Abstract
SPT5 exhibits versatile functions in RNA Pol II promoter proximal pausing, pause release, and elongation in metazoans. However, the mechanism underlying the functional switch of SPT5 during early elongation has not been fully understood. Here, we report that the phosphorylation site-rich domain (PRD)/CTR1 and the prion-like domain (PLD)/CTR2, which are situated adjacent to each other within the C-terminal repeat (CTR) in SPT5, play pivotal roles in Pol II pausing and elongation, respectively. Our study demonstrates that LEDGF/p75 is highly enriched at promoters, especially paused promoters, and prevents the phosphorylation of SPT5 PRD by the super elongation complex (SEC). Furthermore, deletion of LEDGF IBD leads to increased SEC occupancies and SPT5 PRD phosphorylation at promoters and also increased pause release. In sum, our study reveals that LEDGF and SEC function cooperatively on SPT5 distinct domains to ensure proper transcriptional transition from pausing to elongation.
LEDGF and SEC facilitate the functional transition of SPT5 by orchestrating the transcriptional switch from pause to elongation.
INTRODUCTION
RNA polymerase II (Pol II) pauses downstream of the transcription start site at most eukaryotic genes. Promoter-proximal pausing is a key rate-limiting step, which steers Pol II to adopt one of the two states: premature termination or productive elongation (1–4). Pausing deregulation is associated with a broad range of human diseases (5). In vitro transcription or cell-based assays revealed that promoter-proximal pausing and its transition to productive elongation are orchestrated by numerous factors (2, 3). DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF) were demonstrated to stabilize and maintain the paused state (6–8). Accompanied by the phosphorylation of DSIF, NELF, and Pol II C-terminal domains by cyclin-dependent kinase 9 (CDK9) subunit of positive transcription elongation factor b (P-TEFb), NELF dissociates, but DSIF remains associated with Pol II, and the transcription elongation complex is formed to trigger release of Pol II from pausing into productive elongation (8–11). Among them, DSIF is unique with a binary function, not only maintaining promoter-proximal pausing but also promoting transcription elongation (6).
Suppressor of Ty5 homolog (SPT5) was originally identified in S. cerevisiae through a genetic screening for mutations that can restore HIS4 gene expression suppressed by transposon insertion (12). Subsequent studies revealed that SPT5 forms the DSIF complex together with SPT4 to regulate transcription elongation (13). SPT5, the large subunit of DSIF, is evolutionarily conserved from bacteria to humans (14, 15). A NusG N-terminal domain (NGN) domain, mediating the interaction with SPT4, and a following Kyrpides Ouzounis Woese (KOW) domain, designated as KOW1, are common to all the SPT5 orthologous, endowing SPT5 with activity to promote Pol II elongation (6, 14).
To adapt to the high complexity of transcriptional regulation in higher eukaryotes, such as promoter-proximal pausing and release, SPT5, which originated in bacteria and archaea, has evolved to contain more copies of the KOW domain and also includes dual C-terminal repeat (CTR) regions and an N-terminal acidic region (6, 14, 15). Truncation analysis indicated that a region encompassing KOWx-4 and KOW5 could be important for Pol II pausing at the Hsp70 gene in in vitro transcription assay (16). Structural studies revealed that KOWx-4 and KOW5 domains are structurally conserved in yeast and human, making extensive contacts with the rim of the RNA exit tunnel of Pol II (17, 18). Nevertheless, typical promoter-proximal pausing is largely absent in yeast.
In addition, the CTR regions and the flexible linker between KOWx-4 and KOW5 in SPT5 bear phosphorylation hotspots (19, 20). Phosphorylation by P-TEFb appears to be a major regulatory mechanism of SPT5 function. The phosphorylation of the threonine-4 (T4) residues on CTR1 has been demonstrated to be required for its function as a Pol II processivity stimulating factor, and the phosphorylation of S666 located within the KOWx-4/5 linker could potentially promote pause release (10, 11, 20, 21). The most active form of P-TEFb lies in the super elongation complex (SEC) (22). We have previously shown that SEC is able to induce phase transition of SPT5 from the pausing condensates into the elongation droplets (23). However, the mechanism underlying how the function of SPT5 switches to coordinate transcription fate transition from pausing to elongation remains largely elusive.
