Abstract
Background
T cell receptor (TCR)–engineered T cells targeting neoantigens originated from mutations in KRAS gene have demonstrated promising outcomes in clinical trials against solid tumors. However, the challenge lies in developing tumor-specific TCRs that avoid cross-reactivity with self-antigens to minimize the possibility of severe clinical toxicities. Current research efforts have been put towards strategies to eliminate TCR off-target recognition.
Methods
Naive T cell repertoire was used for screening KRASG12D-reactive TCRs. Specific TCRs were subsequently identified and their functionality was assessed using TCR Jurkat cells and TCR T cells. Peptide specificity was evaluated using the X-scan assay. To enhance TCR specificity for KRASG12D and reduce their reactivity to self-peptide SMC1A29-38, mammalian TCR display libraries were employed for the design of modification in the complementarity-determining region (CDR).
Results
HLA-A*11:01-restricted TCRs targeting the KRASG12D epitope were isolated, and TCR1 was characterized with superior functional avidity and specificity. Alongside a robust recognition of endogenous KRASG12D epitope, this TCR displayed cross-reactivity with the SMC1A29-38 epitope. With an approach utilizing structural-guided mutations in the CDR-1A region of TCR1, we obtained an engineered TCR variant (TCR1a7). Functional characterization of TCR1a7 showed that this TCR not only exhibited enhanced specificity towards KRASG12D, but also demonstrated successful elimination of the off-target recognition of SMC1A29-38.
Conclusions
TCRs targeting the KRASG12D peptide could be isolated from naive T cell repertoires. Integrating the TCR-peptide-HLA complex structure with a mammalian TCR library system could serve as a functional strategy to reduce potential TCR cross-reactivity with self-antigens, such as SMC1A29-38. Our findings evidenced an operable method to enhance TCR peptide specificity, while maintaining advanced functional avidity and potent anti-tumor activity.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-025-06094-1.
Keywords: KRAS mutation, T cell receptor, Cross-reactivity, Off-target
Background
Mutations in tumor driver genes can give rise to common neoantigens across various cancer types and patient cohorts. [1–4] These neoantigens, presented by HLA molecules on tumor cell surfaces, play a pivotal role in eliciting T cell-mediated anti-tumor responses. [5] Mutations in KRAS gene stands out as one of the most widespread driver mutations, occurring in 60–70% of pancreatic cancers and 20–30% of colorectal malignancies. [6] KRAS mutations predominantly affect Glycine at the 12th amino acid position, presented as G12C, G12D, or G12V variants. [6] Adoptive transfer of T cell receptor (TCR) T cells has demonstrated the potential to provide durable clinical responses to previously incurable diseases. [7–9] Recent report of a clinical trial with adoptive transfer of TCR-engineered T cells targeting HLA-C*08:02-restricted KRASG12D epitope demonstrated significant regression of metastatic pancreatic cancer [10]. Ongoing efforts are focused on developing TCRs that target KRAS mutants restricted by diverse HLA molecules, to expand the applicability of TCR-based therapies against mutant KRAS across a broader patient population [11–16].
One of the critical threats associated with the application of TCR-T cell therapy were brought by tumor specific TCRs bearing cross-reactivity to self-antigens, especially for those presented in essential normal tissues [17]. Clinical evidence of unexpected cross-reaction of MAGE-A3 TCR with self-antigen expressed by myocardial cells rose warning to potential fatal outcomes in practice [18]. Proactively, rigorous testing has been applied in preclinical studies to comprehensively evaluate potential cross-reactivity of TCRs with therapeutic efficacy, including deconstructing peptide specificity at the proteomics level and integrating diverse strategies to systematically evaluate the presence of off-target polyspecificity [19, 20].
TCR is composed of a heterodimer comprising highly variable α and β chains, which specifically recognize pathogen peptides presented by major histocompatibility complexes (MHCs) [17]. Structural studies of TCR-peptide-MHC complexes have revealed that the TCR CDR-1 and CDR-2 loops primarily interact with the top of MHC helices, whereas the CDR-3 loops predominantly engage with the peptide within the MHC groove [21, 22]. TCR cross-reactivity, or polyspecificity, arises from the conformational adaptability of the TCR-peptide-HLA complex, particularly within the TCR CDR-3 loops [23, 24]. The cross-reactivity of TCRs physiologically empowers the immune system to recognize a vast array of potential antigens. In the context of TCR-based therapies, accurately predicting and assessing this property is crucial to ensuring TCR safety. Proactively, rigorous testing has been applied in preclinical studies to comprehensively evaluate potential cross-reactivity of TCRs with therapeutic efficacy, including deconstructing peptide specificity at the proteomics level and integrating diverse strategies to systematically evaluate the presence of off-target polyspecificity [25, 26]. However, current “pass/fail” determination for further development of TCR candidates based on the presence of detectable cross-reactivity might considerably reduce the number of applicable TCRs, even those with satisfying efficacy of tumor antigenic recognition. So far, there are very few of exploratory studies addressing whether TCR clones identified with off-target specificity can be modified and salvaged for further development towards therapeutic applications.
In this study, we identified an HLA-A11:01-restricted TCR (TCR1) targeting the KRASG12D epitope, demonstrating high functional avidity and effective cytotoxicity against HLA-A11:01+ cancer cells harboring KRASG12D mutation. We investigated potential off-target effects of TCR1 by X-scan assays in conjunction with human proteome scanning, and revealed a self-antigen derived from the SMC1A protein that could be recognized by TCR1. To eliminate this off-target reactivity, we utilized a systematic TCR engineering approach integrating structure-guided design, the mammalian TCR library display system, and multi-level functional screening. This methodology enabled the development of an engineered variant of TCR1 (TCR1a7) that effectively eliminated off-target recognition and enhanced TCR peptide specificity. These findings evidenced for a comprehensive TCR engineering strategy capable of improving peptide specificity, with potential application in advancing TCR-based therapies.
Methods
Cell lines
Cell lines were maintained under specific culture conditions. Jurkat (Clone E6-1, ATCC, TIB-152), K562 (ATCC, CCL-243), AsPC-1 (ATCC, CRL-1682), and SW480 (ATCC, CCL-228) were cultured in RPMI media supplemented with 10% fetal bovine serum (FBS, Gibco), 2 mM L-glutamine (Gibco), and 1X penicillin/streptomycin (Gibco). COS-7 (ATCC, CRL-1651) and Lenti-X 293 T cells (Takara, 632180) were maintained in DMEM media enriched with 10% FBS (Gibco), 2 mM L-glutamine (Gibco), 1X penicillin/streptomycin (Gibco), 1 mM sodium pyruvate (Gibco), and 0.1 mM non-essential amino acids (Gibco). The AGS cell line (ATCC, CRL-1739) was cultured in F-12 K media containing 10% FBS (Gibco), 2 mM L-glutamine (Gibco), and 1X penicillin/streptomycin (Gibco). All cell lines were authenticated, obtained as specified, passaged twice, aliquoted, and cryopreserved for long-term storage. Each aliquot was utilized for up to 15 passages. Cultures were incubated in a humidified environment at 37 °C with 5% CO_2 and routinely tested for mycoplasma contamination.
Generation of transgenic cell lines
CD8+ Jurkat cells: A Jurkat E6.1 human TCRα/β-negative cell line was generated using Cas9 protein and sgRNAs targeting TRAC (5’-AGAGTCTCTCAGCTGGTACA-3’) and TRBC1/TRBC2 (5’-GGAGAATGACGAGTGGACCC-3’) as reported. [35] TCR-negative Jurkat cells were then transduced with lentiviral particles encoding the human CD8α co-receptor to facilitate stable interaction with peptide-HLA (pHLA) complex. HLA-A*11:01 K562 cells: K562 cells were transduced with lentiviral particles expressing HLA-A*11:01. HLA-A*11:01+ tumor cells: AGS and AsPC-1 cells were transduced with lentiviral particles expressing HLA-A*11:01. SW480 cells were sequentially transduced with two distinct lentiviral particles expressing HLA-A*11:01 and the full-length KRASG12D gene, respectively. HLA-A*11:01+ SMC1A+ COS-7 cells: COS-7 cells underwent sequential transduction with two different lentiviral particles expressing HLA-A*11:01 and the full-length SMC1A gene.
Primary cells culture
T cells were cultured in RPMI 1640 complete medium, consisting of RPMI 1640 (Gibco) supplemented with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 1X penicillin/streptomycin (Gibco), 2 mM GlutaMAX(Gibco), 55 μM 2-mercaptoethanol (Sigma-Aldrich), and 25 mM HEPES buffer(Gibco).Monocyte-derived dendritic cells (moDCs) were maintained in RPMI 1640 complete medium supplemented with additional cytokines, including 800 U/mL Granulocyte–Macrophage Colony-Stimulating Factor (GM-CSF, PeproTech) and 800 U/mL Interleukin-4 (IL-4, PeproTech).
Peptides and plasmids
Peptides and peptide libraries were synthesized by Genscript, resuspended at 10 mg/mL in dimethyl sulfoxide (DMSO), and stored at − 80 °C, with sequences detailed in Supplementary Table 1. The TCR lentiviral vectors were constructed by fusing the TCR-alpha chain's variable-joining (V-J) regions and the TCR-beta chain's variable-diversity-joining (V-D-J) regions to their respective mouse TCR constant regions, separated by a P2A linker to create a bicistronic construct. This construct, alongside genes for CD8α, SMC1A and KRASG12D, was cloned into the pWPXL lentiviral vector for lentiviral production. The use of mouse TCR constant regions ensures correct TCR chain pairing and facilitates the identification of transduced T cells via flow cytometry using an APC-anti-mouse TCR β chain Antibody (Biolegend, Catalogue#109212).
