Effects of IL‐8 and CXCR2 Gene Polymorphisms on Breast Cancer Susceptibility and Aggressiveness,” BMC Cancer 10 (2010): 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Song B., et al., “Association of Interleukin‐8 With Cachexia From Patients With Low‐Third Gastric Cancer,” Comparative and Functional Genomics 2009 (2009): 212345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Szoke D., et al., “T−251A Polymorphism of IL‐8 Relating to the Development of Histologica l Gastritis and G‐308A Polymorphism of TNF‐Alpha Relating to the Devel Opment of Macroscopic Erosion,” European Journal of Gastroenterology & Hepatology 20, no. 3 (2017): 191–195. [DOI] [PubMed] [Google Scholar]
- 105. Taguchi A., et al., “Interleukin‐8 Promoter Polymorphism Increases the Risk of Atrophic Gas Tritis and Gastric Cancer in Japan,” Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology 14, no. 11 Pt 1 (2005): 2487–2493. [DOI] [PubMed] [Google Scholar]
- 106. Taheri M., Noroozi R., Dehghan A., Roozbahani G. A., Omrani M. D., and Ghafouri‐Fard S., “Interleukin (IL)‐8 Polymorphisms and Risk of Prostate Disorders,” Gene 692 (2019): 22–25. [DOI] [PubMed] [Google Scholar]
- 107. Theodoropoulos G., Papaconstantinou I., Felekouras E., et al., “Relation Between Common Polymorphisms in Genes Related to Inflammatory Response and Colorectal Cancer,” World Journal of Gastroenterology 12, no. 31 (2006): 5037–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Tsilidis K. K., et al., “Association of Common Polymorphisms in IL10, and in Other Genes Relate d to Inflammatory Response and Obesity With Colorectal Cancer,” Cancer Causes & Control: CCC 20, no. 9 (2009): 1739–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Vairaktaris E., Yapijakis C., Serefoglou Z., et al., “Gene Expression Polymorphisms of Interleukins‐1 Beta, −4, −6, −8, −10, and Tumor Necrosis Factors‐Alpha, −Beta: Regression Analysis of Their Effect Upon Oral Squamous Cell Carcinoma,” Journal of Cancer Research and Clinical Oncology 134, no. 8 (2008): 821–832. [DOI] [PubMed] [Google Scholar]
- 110. Vairaktaris E., et al., “The Interleukin‐8 (−251A/T) Polymorphism Is Associated With Increased Risk for Oral Squamous Cell Carcinoma,” European Journal of Surgical Oncology: Journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology 33, no. 4 (2007): 504–507. [DOI] [PubMed] [Google Scholar]
- 111. Vinagre R. M. D. F., Corvelo T. C. O., Arnaud V. C., Leite A. C. K., Barile K. A. S., and Martins L. C., “Determination of Strains of Helicobacter Pylori and of Polymorphism in the Interleukin‐8 Gene in Patients With Stomach Cancer,” Arquivos de Gastroenterologia 48, no. 1 (2011): 46–51. [DOI] [PubMed] [Google Scholar]
- 112. Vogel U., et al., “Prospective Study of Interaction Between Alcohol, NSAID Use and Polymo Rphisms in Genes Involved in the Inflammatory Response in Relation to Risk of Colorectal Cancer,” Mutation Research 624, no. 1–2 (2007): 88–100. [DOI] [PubMed] [Google Scholar]
- 113. Walczak A., Przybylowska K., Dziki L., et al., “The lL‐8 and IL‐13 Gene Polymorphisms in Inflammatory Bowel Disease and Colorectal Cancer,” DNA and Cell Biology 31, no. 8 (2012): 1431–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wang J.‐l., et al., “Association of Interleukin‐8 Gene Polymorphisms With the Risk of Hepatocellular Carcinoma,” Molecular Biology Reports 41, no. 3 (2014): 1483–1489. [DOI] [PubMed] [Google Scholar]
- 115. Wang M.‐H., et al., “Association of IL10 and Other Immune Response‐ and Obesity‐Related Gen Es With Prostate Cancer in CLUE II,” Prostate 69, no. 8 (2009): 874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wang Y.‐C., et al., “The Contribution of Interleukin‐8 Rs4073 Genotypes to Triple Negative Breast Cancer Risk in Taiwan,” Anticancer Research 42, no. 8 (2022): 3799–3806. [DOI] [PubMed] [Google Scholar]