Lens epithelium–derived growth factor (LEDGF)/p75, also known as PC4- and SF2-interacting protein 1, is a chromatin associated protein having a methyl-lysine reading Pro-Trp-Trp-Pro (PWWP) domain and an integrase binding domain (IBD) (24–26). Relying on protein interactions through PWWP and IBD, LEDGF plays important roles in transcription regulation and DNA repair, and its aberrations are associated with various human diseases, including HIV/AIDS, cancer, and autoimmunity (27). Here, we demonstrated that the phosphorylation site-rich domain (PRD)/CTR1 and the prion-like domain (PLD)/CTR2 adjacent in SPT5-CTR are required for Pol II pausing and elongation, respectively. Mechanistically, IBD in LEDGF co-condensates with PLD in SPT5 and prevents the phosphorylation of PRD by SEC, thus leading to promoter-proximal pausing. Our study highlights the pivotal role of LEDGF and SEC in mediating the functional switch of SPT5, thereby coordinating transcriptional transition from pausing to elongation.
RESULTS
Adjacent PRD and PLD in SPT5 CTR exhibit different phase separation properties
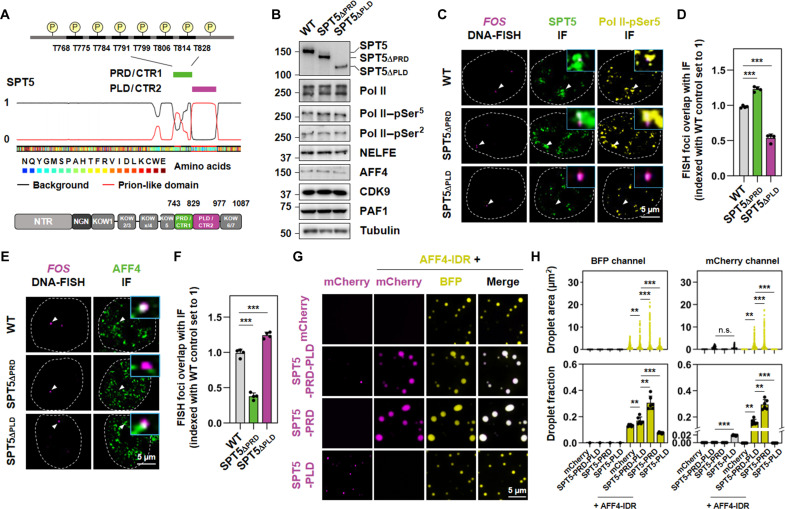
Previous studies have established that SPT5 has a dual function in transcription, serving as both a pause factor that impedes the release of promoter-proximally paused Pol II and an elongation factor that promotes Pol II processivity (3, 6). Nevertheless, the mechanism underlying the regulation of Pol II pausing and elongation by SPT5 has not been fully understood. Evolutionary analysis showed that, in addition to the structured domains NGN and KOW, CTR within SPT5 is also conserved across metazoans (fig. S1, A and B). Amino acid sequence analysis revealed that CTR1 within SPT5-CTR is a PRD, while CTR2 constitutes a PLD (Fig. 1A).
Fig. 1. Adjacent PRD and PLD in SPT5 CTR exhibit different phase separation properties.