Lentiviral vector constructs and production
On the first day, Lenti-X 293 T cells were seeded at a density of 1E6 cells per well in a 6-well plate. After 24 h, the medium was reduced to 1 mL per well, and a DNA transfection complex was added into each well. This complex consisted of 100 µL Xfect Transfection Buffer (Takara), 2.6 µL Xfect Transfection Reagent (Takara), 2.5 µg of psPAX2, 1.25 µg of pMD2.G, and 5 µg of the pWPXL-X lentiviral vector, which contains the genes of interest. During the addition of the transfection complex, gentle agitation of the plate was performed to ensure even distribution. Four hours post-transfection, the medium was replaced with 2 mL of fresh culture medium to support cell growth. Forty-eight hours after transfection, the supernatant containing the viral particles was harvested for cell infection purposes. The psPAX2 (Addgene Plasmid #12260), pMD2.G (Addgene Plasmid #12259) plasmids and pWPXL (Addgene Plasmid #12257), were gifts from Didier Trono.
Generation of KRASG12D reactive T cells
Experiments were performed as reported [36]. Peripheral blood mononuclear cells (PBMCs) were isolated from healthy donors through density-gradient centrifugation using Lymphoprep™ (STEMCELL Technologies) and subsequently cryopreserved in liquid nitrogen until required. To generate autologous monocyte-derived dendritic cells (moDCs), monocytes were then enriched from these PBMCs using the EasySep™ CD14+ Cell Selection Kit (STEMCELL Technologies), according to the manufacturer’s instructions. These monocytes were cultured in dendritic cell (DC) specific media, with fresh complete media added every alternate day. By the sixth day, the moDCs were cryopreserved and stored in liquid nitrogen for future assays.
Naive CD8+ T cells were isolated from PBMCs utilizing the Naive CD8+ T Cell Isolation Kit (STEMCELL Technologies), following the manufacturer’s protocol. These cells were then co-cultured with moDCs that had been pulsed with KRASG12D peptides at a final concentration of 10 μM and activated with lipopolysaccharide (LPS, 10 ng/mL) and interferon-gamma (IFN-γ, 100 U/mL) for 16 h to enhance antigen presentation. The co-culture, set up at a 2:1 ratio of naive CD8+ T cells to moDCs, was conducted in 48-well plates with each well containing 5E5 cells. The culture medium, supplemented with 30 ng/mL IL-21, was refreshed every three days by the addition of IL-7 and IL-15 (5 ng/mL each) to support T cell growth and differentiation over a 10-day period. Then, the stimulated products underwent a second round of stimulation to enhance the activation and expansion of KRASG12D specific T cells. The final products were used to identify KRASG12D specific T cells and TCR cloning.
Isolation and amplification of TCR gene from single T cell
Following the secondary stimulation, samples were stained with Dead Dye (APC-Cy7), BV510-anti-Human CD3 antibody, FITC-anti-human CD8a antibody, PE-KRASG12D Tetramer and APC-KRASG12D Tetramer to identify antigen-specific T cells. Tetramer+ CD8+ T cells were single-cell sorted by FACSAria into a 96-well PCR plate, which was subsequently stored at − 80 °C. Gating strategy was shown in Supplementary Fig. 1. Upon thawing the sorted single T cells on ice, the TCRα and TCRβ chain genes were amplified as previously described [37, 38]. Each well received a 5 μl reaction mixture for reverse transcription and the first round of PCR (RT&1st PCR). This mixture consisted of PrimeScript II Reverse Transcriptase, 2 × PrimeSTAR GC Buffer, RNase-free water, RNase inhibitor, PrimeSTAR HS DNA Polymerase, a dNTP mixture (all reagents sourced from Takara), and primers specific for TCRα and TCRβ chains, to achieve a reaction volume of 5 μl. The RT&1st PCR protocol was as follows: an initial reverse transcription at 45 °C for 45 min, denaturation at 98 °C for 1 min, followed by 30 cycles of 98 °C for 10 s, annealing at 52 °C for 5 s, and elongation at 72 °C for 1 min. Subsequently, the PCR products were diluted 50-fold with water. A 2 μl aliquot of the diluted PCR products served as the template in an 18 μl nested PCR mix. The nested PCR conditions included an initial denaturation at 98 °C for 1 min, 35 cycles of 98 °C for 10 s, 52 °C for 5 s, and a final elongation step at 72 °C for 30 s. The products of the nested PCR were then analyzed through Sanger sequencing. The TCR repertoire was analyzed with the IMGT/V-Quest tool (http://www.imgt.org/).
Preliminary functional analysis of TCRs using TCR Jurkat cells
Generation of TCR-engineered Jurkat cells: CD8+ Jurkat cells were counted and aliquoted at 2 × 10^5 cells per sample prior to centrifugation. Lentiviral particles expressing the TCR of interest were prepared in serial two-fold dilutions. These cells were then resuspended in the lentiviral solution and seeded into 24-well plates for transduction. The medium was completely replaced one day after transduction. Two days post-transduction, the expression of the mouse TCR on Jurkat cells was assessed using flow cytometry. Only those TCR Jurkat cells demonstrating a transduction efficiency between 70 and 80% were selected for further functional analysis, ensuring uniformity and comparability across various TCR constructs. KRASG12D tetramer binding assay: TCR Jurkat cells were stained with APC-anti-mouse TCR β chain antibody and PE-KRASG12D Tetramer for 30 min at 4 °C, protected from light. Following incubation, cells underwent two PBS washes before being subjected to flow cytometric analysis to evaluate tetramer binding capabilities. Functional avidity assay: For the assessment of functional avidity, TCR Jurkat cells were co-cultured with HLA-A*11:01+ K562 cells pre-loaded with a range of KRASG12D peptide concentrations. After incubating for 6 h, cells were stained with APC-anti-human CD69 antibody for 10 min at 4 °C in the dark. Subsequent to two PBS washes, flow cytometry was performed to analyze the expression of the CD69. The half-maximal effective concentration (EC50) values for each TCR were calculated by identifying the peptide concentration that induced 50% of the maximal activation response.
Expression of KRASG12D specific TCRs in primary CD8+ T cells
For the production of TCR T cells: Primary CD8+ T cells were enriched using the EasySep™ Human CD8+ T Cell Enrichment Kit. Then, CD8+ T cells were activated for 24 h using anti-CD3/CD28 T Cell TransAct (Miltenyi Biotec) for 24 h. Subsequently, these activated T cells were transduced with lentiviral vectors encoding the TCRs specific for KRASG12D. The culture medium was refreshed 8 h post-transduction with a medium supplemented with IL-7 and IL-15 at a concentration of 10 ng/ml each. Cultures were maintained with regular medium changes every 3 days. After a cultivation period of 10 days, T cells were stained with APC-anti-mouse TCR β chain antibody. The EasySep™ APC Positive Selection Kit (STEMCELL Technologies) was used for the enrichment of mTCR-positive cells. To further expand the enriched TCR T cells, a secondary expansion phase was initiated. TCR T cells were stimulated with irradiated allogeneic feeder cells, OKT3, and IL-2. The entire processing time was approximately 24 days. For the production of hTCR-KO TCR T cells: 1E6 activated T cells were electroporated with CRISPR/Cas9 ribonucleoprotein (RNP) complexes utilizing the P3 Primary Cell 4D X kit (Lonza: PBP3-02250). 5 μL of RNP complexes were gently mixed with the T cells in 20 μL of electroporation buffer. Immediately after electroporation (Program ID: EH115), the T cells were placed into culture medium containing TCR lentivirus. The culture medium was refreshed after 8 h. The cells underwent similar culture conditions as described above to support their expansion.
Flow cytometry and antibodies
A comprehensive list of antibodies was listed in Supplementary Table 2. Tetramers were generated using the PE QuickSwitch Quant HLA-A*11:01 Tetramer Kit (MBL) and the APC QuickSwitch Quant HLA-A*11:01 Tetramer Kit (MBL), according to the manufacturer's protocols. SARS-CoV-2 NP361–369 tetramer was used as the negative control. hTCR-KO TCR T cells were stained with BV510-anti-human CD8a antibody, PE-anti-mouse TCR β chain antibody, APC-anti-human TCR antibody for 10 min at room temperature in the dark. After washing twice with PBS, flow cytometry was performed to analyze the expression of the human TCR. Samples were analyzed on a BD LSR Fortessa instrument (BD Biosciences). Fluorescence-activated cell sorting (FACS) was conducted using a BD FACSAria III instrument (BD Biosciences). FACSDiva and FlowJo version 10.8.1 (FlowJo, BD Biosciences) were utilized for data collection and analysis, respectively.
TCR functional avidity assay
TCR T cells were co-cultured with HLA-A*11:01+ K562 cells loaded with KRASG12D peptide at varied concentrations for 24 h. Cells were stained with PE-anti-human CD137 antibody for 10 min at room temperature in the dark. After washing twice with PBS, flow cytometry was performed to analyze the expression of the CD137. EC50 values were calculated as described above.