- 117. Wang Y.‐M., Li Z. X., Tang F. B., et al., “Association of Genetic Polymorphisms of Interleukins With Gastric Cancer and Precancerous Gastric Lesions in a High‐Risk Chinese Population,” Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine 37, no. 2 (2016): 2233–2242. [DOI] [PubMed] [Google Scholar]
- 118. Wang Z., et al., “Association of IL‐8 Gene Promoter −251 A/T and IL‐18 Gene Promoter −13 7 G/C Polymorphisms With Head and Neck Cancer Risk: A Comprehensive Me Ta‐Analysis,” Cancer Management and Research 10 (2018): 2589–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wang Z., Liu Q. L., Sun W., et al., “Genetic Polymorphisms in Inflammatory Response Genes and Their Associations With Breast Cancer Risk,” Croatian Medical Journal 55, no. 6 (2014): 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wilkening S., et al., “Interleukin Promoter Polymorphisms and Prognosis in Colorectal Cancer,” Carcinogenesis 29, no. 6 (2008): 1202–1206. [DOI] [PubMed] [Google Scholar]
- 121. Wu C.‐C., Huang Y. K., Chung C. J., et al., “Polymorphism of Inflammatory Genes and Arsenic Methylation Capacity Are Associated With Urothelial Carcinoma,” Toxicology and Applied Pharmacology 272, no. 1 (2013): 30–36. [DOI] [PubMed] [Google Scholar]
- 122. Yamamoto Y., Kiyohara C., Suetsugu‐Ogata S., Hamada N., and Nakanishi Y., “Biological Interaction of Cigarette Smoking on the Association Between Genetic Polymorphisms Involved in Inflammation and the Risk of Lung Cancer: A Case‐Control Study in Japan,” Oncology Letters 13, no. 5 (2017): 3873–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Yang H. P., et al., “Genetic Variation in Interleukin 8 and Its Receptor Genes and Its Infl Uence on the Risk and Prognosis of Prostate Cancer Among Finnish Men i n a Large Cancer Prevention Trial,” European Journal of Cancer Prevention: The Official Journal of the European Cancer Prevention Organisation (ECP) 15, no. 3 (2006): 249–253. [DOI] [PubMed] [Google Scholar]
- 124. Ye B. D., et al., “The Interleukin‐8 −251 A Allele Is Associated With Increased Risk of Noncardia Gastric Adenocarcinoma in Helicobacter pylori ‐Infected Koreans,” Journal of Clinical Gastroenterology 43, no. 3 (2009): 233–239. [DOI] [PubMed] [Google Scholar]
- 125. Zamora‐Ros R., et al., “Dietary Inflammatory Index and Inflammatory Gene Interactions in Relat Ion to Colorectal Cancer Risk in the Bellvitge Colorectal Cancer Case‐ Control Study,” Genes & Nutrition 10, no. 1 (2015): 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zhang J., Dhakal I. B., Lang N. P., and Kadlubar F. F., “Polymorphisms in Inflammatory Genes, Plasma Antioxidants, and Prostate Cancer Risk,” Cancer Causes & Control 21, no. 9 (2010): 1437–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zhang J., Han X., and Sun S., “IL‐8 −251A/T and +781C/T Polymorphisms Were Associated With Risk of Breast Cancer in a Chinese Population,” International Journal of Clinical and Experimental Pathology 10, no. 7 (2017): 7443–7450. [PMC free article] [PubMed] [Google Scholar]
- 128. Zhang L., du C., Guo X., et al., “Interleukin‐8 −251A/T Polymorphism and Helicobacter pylori Infection Influence Risk for the Development of Gastric Cardiac Adenocarcinoma in a High‐Incidence Area of China,” Molecular Biology Reports 37, no. 8 (2010): 3983–3989. [DOI] [PubMed] [Google Scholar]
- 129. Chang Y. W., Oh C. H., Kim J. W., et al., “Combination of Helicobacter Pylori Infection and the Interleukin 8 –251 T > A Polymorphism, but Not the Mannose‐Binding Lectin 2 Codon 54 G > A Polymorphism, Might Be a Risk Factor of Gastric Cancer,” BMC Cancer 17, no. 1 (2017): 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Li J. R., et al., “Relationship Between Serum Interleukin‐6 and Interleukin‐8 Levels and Their Gene Polymorphisms and Non‐small Cell Lung Cancer,” Clinical Medicine of China 31, no. 7 (2015): 581–584. [Google Scholar]
- 131. Wei Y. S., Lan Y., Tang R. G., et al., “Single Nucleotide Polymorphism and Haplotype Association of the Interleukin‐8 Gene With Nasopharyngeal Carcinoma,” Clinical Immunology 125, no. 3 (2007): 309–317. [DOI] [PubMed] [Google Scholar]