(A) Color-coded schematic representation of the amino acid sequence and corresponding prion-like domain disorder propensity plots (red and black traces) for SPT5, generated using PLAAC (prion-like amino acid composition) (56). Y axis of the plot representing prion-like regions (1) and regions of background amino acid composition (0). Posttranslational modification sites in SPT5-PRD. The predicted residuals phosphorylated by P-TEFb are indicated above. Domain organization of SPT5 is indicated below. (B) Western blot analysis of SPT5, Pol II, Pol II–pSer5, Pol II–pSer2, NELFE, AFF4, CDK9, PAF1 in wild-type (WT), SPT5ΔPRD, and SPT5∆PLD HCT 116 cells. α-Tubulin was used as a loading control. (C) Confocal imaging of FOS DNA fluorescence in situ hybridization (FISH) with concurrent SPT5 and Pol II–pSer5 IF in WT, SPT5∆PRD, and SPT5∆PLD HCT 116 cells after serum starvation. Zoomed-in views of the merged regions are indicated by white arrows. (D) Fraction of overlap of FISH foci with IF puncta in (C). Two-tailed, unpaired Student’s t test was performed. ***P < 0.001. (E) Confocal imaging of FOS DNA FISH with concurrent AFF4 IF in WT, SPT5∆PRD, and SPT5∆PLD HCT 116 cells after serum stimulation. Zoomed-in views of the merged regions are indicated by white arrows. (F) Fraction of overlap of FISH foci with IF puncta in (E). Two-tailed, unpaired Student’s t test was performed. ***P < 0.001. (G) Fluorescence microscopy images showing individual- and co-phase–separated droplets formed in a buffer containing 150 mM NaCl and 2 μM BFP-AFF4-IDR, mCherry-SPT5-PRD-PLD, mCherry-SPT5-PRD, or mCherry-SPT5-PLD with 10% PEG-8000. (H) Dot plot (top) or bar plot (bottom) showing the droplet area or fraction in (G). Fields per condition n = 5. Two-tailed, unpaired Student’s t test was performed. **P < 0.01 and ***P < 0.001. n.s., not significant.
We have recently found that the intrinsic C-terminal disordered regions within SPT5 are essential for the formation of phase-separated transcription clusters and its role in gene activation (23). To investigate the direct impact of PRD and PLD in SPT5 on the transcription cluster formation, we used CRISPR-Cas9–mediated genomic editing to remove PRD and PLD from SPT5 endogenously, generating SPT5∆PRD and SPT5∆PLD cell lines, respectively (Fig. 1B and fig. S1C). We found that removing PRD or PLD from SPT5 did not notably alter the protein levels of Pol II, as well as its initiating/paused (Pol II–pSer5) and elongating (Pol II–pSer2) forms (Fig. 1B). Co-immunostaining of SPT5 together with Pol II–pSer5, negative elongation factor E (NELFE), and AF4/FMR2 family member 4 (AFF4) revealed that deletion of PLD, but not PRD, compromised formation of the pausing clusters represented by SPT5-NELFE or SPT5–Pol II–pSer5 heterotypic condensates but augmented the formation of elongation condensates represented by SPT5-AFF4 heterotypic condensates (fig. S2, A to F). These findings suggest that PLD prevents SPT5 from transitioning from a paused Pol II cluster to an elongation droplet, whereas PRD exerts the opposite effect. Furthermore, combined DNA fluorescence in situ hybridization (FISH) and immunofluorescence (IF) analyses further confirmed that the levels of the pausing clusters were substantially reduced at the FOS gene in SPT5∆PLD cells but increased in SPT5∆PRD cells after serum starvation (Fig. 1, C and D). In SPT5∆PLD cells, the levels of AFF4 condensates were increased at the FOS locus upon rapid activation of the FOS gene by serum stimulation, whereas, in SPT5∆PRD cells, the levels of AFF4 condensates were decreased (Fig. 1, E and F).
Moreover, compared to wild-type (WT) SPT5 and SPT5∆PRD, the SPT5∆PLD containing pausing clusters were more sensitive to 1,6-hexanediol treatment (fig. S2, G and H). In vitro analysis also indicated that, compared to SPT5-PRD, SPT5-PLD was able to readily undergo phase separation possibly due to its prion-like property (Fig. 1, G and H, and fig. S3A) (28). Thus, PLD in SPT5 might be essential for maintaining the biophysical properties of the pausing clusters. SPT5 can be relocated from the paused clusters to the elongation droplets by SEC via the IDR of the SEC scaffold AFF4 (23, 29). Consistently, SPT5-PRD and SPT5-PRD-PLD were capable of forming the heterotypic condensates with AFF4-IDR, while the SPT5-PLD condensates were dissolved in the presence of AFF4-IDR (Fig. 1, G and H, and fig. S3A).
Adjacent PLD and PRD in SPT5 CTR are required for Pol II pausing and elongation, respectively
We then wondered how deletion of PRD or PLD in SPT5 affects transcription. To address this question, we first investigated the impact of removing PRD or PLD from SPT5 on the synthesis of nascent RNA using the 5-ethynyl uridine (EU) incorporation assay. Our results revealed that SPT5∆PRD cells displayed a global reduction in nascent RNA synthesis, whereas SPT5∆PLD cells exhibited an increase in comparison to WT cells (fig. S3, B and C).