IFN-γ ELISA assay
IFN-γ Cytokine quantification was conducted using the suggested antibody pairs, including IFN-γ capture antibody (Clone: MD-1, Biolegend) and IFN-γ detection antibody (Clone: 4S.B3, Biolegend). ELISA plates were coated overnight at 4 °C with 50 μl/well of the capture antibody at a concentration of 2 μg/ml. Following coating, plates were washed thrice with PBS containing 0.05% Tween-20 (PBS-T) and blocked with 3% bovine serum albumin (BSA) in PBS for 1 h at room temperature to prevent nonspecific binding. Supernatants collected from co-cultures of T cells and target cells were added to the prepared plates at a volume of 50 μl per well and incubated for 2 h at room temperature to allow cytokine binding. After incubation, plates were washed three times with PBS-T to remove unbound substances. The detection antibody, diluted to 1 μg/ml, was then added at 50 μl/well and incubated for 1 h at room temperature. Following another series of five washes, streptavidin–alkaline phosphatase (Mabtech) diluted 1:1000 in PBS was added to each well and incubated for 1 h in the dark at room temperature. Post incubation, plates were washed six times, and p-nitrophenyl phosphate (pNPP) substrate(Thermo) was added at 50 μl/well. The enzymatic reaction was allowed to develop for 30 min at 37 °C in the dark, after which the reaction was stopped by adding 50 μl of stop solution to each well. The plate was read at 405 nm (OD405) by the Varioskan LUX Multi-mode Microplate Reader (Thermo). The amount of IFN-γ in the samples was determined by comparing the optical densities of the samples to those of a standard curve prepared with known concentrations of recombinant human IFN-γ.
Cytotoxicity assay
The capacity of TCR T cells to lyse tumor cells was quantified employing the OneLumi™ Firefly Luciferase Assay Kit (Beyotime), in accordance with the manufacturer's protocol. Initially, target cells were seeded at a density of 1E4 cells per well in a 96-well plate and incubated overnight to achieve adherence. Subsequently, TCR T cells, at varying effector-to-target (E:T) ratios (10:1, 5:1, 1:1, 1:2, and 1:10), were introduced to the co-culture and incubated for a period of 24 h. Following incubation, The cell culture plate was placed at room temperature for 10 min, followed by the addition of 100 μl of OneLumi™ detection reagent to each well. It was then incubated at room temperature for 5 min to allow the luminescent signal to stabilize. Luminescence, indicative of cell viability, was quantified as Relative Luminescence Units (RLUs) using the Varioskan LUX Multimode Microplate Reader. Cell viability and cytotoxicity were quantitatively assessed based on luminescence measurements. Luminescence readings from wells devoid of T cells served as the baseline for 100% survival. The survival rate of tumor cells in the presence of TCR T cells was determined by comparing the luminescence of treated wells to that of the control wells. The cytotoxicity rate was determined as 1 minus the survival rate. Each condition was replicated in triplicate.
Statistics and reproducibility
Appropriate statistical tests were used to analyze data, as described in each figure legend. Statistical analyses were performed with Prism 9.0 (GraphPad). Standard deviation (SD) is depicted as error bars in graphical representations. Significance levels were annotated as follows: *p < 0.05, **p < 0.01, and ***p < 0.001 and NS indicates a non-significant difference. All data presented are representative of two or more independent experiments, ensuring the reproducibility of findings. No data were excluded from any analysis.
Results
Identification and functional profiling of HLA-A*11:01-restricted TCRs against KRASG12D epitope
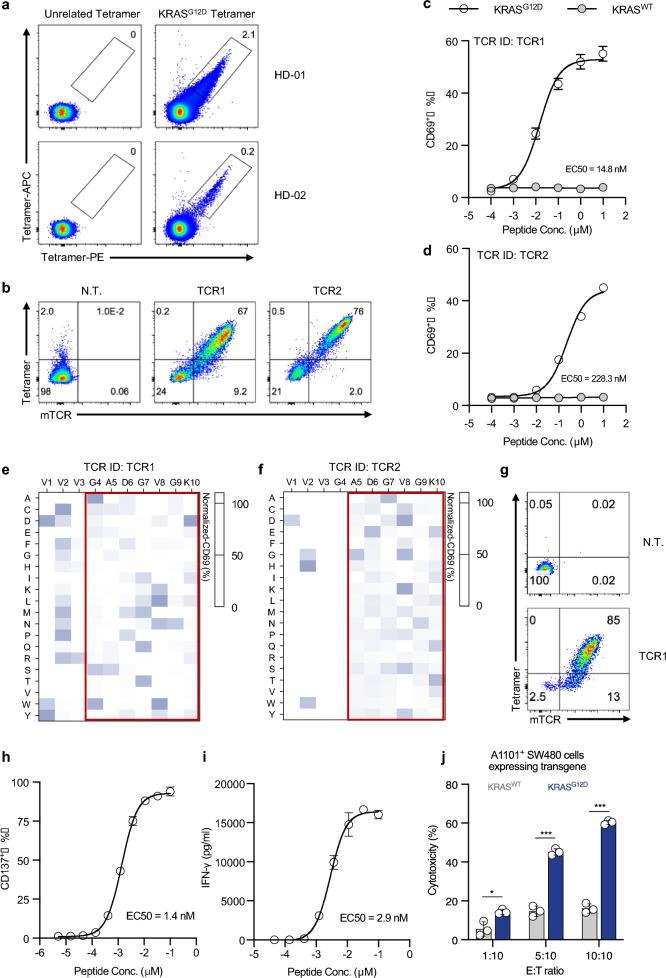
To isolate TCRs specifically recognizing KRASG12D epitope, naive T cells from HLA-A*11:01+ health donors were co-cultured with autologous monocyte-derived dendritic cells (mo-DCs), which were pre-loaded with KRASG12D peptide. We identified T cells derived from two individual donors that could bind to the KRASG12D tetramer (Fig. 1a). The KRASG12D specific T cells were sorted as single cells for the identification of TCR gene sequences. DNA sequencing data revealed that specific T cells from each sample contained one unique TCR clonotype, termed TCR1 and TCR2 (Supplementary Table 3). To conduct the preliminary functional evaluation of the TCRs, the CD8+ Jurkat cell model expressing recombinant TCR1/2 with mouse TCR constant region (mTCR) were established. Both TCRs were capable of binding to the KRASG12D tetramer (Fig. 1b). After co-culturing with HLA-A*11:01+ K562 cells that had been loaded with serially diluted KRASG12D peptide, both TCR clones expressed the activation marker CD69, whereas no activation was detected in both TCR clones stimulated by the wild-type KRAS peptide (KRASWT) (Fig. 1c and d). The functional avidity of each TCR was measured to assess their relative antigenic sensitivity, and the EC50 values of TCR1 and TCR2 were found to be 14.8 nM and 228.3 nM, respectively (Fig. 1c and d). This result suggested that TCR1 had advanced functional avidity relative to TCR2 in targeting HLA-A*11:01+ restricted KRASG12D epitope.
Fig. 1.
Discovery and functional characterization of HLA-A*11:01-restricted KRASG12D specific TCR clones. a KRASG12D-specific T cells were assessed using KRASG12D tetramer, showing live+ CD3+ CD8+ T cells. b TCR Jurkat cells were evaluated for TCR expression and KRASG12D tetramer binding. c, d TCR Jurkat cells were co-cultured with HLA-A11:01+ K562 cells loaded with KRASG12D peptide and functional avidity (EC50) determined by CD69 expression. Wild-type KRAS peptide served as negative control. Peptide specificity of TCR1 e and TCR2 f was assessed by X-scan assay. Normalized data are displayed as heatmaps. g TCR1 expression on CD8+ T cells was measured; TCR1 T cells were co-cultured with HLA-A11:01+ K562 cells and analyzed for CD137 (h) and IFN-γ release (i). j TCR1 T cells were co-cultured with HLA-A*11:01+ SW480 cells expressing KRASWT or KRASG12D and the cytotoxicity were assessed after 24 h. Data are presented as mean ± SD and all panels represent data from two independent experiments. HD healthy donor. N.T. non-transduced
To analyze the epitope-recognition specificity of these TCRs, a peptide library was designed (referred to as KRASG12D-pep-Lib), and corresponding peptides were synthesized by systematically substituting each amino acid of the KRASG12D peptide with the other 19 common amino acids (Supplementary Table 4). HLA-A*11:01+ K562 clones, each loaded with an individual peptide from the KRASG12D-pep-Lib, were co-cultured with TCR Jurkat cells, and CD69 expression was measured as a quantitative indicator of the TCR recognition capability. The percentage of CD69 expression was converted into a residue scanning heatmap for visualized comparison. The recognition pattern of TCR for the 10mer KRASG12D epitope was characterized. Mutations in the KRASG12D peptide at positions 4 to 9 impaired TCR1 activation (Fig. 1e), while mutations at positions 5 to 9 had the detrimental effect on TCR2 activation (Fig. 1f). These results indicated that TCR1 interacts with a broader region of the peptide compared to TCR2. These findings demonstrated that TCR1 exhibited advanced functional importance as a TCR candidate and was thus chosen for further evaluation for potential antitumor immunotherapy of T cells targeting KRASG12D.