- 132. Elsamanoudy A. Z., et al., “Study of Interleukin‐8 Gene Polymorphisms in Egyptian Hepatocellular Carcinoma Patients and Association With Insulin Resistance State,” International Journal of Advanced Research 3, no. 1 (2015): 216–226. [Google Scholar]
- 133. Shirai K., Ohmiya N., Taguchi A., et al., “Interleukin‐8 Gene Polymorphism Associated With Susceptibility to Non‐cardia Gastric Carcinoma With Microsatellite Instability,” Journal of Gastroenterology and Hepatology 21, no. 7 (2006): 1129–1135. [DOI] [PubMed] [Google Scholar]
- 134. Vogel U., Christensen J., Wallin H., et al., “Polymorphisms in Genes Involved in the Inflammatory Response and Interaction With NSAID Use or Smoking in Relation to Lung Cancer Risk in a Prospective Study,” Mutation Research 639, no. 1–2 (2008): 89–100. [DOI] [PubMed] [Google Scholar]
- 135. van der Kuyl A. C., Polstra A. M., Weverling G. J., Zorgdrager F., van den Burg R., and Cornelissen M., “An IL‐8 Gene Promoter Polymorphism Is Associated With the Risk of the Development of AIDS‐Related Kaposi's Sarcoma: A Case‐Control Study,” AIDS 18, no. 8 (2004): 1206–1208. [DOI] [PubMed] [Google Scholar]
- 136. McCarron S. L., et al., “Influence of Cytokine Gene Polymorphisms on the Development of Prostate Cancer,” Cancer Research 62, no. 12 (2002): 3369–3372. [PubMed] [Google Scholar]
- 137. Higgins J. P. and Thompson S. G., “Quantifying Heterogeneity in a Meta‐Analysis,” Statistics in Medicine 21, no. 11 (2002): 1539–1558. [DOI] [PubMed] [Google Scholar]
- 138. DerSimonian R Fau‐Laird N. and Laird N., “Meta‐analysis in clinical trials,” 197–2456.
- 139. Mantel N., Haenszel W., and Haenszel W., “Statistical aspects of the analysis of data from retrospective studies of disease,” 719–748. [PubMed]
- 140. Hayashino Y., Noguchi Y., and Fukui T., “Systematic Evaluation and Comparison of Statistical Tests for Publication Bias,” Journal of Epidemiology 15, no. 6 (2005): 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Shao H. B., Ren K., Gao S. L., et al., “Human Methionine Synthase A2756G Polymorphism Increases Susceptibility to Prostate Cancer,” Aging 10, no. 7 (2018): 1776–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Castro F. A., et al., “Inflammatory gene variants and the risk of biliary tract cancers and stones: a population‐based study in China,” 1471–2407. [DOI] [PMC free article] [PubMed]
- 143. Miller K. A. ‐O., et al., Cancer statistics for adolescents and young adults, (2020). (1542–4863). [DOI] [PubMed]
- 144. Araki S., et al., “Interleukin‐8 is a molecular determinant of androgen independence and progression in prostate cancer,” 6854–6862. [DOI] [PubMed]
- 145. Farbod M., Dastgheib S. A., et al., “Association of IL‐8 −251T>A and IL‐18 ‐607C>A Polymorphisms With Susceptibility to Breast Cancer—A Meta‐Analysis,” 1802–5307. [DOI] [PubMed]
- 146. Wang X., et al., “The roles of IL‐6, IL‐8 and IL‐10 gene polymorphisms in gastric cancer: A meta‐analysis,” 230–236. [DOI] [PubMed]
- 147. Antikchi M. H., et al., “Cumulative Evidence for Association Between IL‐8 −251T>A and IL‐18 ‐607C>A Polymorphisms and Colorectal Cancer Susceptibility: A Systematic Review and Meta‐analysis,” 1941–6636. [DOI] [PubMed]
- 148. Gao J., et al., “Certain interleukin polymorphisms might influence predisposition to lung cancer: A meta‐analysis of 35 published studies,” 1521–6551. [DOI] [PubMed]
- 149. Rezaei F., et al., “Association Between IL‐8 (−251T/A) and IL‐6 (−174G/C) Polymorphisms and Oral Cancer Susceptibility: A Systematic Review and Meta‐Analysis,” Medicina (2021): 1648–9144, 10.3390/medicina57050405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and material in this study were available. In addition, the present work has not been published and not in consideration elsewhere. Also, the current work has been published as preprint https://www.researchsquare.com/article/rs‐3348999/v1.