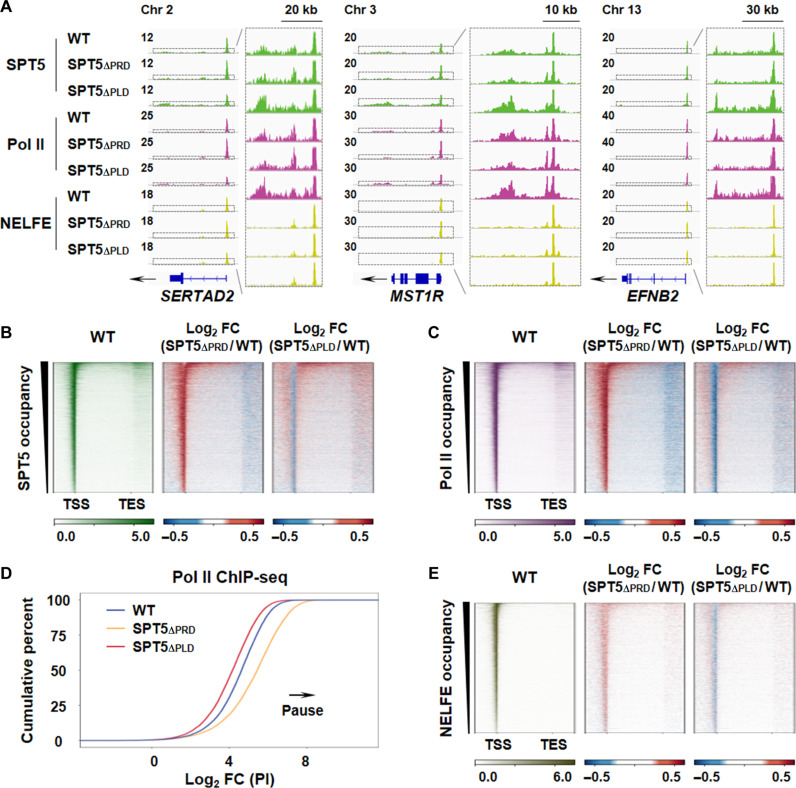
SPT5 chromatin immunoprecipitation sequencing (ChIP-seq) analysis revealed that deletion of either PRD or PLD from SPT5 altered the binding profile of SPT5 across the transcribed regions. PRD removal resulted in a notable increase in SPT5 occupancy at the promoter-proximal regions but a decrease at the gene bodies (Fig. 2, A and B). Conversely, PLD deletion reduced SPT5 occupancy at the promoter-proximal regions but increased occupancy at the gene bodies (Fig. 2, A and B). To investigate the roles of PRD and PLD in SPT5 in transcription regulation, we further examined Pol II occupancy change in SPT5∆PRD and SPT5∆PLD cells. Similar to the observations for SPT5 occupancy, Pol II occupancy at the promoter-proximal regions was elevated in SPT5∆PRD cells but reduced in SPT5∆PLD cells, while occupancy at the gene bodies was decreased in SPT5∆PRD cells but increased in SPT5∆PLD cells (Fig. 2, A and C). Accordingly, an increase and decrease in the pausing index, signifying the ratio of Pol II occupancy in the proximal promoter region to that in the gene body, were observed in SPT5∆PRD and SPT5∆PLD cells, respectively (Fig. 2D and fig. S3D).
Fig. 2. Adjacent PLD and PRD in SPT5 CTR are required for Pol II pausing and elongation, respectively.
(A) Representative gene examples of SPT5, Pol II, and NELFE chromatin immunoprecipitation sequencing (ChIP-seq) in WT, SPT5∆PRD, and SPT5∆PLD HCT 116 cells. Y axis representing reads per million (RPM). (B) Heatmaps of SPT5 occupancy showing RPM and log2 fold change (FC) on scaled genes ranked by decreasing occupancy in WT, SPT5∆PRD, and SPT5∆PLD HCT 116 cells. n = 6732 genes. (C) Heatmaps of Pol II occupancy showing RPM and log2 FC on scaled genes ranked by decreasing occupancy in WT, SPT5∆PRD, and SPT5∆PLD HCT 116 cells. n = 6732 genes. (D) Empirical cumulative distribution function plots of the Pol II pause index (PI) in WT, SPT5∆PRD, and SPT5∆PLD HCT 116 cells. (E) Heatmaps of NELFE occupancy showing RPM and log2 FC on scaled genes ranked by decreasing occupancy in WT, SPT5∆PRD, and SPT5∆PLD HCT 116 cells. n = 6732 genes.