To further characterize the functionality of TCR1, it was expressed in CD8+ T cells derived from healthy donors. We found that 85% T cells could bind to KRASG12D Tetramer (Fig. 1g). The functional avidity of TCR1 was confirmed by co-culturing TCR1 T cells with HLA-A*11:01+ K562 cells loaded with serially diluted KRASG12D peptides, and the expression of the activation marker CD137 and IFN-γ production were measured after 24 h. The results showed that both the proportion of CD137+ T cells and IFN-γ secretion increased in exposure to the KRASG12D peptide in a dose-dependent manner (Fig. 1h and i). The functional avidity (EC50 value) calculated by CD137 and IFN-γ measurements were 1.4 nM and 2.9 nM, respectively, suggesting that TCR1 possesses sharp KRASG12D peptide sensitivity. Furthermore, we assessed the recognition and killing capability of TCR1 T cells against tumor cells carrying KRASG12D mutation. CD8+ TCR1 T cells was co-cultured with HLA-A*11:01+ SW480 cells expressing KRASG12D or KRASWT. Compared to the KRASWT control group, the KRASG12D group demonstrated significantly higher levels of cell death to TCR1 T cell-mediated killing at different E:T ratios (Fig. 1j). These results showed that TCR1 exhibited high functional avidity in CD8+ T cells and mediated effective cytotoxicity against tumor cells expressing KRASG12D mutation.
Assessment of the cross-reactivity potential for TCR1
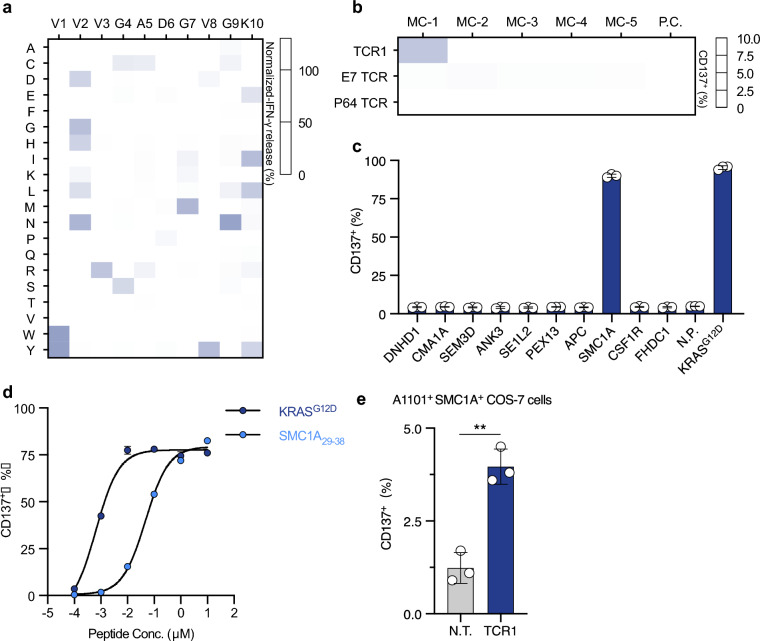
To assess potential off-target recognition of TCR1, we employed X-scan analysis to profile TCR peptide specificity. Initially, TCR1 T cells were co-cultured with HLA-A*11:01+ K562 cells loaded with the KRASG12D-pep-Lib, followed by IFN-γ release measurement after 24 h. Specificity profiling showed that most KRASG12D peptide mutations at positions 4, 5, 6 and 9 were detrimental for TCR1 activation (Fig. 2a). Based on these results, we revealed the TCR1 recognition motif (Supplementary Table 5), with 20% reactivity to the KRASG12D peptide as cutoff. Scanning this motif against the Human reference proteome (Proteome ID: UP000005640) by ScanProsite identified a total of 48 TCR1 cross-recognized candidate peptides (Supplementary Table 6). We designed a total of 5 minigene-clusters (MC-1 to MC-5), encoding up to ten of the above homologous peptides each (Supplementary Fig. 2a). In accordance, 5 groups of HLA-A*11:01+ K562 cells expressing one MC per group (Supplementary Fig. 2b) were co-cultured with TCR1 T cells. Flow cytometric analysis of CD137+ cell proportions showed that only MC-1 expressing K562 cells moderately induced TCR1 T cell activation (Fig. 2b). Subsequently, the ten short peptides encoded in MC-1 were synthesized and individually loaded to HLA-A*11:01+ K562 cells. Co-culture examination with TCR1 T cells showed that a homologous peptide derived from protein SMC1A induced robust elevation of CD137+ T cell proportions (Fig. 2c and d). This peptide corresponds to the N-terminal 29th to 38th amino acids of the protein SMC1A (namely SMC1A29–38), which was conserved across different mammalian species (Supplementary Fig. 2c). Then, the full-length cDNA of SMC1A was transduced into HLA-A*11:01+ COS-7 cells. Co-culture experiment with TCR1 T cells showed that HLA-A*11:01+SMC1A+ COS-7 cells significantly increased the proportion of CD137+ T cells, comparing to the vector transduced control group (Fig. 2e). This result confirmed that the SMC1A29–38 epitope could be endogenously processed and presented on the cellular membrane by HLA-A*11:01. Collectively, these results showed that TCR1 was a functional TCR that could recognize the KRASG12D epitope, however, it exhibited cross-reactivity to a self-antigen SMC1A29–38 epitope.
Fig. 2.
Peptide specificity profiling of TCR1 T cells for identifying potential off-target peptides. a TCR1 peptide specificity was assessed by X-scan assay and shown as a heatmap. b TCR T cells were co-cultured with HLA-A11:01+ K562 cells with Minigene-cluster constructs; CD137 expression was analyzed after 24 h. E7 TCR and P64 TCR served as negative and positive controls, respectively. c Peptides from Minigene-cluster 1 were loaded onto HLA-A11:01+ K562 cells and co-cultured with TCR1 T cells; CD137 expression was measured. d TCR1 T cells were co-cultured with HLA-A11:01+ K562 cells loaded with SMC1A29–38 peptide. CD137 expression was analyzed. e TCR1 T cells were co-cultured with HLA-A11:01+ COS-7 cells expressing full-length SMC1A; CD137 expression was measured. Data are presented as mean ± SD and all panels represent data from two independent experiments. N.T. non-transduced T cells, P.C. positive control, N.C. negtive control, N.P. Non-peptide
To examine whether SMC1A29–38 was a shared off-target self-antigen for different HLA-A11:01-restricted KRASG12D specific TCR clones, two previously reported KRASG12D-specific TCRs, including JDI [14] and TIL4373 [13], were expressed in the CD8+ Jurkat cell (Supplementary Table 3). The TCR clones were co-cultured with HLA-A11:01+ K562 cells loaded with serially diluted KRASG12D peptide, and the functional avidity of TCR1, JDI, and TIL4373 was shown to be 14.8 nM, 115 nM and 5 nM, respectively (Supplementary Fig. 3a). In the parallel off-target examination, TCR clones were co-cultured with HLA-A*11:01+ K562 cells loaded with serially diluted SMC1A29–38 peptide. The results showed that CD69 expression in TIL4373 T cells was induced by SMC1A29–38 at a concentration of 1 μM, while JDI TCR remained in an inactive state even at the concentration of 10 μM (Supplementary Fig. 3b). These results suggested that both TCR1 and TIL4373 exhibit off-target recognition of the SMC1A29–38 peptide.
Comparison of peptide specificity of the three KRASG12D epitope specific TCR clones
To compare the characteristic of TCR recognition, peptide specificity profiling of TCR clones were conducted. A comparison analysis of X-scan result between TCR1 Jurkat cells and TCR T cells showed the good consistency (R2 = 0.8893, Supplementary Fig. 3c), suggesting for the feasibility of using TCR Jurkat cells for all following X-scan analysis. Then, we performed X-scan analysis on the 3 clone KRASG12D-specific TCR Jurkat cells using the KRASG12D-pep-Lib (Supplementary Fig. 3d–f). The result showed that KRASG12D peptide mutations at positions 4 to 8 were detrimental for TIL4373 TCR activation, and had the distinctly different peptide recognition pattern with TCR1 (Supplementary Fig. 3f). Interestingly, specificity profiling showed that most KRASG12D peptide mutations at positions 4 to 9 were detrimental for JDI TCR activation, which was the similar recognition pattern with TCR1 (Supplementary Fig. 3d and 3e). Furthermore, JDI TCR showed a lower percentage of T cell activation to the majority of KRASG12D-pep-Lib members (Supplementary Fig. 3 g). These findings indicated that JDI with TCR1 had similar peptide specificity, despite its inability to recognize the SMC1A29–38 peptide.
Further, the amino acid sequences of TCR1 and JDI TCR showed high sequence conservation in multiple CDR regions, including CDR-1A, CDR-2A and CDR-3B (Supplementary Fig. 4a). Structural analysis of the reported JDI TCR-KRASG12D peptide-HLA complex (pMut-HLA) showed that the CDR-1A, CDR-3A and CDR-3B of JDI TCR all formed hydrogen bonds with the pMut-HLA, with CDR-3B predominantly engaging in peptide interaction and CDR-1A and CDR-3A primarily interacting with HLA (Supplementary Fig. 4b and 4c). Based on these data, we hypothesized that the identical CDR-3B region in TCR1 might exhibit similar interactions with the KRASG12D peptide, thus leading to similar KRASG12D-pep-Lib reactivity between the two TCRs. To confirm this hypothesis, we conducted point mutation analysis on the shared CDR-1A T30 amino acid residue of TCR1, which formed hydrogen bonds with HLA in JDI TCR (Supplementary Fig. 4d and supplementary Table 7). The T30Q mutation decreased the pMut-HLA binding positivity rate from 91 to 60%, and reduced the functional avidity of TCR1 for KRASG12D from 15 to 53 nM (Supplementary Fig. 4e). These results revealed the similarity in the recognition pattern of TCR to pMut-HLA between TCR1 and JDI TCR, suggesting that the known JDI TCR pMut-HLA complex structural information could serve as good reference for the modification design to alternate TCR1 peptide specificity.
The design of a structure-guided TCR library and the functional screening of TCRs absent of SMC1A29–38 recognition
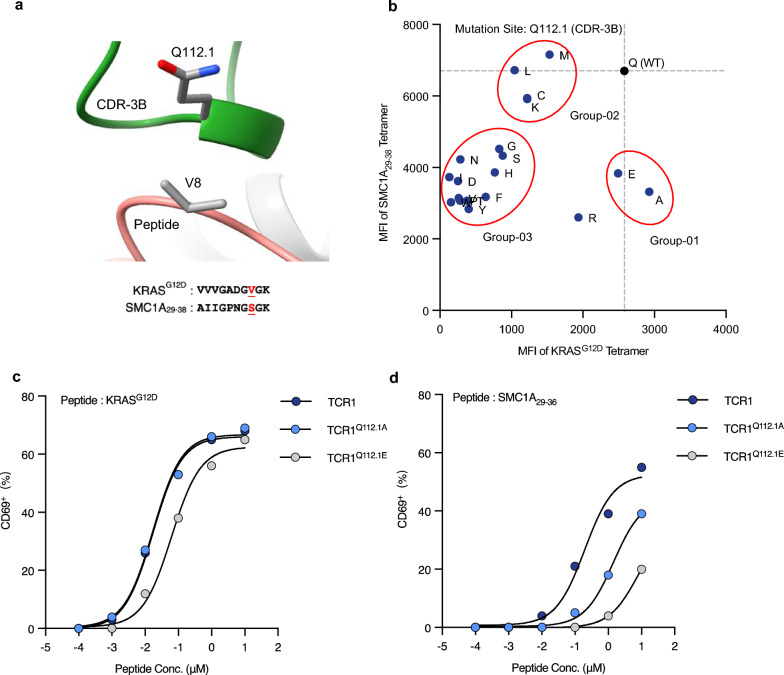
To eliminate the recognition of the SMC1A29–38 peptide, the primary approach focused on altering amino acid residues in the CDR-3B region of TCR1. Structural information suggested that the TCR CDR-3B was in proximity to the amino acids at positions 7 to 9 of the KRASG12D peptide (Fig. 3a). We found that KRASG12D and SMC1A29–38 differed at the 8th amino acid residue, which was Valine (V) and Serine (S), respectively (Fig. 3a). Due to the structural correspondence between the amino acid Q112.1 of TCR CDR-3B and the 8th position of the KRASG12D peptides, a single-point saturation mutation profiling was performed on this residue. Flow cytometric assessment of HLA binding affinity with KRASG12D and SMC1A29–38 tetramers was conducted on the 19 TCR mutant clones. As shown in Fig. 3b, the mutant TCR clones were categorized into three groups, relative to the wild-type TCR1: Group-01 (Q to A or E) displayed a decrease in the MFI value of the SMC1A29–38 tetramer binding by half, while the MFI value for KRASG12D tetramer binding remained unchanged; Group-02 (Q to C, K, L or M) showed no alteration in the off-target MFI value, but the MFI value for target tetramer binding decreased by half; and Group-03 (Q to each of the other 13 amino acids) decreased the off-target recognition MFI value by half, while the KRASG12D tetramer binding MFI value dropped two-fold. These results indicated that the mutant TCR clones in Group-01 exhibited reduced binding affinity to SMC1A29–38 without affecting their recognition of KRASG12D. Two TCR clones from Group-01 were subsequently selected for a functional avidity assay against the SMC1A29–38 and KRASG12D peptides. The functional avidity of TCR1, TCR1Q112.1A, and TCR1Q112.1E were 17 nM, 17 nM, and 62 nM to KRASG12D, respectively (Fig. 3c), and the minimum SMC1A29–38 peptide concentrations for TCR activation were 10 nM, 100 nM, and 1 μM, respectively (Fig. 3d). This data showed that the Q112.1A mutation maintained the functional avidity for KRASG12D and eliminated the recognition ability of SMC1A29–38 at the concentration of 10 nM. Meanwhile, the Q112.1E mutation resulted in the complete removal of off-target recognition at the maximum physiological concentration (100 nM), but also showed slightly reduced functional avidity for KRASG12D. These results indicated that the Q112.1 mutation alone could not fully eliminate SMC1A29–38 recognition without compromising KRASG12D recognition. Nonetheless, these data supported the advanced modifications of TCR1’s residues to enhance its peptide specificity.
Fig. 3.
Diminishing TCR1 recognition of SMC1A29-38 peptide through Q112.1 residue mutation in the CDR-3B region. a Analysis of the TCR and peptide interaction region of the JDI/pHLA complex. The AA sequences of KRASG12D and SMC1A29-38 are shown. b Q112.1 residue was mutated into 19 other AAs. These mutant TCR clones were stained with KRASG12D Tetramer and SMC1A29–38 Tetramer. Functional avidity of TCR1Q112.1A clone and TCR1Q112.1E clone towards KRASG12D peptide c and SMC1A29–38 peptide d were assessed. All panels represent data from two independent experiments. AA amino acid
We broadened the modified residues range to construct two TCR libraries involving the TCR-pHLA interacting region. TCR1 Lib1 consisted of a saturation mutation library formed by the four consecutive sites in close contact (5 Å) with the KRASG12D peptide (CDR-3B: G111 to N113) (Supplementary Fig. 5a), and TCR1 Lib2 comprised a saturation mutation library formed by the three consecutive sites closest to the HLA interaction (CDR-1A: D29 to T36) (Supplementary Fig. 5b). After expressing the TCR libraries in CD8+ Jurkat cells, four rounds of flow cytometric sorting were conducted to enrich TCR clones that could bind to the KRASG12D tetramer and exhibited activation upon exposure to the KRASG12D peptide (Supplementary Fig. 5c). After 4 rounds of enrichment, TCR libraries were stained with SMC1A29–38 tetramer and KRASG12D tetramer, and TCR clones demonstrated sole binding to the KRASG12D tetramer were collected (Supplementary Fig. 5c). Through cloning the single-cell TCR genes, a total of 24 TCR clones were obtained (Supplementary Table 8). Sequence profiling revealed that TCR Lib1 had more notable enrichment trend comparing to Lib2, with a particular increase in the frequency of histidine at N112 in the CDR-3B (Supplementary Fig. 5d and e). This highlighted the possibility that the N112H substitution might be a determinant of TCR functional avidity. Those results suggested that the directed modification of TCR-pHLA interacting region can generate the novel KRASG12D peptide specific TCR clones.
Functional assessment and specificity profiling of candidate TCR clones
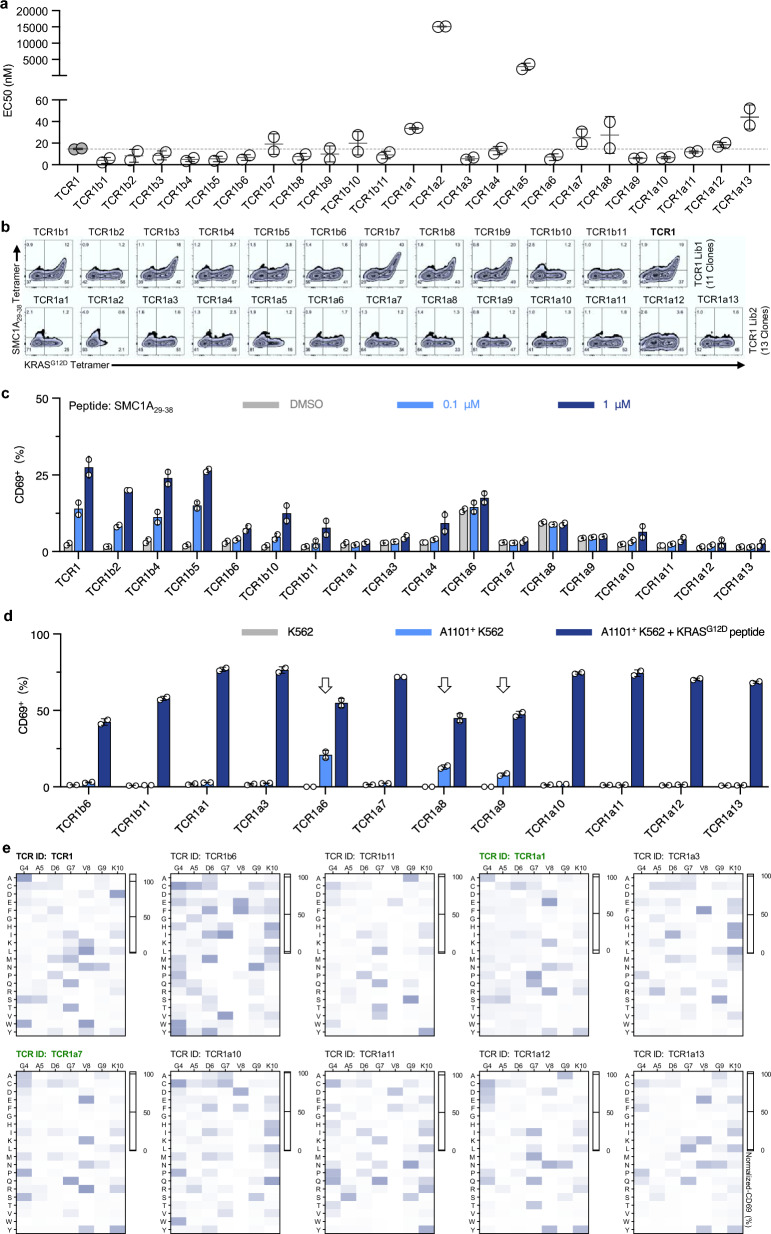
So far, we have successfully established 24 novel TCR clones from the two TCR1 libraries, and their characteristics and functionality were subjected to a four-step evaluation. Firstly, the functional avidity of all TCR clones to KRASG12D peptide was analyzed, and majority of these TCR clones exhibited the lower in EC50 value, suggesting for broadly enhanced functional avidity (Fig. 4a). Two TCR clones, TCR1a2 and TCR1a5 showed over ten-fold increase in EC50, thus were eliminated from future analysis. Secondly, the binding capability of the 22 TCR clones was examined with the SMC1A29–38 tetramer. The analysis revealed that 6 TCR clones from TCR1 Lib1 and all clones from Lib2 display dismissible level of tetramer binding (Fig. 4b). Thirdly, a functional confirmation was performed to study TCR activation in response to off-target stimulation, and the result showed that 12 TCR clones remained inactivated when stimulated by 100 nM SMC1A29–38 peptide (Fig. 4c). Lastly, these artificially engineered TCR clones were examined for any peptide-independent HLA-reactivity. All remaining clones were subjected to co-culture with K562, HLA-A*11:01+ K562, or HLA-A*11:01+ K562 loaded with the KRASG12D peptide. Three TCR clones showed substantial increase in CD69+ proportions when stimulated by HLA-A*11:01+ K562 alone, thus were eliminated from further testing (Fig. 4d). Through this four-step functional assessment, a total of 9 TCR clones (37.5%) were found with sufficient KRASG12D recognition sensitivity and minimal reactivity to SMC1A29–38 peptide.