The destabilization of paused Pol II signals in SPT5∆PLD is reminiscent of the pause-regulating factor NELF, which maintains Pol II pausing (8). We next measured the genomic occupancy of NELFE, a core subunit of NELF, using ChIP-seq, and found a pronounced loss of NELFE at the promoter-proximal regions in SPT5∆PLD cells (Fig. 2, A and E). Conversely, NELFE occupancy was increased in SPT5∆PRD cells (Fig. 2, A and E). Together, these results suggested that PLD in SPT5 might be essential for maintaining paused Pol II, while PRD could be necessary for effective Pol II elongation.
SPT5-PLD promotes transcriptional pausing by inhibiting PRD phosphorylation
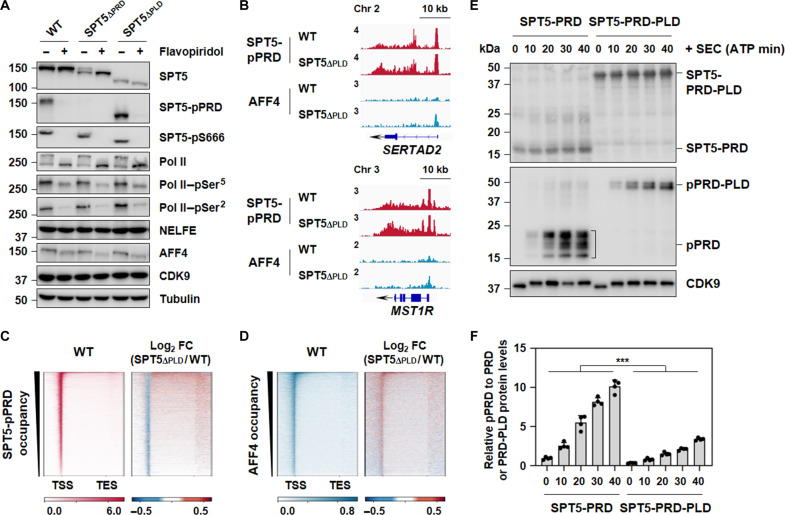
Phosphorylation of SPT5 has been linked to pause release and enhanced transcription elongation (14). We speculated that phosphorylation status of SPT5 might be altered after PLD removal. Phosphorylation of SPT5 at CTR, more specifically at PRD, was substantially enhanced in SPT5∆PLD cells, whereas phosphorylation of SPT5 at S666, located within the KOWx-4/5-linker, remained unchanged after either PLD or PRD removal (Fig. 3A). ChIP-seq analysis further revealed that phosphorylation of SPT5 at PRD was elevated downstream of the promoter regions at the genome-wide scale in SPT5∆PLD cells (Fig. 3, B and C, and fig. S4A). Collectively, these results implied that SPT5-PLD could maintain Pol II pausing by obstructing the phosphorylation of SPT5 at PRD.
Fig. 3. SPT5-PLD maintains pausing by inhibiting PRD phosphorylation.
(A) Western blot analysis of SPT5, SPT5-pPRD, SPT5-pS666, Pol II, Pol II–pSer5, Pol II–pSer2, NELFE, AFF4, and CDK9 in WT, SPT5∆PRD, and SPT5∆PLD HCT 116 cells before or after flavopiridol (FP; 500 nM) exposure for 30 min. α-Tubulin was used as a loading control. (B) Representative gene examples of SPT5-pPRD and AFF4 ChIP-seq in WT and SPT5∆PLD HCT 116 cells. Y axis representing RPM. (C) Heatmaps of SPT5-pPRD occupancy showing RPM and log2 FC on scaled genes ranked by decreasing occupancy in WT and SPT5∆PLD HCT 116 cells. n = 6732 genes. (D) Heatmaps of AFF4 occupancy showing RPM and log2 FC on scaled genes ranked by decreasing occupancy in WT and SPT5∆PLD HCT 116 cells. n = 6732 genes. (E) In vitro kinase assay of SPT5-PRD or SPT5-PRD-PLD in the presence of adenosine 5′-triphosphate and subjected to Western blot analyses with antibodies specific to SPT5-CTR, SPT5-pPRD, and CDK9. (F) Quantification of relative density of SPT5-pPRD compared to that of SPT5-PRD or SPT5-PRD-PLD in (E). n = 4. Two-tailed, unpaired Student’s t test was performed. ***P < 0.001.
The inhibition of P-TEFb kinase activity by flavopiridol (FP) not only prevented SPT5 phosphorylation in WT cells but also abolished the excessive phosphorylation of SPT5 in SPT5∆PLD cells (Fig. 3A). SEC is one of the most active forms of the P-TEFb complexes (22). Western blot analysis showed that treatment with KL-1, a specific inhibitor of SEC, affected the phosphorylation of SPT5-PRD (fig. S4, B to D), indicating that SEC plays a role in SPT5-PRD phosphorylation. In contrast, inhibiting BRD4, another component of the active P-TEFb complex, either through treatment with JQ1, a small-molecule inhibitor of BRD4, or by knocking down BRD4, did not notably change SPT5-PRD phosphorylation levels (fig. S4, E and F). These results suggest that BRD4 may not be essential for this phosphorylation process. To investigate whether SEC is involved in SPT5 hyper-phosphorylation after PLD removal, we performed ChIP-seq of AFF4, the scaffold of SEC, and found that AFF4 occupancy was elevated at the genome-wide level in SPT5∆PLD cells (Fig. 3, B and D, and fig. S4A). In vitro kinase assay further validated that phosphorylation of SPT5 at PRD by purified SEC was compromised in the presence of PLD (Fig. 3, E and F, and fig. S4G), suggesting a direct role of PLD in inhibiting SEC-mediated PRD phosphorylation. Together, our results demonstrated that PLD in SPT5 might prevent genomic recruitment of SEC and thus hyper-phosphorylation of SPT5 at PRD.
LEDGF cooperates with SPT5-PLD in inhibition of SPT5-PRD phosphorylation
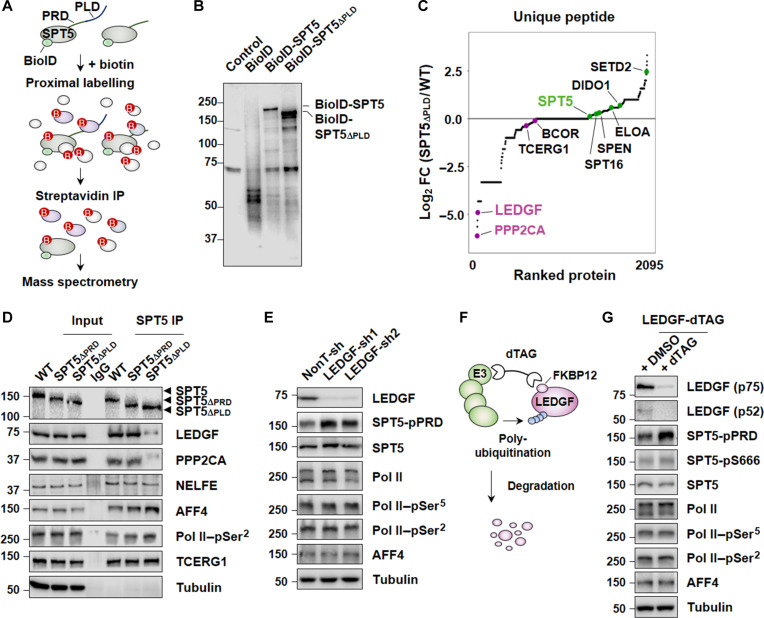
To explore the mechanism underlying the role of PLD in inhibiting SEC-mediated SPT5 phosphorylation at PRD, we sought to identify the proteins that specifically interact with SPT5-PLD by using biotin identification (BioID) (30) proximity labeling proteomics (Fig. 4A and fig. S5A). SPT5 or SPT5∆PLD-associated proteins were purified and subjected to mass spectrometry analyses (Fig. 4B), which revealed transcriptional regulators and chromatin remodelers as top candidates for interaction with SPT5 (Fig. 4C). We found that, compared to the WT SPT5, the absence of PLD led to an increase in the abundance of multiple proteins that associated with Pol II–pSer2, such as spen family transcription repressor (SPEN) (31), elongin A (ELOA) (32), death-inducer obliterator 1 (DIDO1) (31), and SET domain containing 2 (SETD2) (33, 34) (Fig. 4C). This finding aligned with the observation that the absence of PLD from SPT5 promoted pause release.