Fig. 4.
Functional evaluation and peptide specificity profiling of TCR clones derived from TCR1 libraries. a Functional avidity (EC50) of 24 TCR clones was assessed by co-culturing with HLA-A11:01+ K562 cells loaded with KRASG12D peptide, measuring CD69 expression after 6 h. Dashed line represents TCR1 EC50. b TCR clones were stained with SMC1A29-38 and KRASG12D tetramers. c TCR clones were co-cultured with HLA-A11:01+ K562 cells loaded with 0.1 µM and 1 µM SMC1A29–38 peptides, and CD69 expression was analyzed. HLA-A11:01+ K562 cells loaded with DMSO as negative control. d TCR activation was assessed with K562 cells under three conditions: unmodified, HLA-A11:01+ without peptide, and HLA-A11:01+ with KRASG12D peptide. e Peptide specificity of TCR clones was profiled using X-scan assay and results are shown as heatmaps. Data are presented as mean ± SD and all panels represent data from two independent experiments. AA amino acid
The peptide specificity profiles of these 9 TCR clones were determined by X-scan analysis. Given that the first three amino acids at the N-terminus of KRASG12D peptide could not be distinguished by TCR1 (Fig. 2a), we selected a sub-library of KRASG12D peptide mutants consisting of alteration from the 4th to the 10th amino acids only (VVVGACGVGK). Peptide specificity profiling showed that 7 TCR clones exhibited elevated CD69+ proportions with mutations of the 8th amino acid, comparing to the wild-type TCR1 (Fig. 4e and Supplementary Table 9). Importantly, two clones, TCR1a1 and TCR1a7, demonstrated comparable recognition specificity to that of TCR1, with enhanced discrimination capability of the 7th amino acid of KRASG12D peptide (Supplementary Fig. 6). These results showed that two TCR clones were successfully obtained through optimized structural engineering, which exhibited improved peptide specificity and minimum recognition of the SMC1A29–38 peptide.
Functional characterization and safety assessment of engineered KRASG12D-specific TCRs
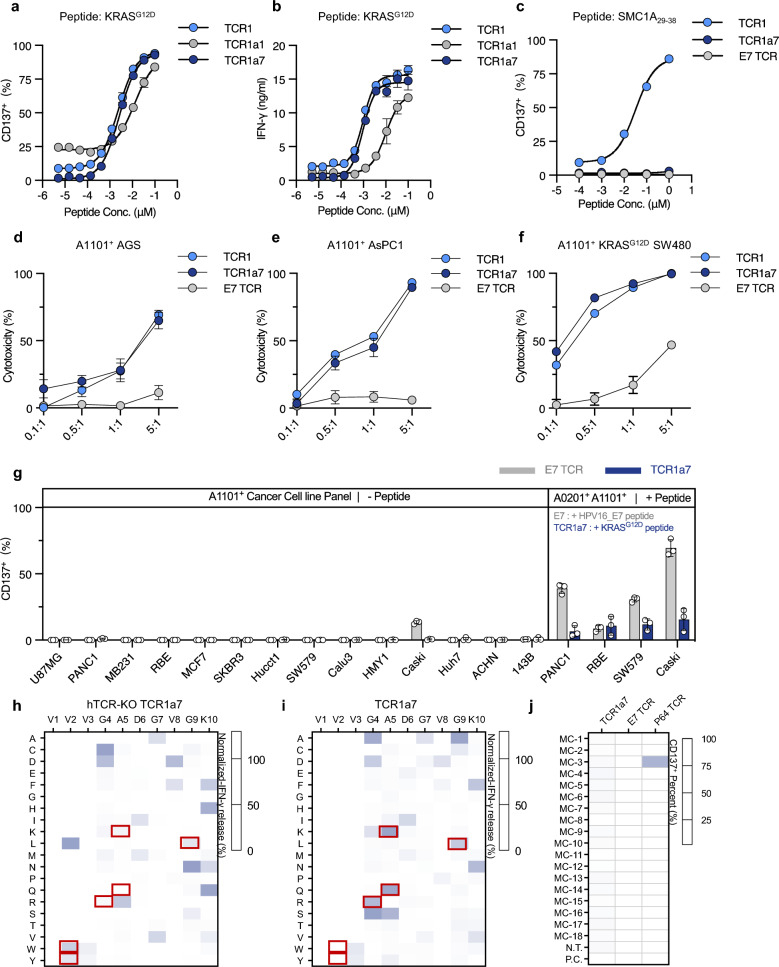
To systematically characterize the functionality of selected TCR clones, we engineered CD8+ T cells obtained from healthy donors with stable expression of TCR1a1, TCR1a7 or TCR1 after endogenous TCR knockout (referred to as hTCR-KO TCR T cells). The efficiency of TCR expression was confirmed (Supplementary Fig. 7a and 7b). The functional avidity analyses showed that the EC50 of TCR1a7 was 2.6 nM as determined by the proportion of CD137+ T cells, and 1.0 nM as indicated by the level of IFN-γ release (Fig. 5a and b). These levels were comparable to those of TCR1 (Fig. 5a and b). TCR1a1 demonstrated reduced functional avidity, with EC50 of 12.8 nM and 10.0 nM as determined by CD137+ proportions and IFN-γ secretion, respectively (Fig. 5a and b). Given the apparent functional advance of TCR1a7, we further tested its recognition of SMC1A29–38 peptide. A concentration-dependent increase of CD137+ TCR1 T cell proportion was observed in response to the SMC1A29–38 peptide (Fig. 5c). In contrast, no activation signals were detected in TCR1a7 T cells in response to this peptide at the physiological concentrations (≤ 100 nM), which was similar to the results observed in the HPV16 E7 specific TCR (E7 TCR) as negative control group (Fig. 5c). These results were further confirmed using HLA-A*11:01+ SMC1A+ COS-7 cells expression full length of SMC1A cDNA. We found that the proportion of CD137+ TCR1a7 was at a level comparable to that of E7 TCR control, whereas TCR1 demonstrated significantly higher level of CD137+ proportions (Supplementary Fig. 7c). Furthermore, we assessed the cytotoxic capabilities of TCRs against tumors harboring the KRASG12D mutation. Co-culture experiments were conducted with tumor cells of various cancer types, including colorectal cancer (SW480 cells), pancreatic cancer (AsPC-1 cells) and gastric cancer (AGS cells). TCR1a7 and TCR1 both exhibited enhanced cytotoxicity with increased effector-to-target (E:T) ratios, whereas minimum cytotoxicity was detected in the negative control group (E7 TCR) (Fig. 5d–f). These results evidenced that TCR1a7 exhibit efficient functional avidity and potent anti-tumor activity, with successful removal of SMC1A29–38 recognition.
Fig. 5.