Fig. 4. LEDGF cooperates with SPT5-PLD in inhibition of SPT5-PRD phosphorylation.
(A) Schematic of the BioID-tagged proximity labeling in FLAG-BioID-SPT5 or FLAG-BioID-SPT5∆PLD cell lines. (B) Western blot analysis of streptavidin pulldown products from cells expressing FLAG-BioID-SPT5 or FLAG-BioID-SPT5∆PLD. (C) Dot plot depicting log2 FC of SPT5 and SPT5∆PLD unique peptides labeled with BioID. (D) Endogenous IPs showing the interaction of WT SPT5, SPT5∆PRD, or SPT5∆PLD with LEDGF, PPP2CA, NELFE, AFF4, Pol II–pSer2, and TCERG1. α-Tubulin was used as a control. (E) Western blot analysis of LEDGF, SPT5-pPRD, SPT5, Pol II, Pol II–pSer5, Pol II–pSer2, and AFF4 in LEDGF knockdown HCT 116 cells. α-Tubulin was used as a loading control. (F) Schematic of the generation of LEDGF-dTAG HCT 116 cells. (G) Western blot analysis of LEDGF, SPT5-pPRD, SPT5-pS666, SPT5, Pol II, Pol II–pSer5, Pol II–pSer2, and AFF4 in LEDGF-dTAG HCT 116 cells treated with 3 hours dTAG or not. α-Tubulin was used as a loading control. DMSO, dimethyl sulfoxide.
In contrast, mass spectrometry analysis suggested that the interaction of SPT5 with the protein phosphatase 2A catalytic subunit α (PPP2CA), LEDGF, and the transcription elongation regulator 1 (TCERG1) could be affected after removal of PLD from SPT5 (Fig. 4C). Further endogenous co-immunoprecipitation experiments showed that, while the interaction between SPT5 and TCERG1 was not affected, the interactions between SPT5 and PPP2CA, as well as SPT5 and LEDGF, were reduced in SPT5∆PLD cells (Fig. 4D and fig. S5B).
In line with previous reports (35), knockdown of the PPP2CA by short hairpin RNA (shRNA) potentiated phosphorylation of SPT5 at PRD (fig. S5C). In addition, we found that knockdown of LEDGF, but not TCERG1, SR-related CTD-associated factor 1 (SCAF1), and BCL6 corepressor (BCOR) identified from the mass spectrometry analysis, also led to an increase in the phosphorylation levels of SPT5 at PRD (Fig. 4E and fig. S5C). To further investigate the role of LEDGF, we generated LEDGF rapid degradation cell lines by using the degradation tag (dTAG) technology (36) to integrate the FLAG-FKBP12F36V tag to the N terminus of LEDGF (Fig. 4F). Immunoprecipitation analysis indicated that the interaction between PPP2CA and SPT5 was reduced upon acute depletion of LEDGF (fig. S5D). Similar to what was observed after PLD removal from SPT5 (Fig. 3A and fig. S3, B and C), the rapid depletion of LEDGF led to a profound increase in the phosphorylation levels of SPT5 at PRD, along with a notable increase in nascent RNA synthesis (Fig. 4G, and fig. S6, A and B). Together, our results indicated that LEDGF could interact with PLD in SPT5 and inhibit PRD phosphorylation.