Functional characterization and specificity profiling of engineered TCR1a7 T cells. hTCR-KO TCRs T cells were co-cultured with HLA-A11:01+ K562 cells loaded with KRASG12D peptide. After 24 h, CD137 expression a and IFN-γ release b were analyzed. c hTCR-KO TCR T cells were co-cultured with HLA-A11:01+ K562 cells loaded with SMC1A29-38 peptide and CD137 expression was measured. E7 TCR-T cells served as N.C. d–f TCR1a7 T cells were co-cultured with HLA-A11:01+ AGS, AsPC-1, or KRASG12D SW480 cells, and cytotoxicity was assessed. g CD137 expression on TCR1a7 T cells co-cultured with HLA-A11:01+ cancer cell lines was analyzed. h, i Peptide specificity profiling of TCR1a7 was performed by X-scan assay. j TCR1a7 T cells were co-cultured with HLA-A*11:01+ K562 cells expressing Minigene-cluster constructs, and CD137 expression was analyzed. E7 TCR-T cells served as N.C. and P64 TCR-T cells as P.C. HLA+ K562 cells without constructs were N.T., and those with antigen peptide were P.C. Data are presented as mean ± SD and all panels represent data from two independent experiments. N.T. non-transduced, N.C. negative control, P.C. positive control
Furthermore, potential new off-target recognition of TCR1a7 was evaluated. The hTCR-KO TCR1a7 T cells were co-cultured with a panel of HLA-A*11:01+ cancer cell lines that did not harbor KRASG12D mutation. TCR1a7 T cells activation was not detected in presence of cancer cells either naturally expressing or genetically engineered to express HLA-A*11:01, contrary to the robust T cell activation when target cells were loaded with the relevant peptide (Fig. 5g). Moreover, X-scan analysis was conducted on hTCR-KO TCR1a7 and TCR1 T cells. Peptide specificity comparison between TCR1a7 and TCR1 revealed that the most outstanding change was the enhanced discrimination ability of TCR1a7 to recognize the 7th amino acid of the KRASG12D peptide (Supplementary Fig. 7d), which was consistent with earlier results obtained from TCR1a7 Jurkat cells (Supplementary Fig. 6). Compared with TCR1a7 T cells expressing endogenous hTCR, hTCR-KO TCR1a7 T cells exhibited marked improvement of peptide specificity on the 2nd, 4th, 5th and 9th amino acids of the KRASG12D peptide (Fig. 5h and i). In addition, potential cross-reactivity peptides were predicted using the TCR recognition motif (Supplementary Table 5, Supplementary Table 10 and supplemental Table 11), and the result showed that the removal of hTCR reduced the number of predicted cross-recognition by 2.8-fold (493 versus 174, Supplementary Fig. 7e). These results indicated that the knockout of endogenous hTCR improved the peptide specificity of TCR1a7. Accordingly, the 174 potential homologous peptides for hTCR-KO TCR1a7 were constructed into 18 minigene-cluster constructs (Supplementary Table 11), and tested by TCR cross-reactivity examination. The results showed that none of the 18 constructs stimulated an activation of hTCR-KO TCR1a7 T cells, suggesting that the engineered TCR1a7 did not introduce new off-targets (Fig. 5j). Taken together, these data demonstrated successful TCR modification via structure-guided engineering could remove TCR cross-reactivity and enhance the peptide specificity. With this approach, we successfully obtained a TCR (TCR1a7) that demonstrated advanced functional avidity to HLA-A*11:01+-restricted KRASG12D peptide with potent anti-tumor activity.
Discussion
In the present study, two HLA-A11:01-restricted KRASG12D-specific TCR clones were originally obtained from the human naive T cell repertoire, from which TCR1 exhibited advantageous functional avidity and peptide specificity. TCR1 specifically responded to tumor cells expressing KRASG12D mutation, suggesting for the recognition capability of TCR1 to endogenously processed KRASG12D epitope. However, we found that TCR1 also recognized a self-antigen SMC1A29–38. Rather than disqualifying it as a typical course of action, we proceeded to test whether this off-target recognition could be eliminated through TCR engineering. We took an initial step of determining whether SMC1A29–38 was a shared target for other reported HLA-A11:01-restricted KRASG12D-specific TCRs, and found that TIL4373, but not JDI TCR, also recognized the SMC1A29–38 peptide. Further analysis clarified that TIL4373 could only distinguish the central region of the KRASG12D peptide, and its recognition of SMC1A29–38 was a possible consequence of its comparatively broader peptide specificity than the other KRASG12D-specific TCRs. These resluts supported that SMC1A29–38 was likely a private off-target antigen for TCR1.
Public TCRs are known as TCRs with identical CDR-3B sequences from multiple individuals targeting the same antigen epitope, which were mainly reported in viral infections and autoimmune diseases but were less common in tumor antigens. [20, 27, 28] TCR1 and JDI TCR shared identical CDR-3B sequence, the X-scan analysis revealed an evident similarity in peptide specificity between them. Therefore, our results might provide evidence for the existence of public TCRs specific to the KRASG12D epitope.
To eliminate the off-target recognition of the SMC1A29–38 peptide, a display library with specific screening pressures was applied for the modification design of TCR1. Unlike phage or yeast display libraries, which display limited TCR TRAV/TRBV allele and inconsistency between TCR binding affinity and TCR functional avidity, mammalian display systems using TCR-Jurkat cells have proper post-translational modifications and measurable activation markers [29, 30]. Therefore, we chose the TCR-Jurkat system to display TCR1 and facilitate the efficient capture of TCR clones with functional activity [30]. To properly fit the capacity of this mammalian display library, we utilized available TCR-pHLA structure data to guide TCR library design, focusing on the residues involved in the TCR-pHLA interaction. After five rounds of TCR functional screening, we obtained 24 TCR clones, 15 out of which exhibited higher functional avidity than the wild-type TCR1. This indicated that our screening strategy is applicable for improving TCR functional avidity. To be noted, among the 12 TCR clones that successfully eliminated the off-target recognition of SMC1A29–38, only 2 clones were derived from modification of the TCR-peptide interaction region, while the other 10 clones were from modification of the TCR-HLA interaction region. It was speculated that the high spatial similarity at the C-terminal of the two peptides rendered it difficult to distinguish between KRASG12D and SMC1A29–38 through engineering modifications in the TCR-peptide interaction region. Our findings shed light on a comprehensive view of TCR library design for TCR peptide specificity to sight both the TCR-peptide and TCR-HLA interaction regions.
Our peptide specificity profiling of 7 TCR clones derived from the TCR-HLA interaction region indicated that TCR modifications at the HLA interaction region only modulated the TCR's recognition capability for specific sites, while generated minimal impact on overall peptide recognition specificity. Since these modifications do not drastically affect the peptide specificity of TCR1, the engineered TCRs ware likely to preserve the parental TCR's potential safety properties acquired during thymic development. However, it is important to note that TCR modification at the HLA interaction region can produce peptide-independent HLA-reactive TCR clones, which are ought to be removed by the corresponding functional evaluation. Our TCR engineering strategy successfully generated TCR1a7 from TCR1, which was shown with efficient functional avidity and potent anti-tumor activity with enhanced peptide specificity in further functional assessment using TCR T cells. These findings evidenced for a practical value of TCR1a7, although further anti-tumor activity assessments are necessary prior to advancing TCR1a7 to clinical research, with proper testing in tumor models and safety evaluations utilizing human-derived organoids and HLA-transgene mice.
The potential effect of endogenous TCR has long been debated for disturbing engineered TCR peptide specificity [31]. To minimize potential influence of endogenous TCR, We utilized TCR T cells with endogenous TCRs knocked out, while employing mouse TCR constant regions to replace those of human TCRs [31, 32]. Consequently, a distinguishable decrease in the response intensity of TCR1a7 to KRASG12D-pep-Lib at certain sites was captured, which corresponded with altered TCR recognition motifs and a reduced number of predicted potential off-targets. These findings underscored that endogenous TCRs did project certain level of impact on the peptide specificity of foreign engineered TCRs, and the current strategy of TCR modification did not completely eliminate these spontaneous interference that would be undoubtedly presented in actual TCR T engineering procedure. Another concern risen on top of this is the current increasing focus on enhancing T cell functionality or attenuating T cell exhaustion by targeting genes related to TCR signaling pathways [32–34]. These interventions could potentially alter the activation threshold of TCR signaling, thereby changing TCR recognition motifs and leading to unexpected off-target effects.
Conclusions
In summary, we identified a TCR specific for the KRASG12D peptide and TCR1 transduced T cells displayed cross-reactivity to a self-antigen SMC1A29–38. Through combining TCR-pHLA structure with the mammalian TCR library display system, we obtained a clone TCR1a7 with enhanced peptide specificity and eliminated off-target recognition. Our study provides an effective approach to improve TCR functional avidity and peptide specificity, which is critical for the development of TCR-based therapeutics.
Supplementary Information
Acknowledgements
We thank all healthy individuals participated in this study.
Author contributions
A.S.J. and M.Y.S. conceived and designed the study. S.Y.C., T.C., L.Z., J.J.H. and L.L. were responsible for collecting samples. X.J.H., X.X.H., Y.N.H. and B.Z.W performed the experiments. X.J.H., X.X.H and M.Y.S. performed data analysis and incorporation. A.S.J., W.W. and X.J.H. wrote the manuscript.
Funding
This study was supported by National Natural Science Foundation, China (Grant no. 82350120 and Grant no.81872329) and Chongqing Medical University fund (X4457) with the donation from Mr. Yuling Feng.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of The First Affiliated Hospital of Chongqing Medical University ( the approval number: 2023–234). All individuals signed an informed consent form.
Consent for publication
Not applicable.
Competing interests
Patent has been filed for KRASG12D specific TCRs presented here. All other authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaojian Han and Xiaxia Han have contributed equally to this work.
Contributor Information
Aishun Jin, Email: aishunjin@cqmu.edu.cn.
Meiying Shen, Email: meiying@hospital.cqmu.edu.cn.
References
- 1.Martinov T, Greenberg PD: Targeting Driver Oncogenes and Other Public Neoantigens Using T Cell Receptor-Based Cellular Therapy. Annu Rev Cancer Biol. 2023:7:331–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, Gartner JJ, Zheng Z, Li YF, Ray S, et al: Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350:1387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, et al: T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N Engl J Med. 2016;375:2255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandran SS, Ma J, Klatt MG, Dundar F, Bandlamudi C, Razavi P, Wen HY, Weigelt B, Zumbo P, Fu SN, et al: Immunogenicity and therapeutic targeting of a public neoantigen derived from mutated PIK3CA. Nat Med. 2022;28:946–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang K, Halima A, Chan TA: Antigen presentation in cancer - mechanisms and clinical implications for immunotherapy. Nat Rev Clin Oncol. 2023;20:604–23. [DOI] [PubMed] [Google Scholar]
- 6.Lee JK, Sivakumar S, Schrock AB, Madison R, Fabrizio D, Gjoerup O, Ross JS, Frampton GM, Napalkov P, Montesion M, et al: Comprehensive pan-cancer genomic landscape of KRAS altered cancers and real-world outcomes in solid tumors. NPJ Precis Oncol. 2022;6:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robbins PF, Morgan RA, Feldman SA, Yang JC, Sherry RM, Dudley ME, Wunderlich JR, Nahvi AV, Helman LJ, Mackall CL, et al: Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol. 2011;29:917–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagarsheth NB, Norberg SM, Sinkoe AL, Adhikary S, Meyer TJ, Lack JB, Warner AC, Schweitzer C, Doran SL, Korrapati S, et al: TCR-engineered T cells targeting E7 for patients with metastatic HPV-associated epithelial cancers. Nat Med. 2021;27:419-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong DS, Van Tine BA, Biswas S, McAlpine C, Johnson ML, Olszanski AJ, Clarke JM, Araujo D, Blumenschein GR, Jr., Kebriaei P, et al: Autologous T cell therapy for MAGE-A4+ solid cancers in HLA-A*02+ patients: a phase 1 trial. Nat Med. 2023;29:10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leidner R, Sanjuan Silva N, Huang H, Sprott D, Zheng C, Shih YP, Leung A, Payne R, Sutcliffe K, Cramer J, et al: Neoantigen T-Cell Receptor Gene Therapy in Pancreatic Cancer. N Engl J Med. 2022;386:2112-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi J, Goulding SP, Conn BP, McGann CD, Dietze JL, Kohler J, Lenkala D, Boudot A, Rothenberg DA, Turcott PJ, et al: Systematic discovery and validation of T cell targets directed against oncogenic KRAS mutations. Cell Rep Meth. 2021;1:100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bear AS, Blanchard T, Cesare J, Ford MJ, Richman LP, Xu C, Baroja ML, McCuaig S, Costeas C, Gabunia K, et al: Biochemical and functional characterization of mutant KRAS epitopes validates this oncoprotein for immunological targeting. Nat Commun. 2021;12:4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin N, Paria BC, Vale NR, Yossef R, Lowery FJ, Parkhurst MR, Yu Z, Florentin M, Cafri G, Gartner JJ, et al: Identification and Validation of T-cell Receptors Targeting RAS Hotspot Mutations in Human Cancers for Use in Cell-based Immunotherapy. Clin Cancer Res. 2021;27:5084–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poole A, Karuppiah V, Hartt A, Haidar JN, Moureau S, Dobrzycki T, Hayes C, Rowley C, Dias J, Harper S, et al: Therapeutic high affinity T cell receptor targeting a KRASG12D cancer neoantigen. Nat Commun. 2022;13:5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu D, Chen Y, Jiang M, Wang J, Li Y, Ma K, Sun W, Zheng X, Qi J, Jin W, et al: KRAS G12V neoantigen specific T cell receptor for adoptive T cell therapy against tumors. Nat Commun. 2023;14:6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douglass J, Hsiue EH, Mog BJ, Hwang MS, DiNapoli SR, Pearlman AH, Miller MS, Wright KM, Azurmendi PA, Wang Q, et al: Bispecific antibodies targeting mutant RAS neoantigens. Sci Immunol. 2021;6. [DOI] [PMC free article] [PubMed]
- 17.Klebanoff CA, Chandran SS, Baker BM, Quezada SA, Ribas A: T cell receptor therapeutics: immunological targeting of the intracellular cancer proteome. Nat Rev Drug Discov. 2023;22:996-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cameron BJ, Gerry AB, Dukes J, Harper JV, Kannan V, Bianchi FC, Grand F, Brewer JE, Gupta M, Plesa G, et al: Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci Transl Med. 2013;5:197ra103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, Ozkan E, Davis MM, Wucherpfennig KW, Garcia KC: Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157:1073-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X, Garner LI, Zvyagin IV, Paley MA, Komech EA, Jude KM, Zhao X, Fernandes RA, Hassman LM, Paley GL, et al: Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides. Nature. 2022;612:771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudolph MG, Stanfield RL, Wilson IA: How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–66. [DOI] [PubMed] [Google Scholar]
- 22.Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, Ji X, Han A, Krams SM, Pettus C, et al: Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547:94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riley TP, Hellman LM, Gee MH, Mendoza JL, Alonso JA, Foley KC, Nishimura MI, Vander Kooi CW, Garcia KC, Baker BM: T cell receptor cross-reactivity expanded by dramatic peptide-MHC adaptability. Nat Chem Biol 2018;14:934–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin Y, Mariuzza RA: The multiple mechanisms of T cell receptor cross-reactivity. Immunity 2009;31:849–851. [DOI] [PubMed] [Google Scholar]
- 25.Foldvari Z, Knetter C, Yang W, Gjerdingen TJ, Bollineni RC, Tran TT, Lund-Johansen F, Kolstad A, Drousch K, Klopfleisch R, et al: A systematic safety pipeline for selection of T-cell receptors to enter clinical use. NPJ Vaccines. 2023;8:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishii K, Davies JS, Sinkoe AL, Nguyen KA, Norberg SM, McIntosh CP, Kadakia T, Serna C, Rae Z, Kelly MC, Hinrichs CS: Multi-tiered approach to detect autoimmune cross-reactivity of therapeutic T cell receptors. Sci Adv. 2023;9:eadg9845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H, Wang C, Rubelt F, Scriba TJ, Davis MM: Analyzing the Mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening. Nat Biotechnol. 2020;38:1194–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiou SH, Tseng D, Reuben A, Mallajosyula V, Molina IS, Conley S, Wilhelmy J, McSween AM, Yang X, Nishimiya D, et al: Global analysis of shared T cell specificities in human non-small cell lung cancer enables HLA inference and antigen discovery. . Immunity 2021;54:586-602 e588. [DOI] [PMC free article] [PubMed]
- 29.Zhao X, Kolawole EM, Chan W, Feng Y, Yang X, Gee MH, Jude KM, Sibener LV, Fordyce PM, Germain RN, et al: Tuning T cell receptor sensitivity through catch bond engineering. Science. 2022;376:eabl5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vazquez-Lombardi R, Jung JS, Schlatter FS, Mei A, Mantuano NR, Bieberich F, Hong KL, Kucharczyk J, Kapetanovic E, Aznauryan E, et al: High-throughput T cell receptor engineering by functional screening identifies candidates with enhanced potency and specificity. Immunity. 2022;55:1953-1966 e1910. [DOI] [PubMed] [Google Scholar]
- 31.Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA: Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res 2006;66:8878-8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stadtmauer EA, Fraietta JA, Davis MM, Cohen AD, Weber KL, Lancaster E, Mangan PA, Kulikovskaya I, Gupta M, Chen F, et al: CRISPR-engineered T cells in patients with refractory cancer. Science 2020;367. [DOI] [PMC free article] [PubMed]
- 33.Schmidt R, Steinhart Z, Layeghi M, Freimer JW, Bueno R, Nguyen VQ, Blaeschke F, Ye CJ, Marson A: CRISPR activation and interference screens decode stimulation responses in primary human T cells. Science. 2022;375:eabj4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt R, Ward CC, Dajani R, Armour-Garb Z, Ota M, Allain V, Hernandez R, Layeghi M, Xing G, Goudy L, et al: Base-editing mutagenesis maps alleles to tune human T cell functions. Nature. 2024;625:805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller TR, Jarosch S, Hammel M, Leube J, Grassmann S, Bernard B, Effenberger M, Andra I, Chaudhry MZ, Kauferle T, et al: Targeted T cell receptor gene editing provides predictable T cell product function for immunotherapy. Cell Rep Med. 2021;2:100374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfl M, Greenberg PD: Antigen-specific activation and cytokine-facilitated expansion of naive, human CD8+ T cells. Nat Protoc. 2014;9:950-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi E, Mizukoshi E, Kishi H, Ozawa T, Hamana H, Nagai T, Nakagawa H, Jin A, Kaneko S, Muraguchi A: A new cloning and expression system yields and validates TCRs from blood lymphocytes of patients with cancer within 10 days. Nat Med. 2013;19:1542-1546. [DOI] [PubMed] [Google Scholar]
- 38.Hamana H, Shitaoka K, Kishi H, Ozawa T, Muraguchi A: A novel, rapid and efficient method of cloning functional antigen-specific T-cell receptors from single human and mouse T-cells. Biochem Biophys Res Commun 2016;474:709-714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.