Rapid degradation of LEDGF leads to increased initiating and elongating Pol II
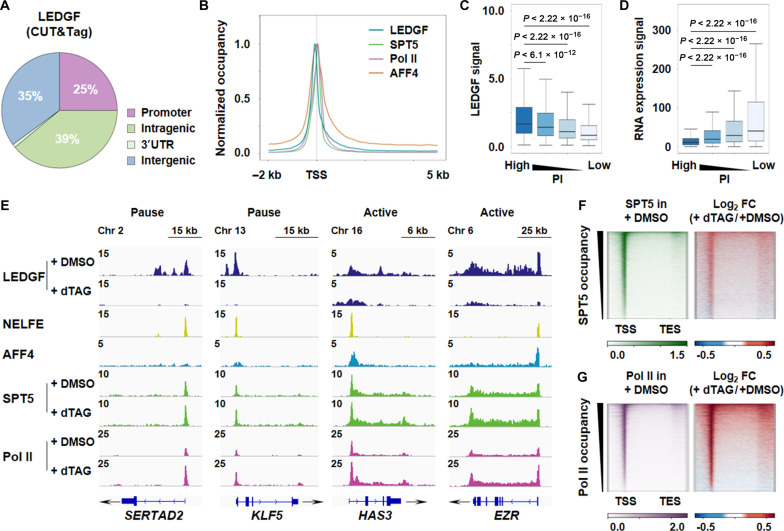
To explore the potential role of LEDGF in transcriptional pause release, we first conducted LEDGF genomic occupancy analysis in human colon cancer cell line 116 (HCT 116) cells using cleavage under targets and tagmentation (CUT&Tag) sequencing. A total of 43,687 highly confident LEDGF peaks (P < 10−7) were identified, with a large portion of sharp peaks located at promoter and intergenic regions, while broad peaks were mostly observed at transcribing units of active genes. Approximately 64% of LEDGF peaks were predominantly detected in genic regions, encompassing both the promoters and intragenic regions (Fig. 5, A and B). Promoters of paused genes, exemplified by SERTAD2, KLF5, UGCG, and LDLR, tended to have stronger LEDGF enrichment compared to active genes, including HAS3, EZR, SRSF1, and GAPDH (Fig. 5, C to E). The specificity of the LEDGF CUT&Tag signals was further confirmed using an alternative LEDGF antibody, and the signals were also validated after rapid LEDGF degradation (Fig. 5E and fig. S6, C to F). Next, we examined the genomic occupancies of SPT5 and Pol II following LEDGF rapid degradation. Notable increases in their enrichment were observed at both the promoters and transcribing units (Fig. 5, E to G, and fig. S6F). For instance, after LEDGF rapid degradation, HAS3 and EZR exhibited increases in the levels of initiating and elongating Pol II, as well as SPT5; SERTAD2 and KLF5 displayed similar trends, albeit with a somewhat lesser increase across the gene bodies (Fig. 5E and fig. S6F).
Fig. 5. Rapid degradation of LEDGF leads to increased initiating and elongating Pol II.
(A) The genomic distribution of LEDGF in HCT 116 cells. 3′UTR, 3′ untranslated region. (B) Metaplot showing LEDGF, SPT5, Pol II, and AFF4 average occupancy across genes in HCT 116 cells. (C) Analysis of LEDGF CUT&Tag signals at promoter sites of highly paused, low paused, low active, and highly active genes in HCT 116 cells. (D) RNA sequencing signals of highly paused, low paused, low active, and highly active genes in HCT 116 cells. (E) Representative gene examples of NELFE, AFF4 ChIP-seq in HCT 116 cells, and SPT5, Pol II ChIP-seq, or LEDGF CUT&Tag in LEDGF-dTAG HCT 116 cells treated with dTAG for 3 hours or not. Y axis representing RPM. (F) Heatmaps of SPT5 occupancy showing RPM and log2 FC on scaled genes ranked by decreasing occupancy in LEDGF-dTAG HCT 116 cells treated with 3 hours dTAG or not. n = 6732 genes. (G) Heatmaps of Pol II occupancy showing RPM and log2 FC on scaled genes ranked by decreasing occupancy in LEDGF-dTAG HCT 116 cells treated with 3 hours dTAG or not. n = 6732 genes.
LEDGF/p75 interacts with SPT5 PLD through its IBD
The LEDGF gene encodes two isoforms, p52 and p75. Both isoforms have a common N-terminal PWWP domain, which specifically binds to H3K36me3, while the C-terminal IBD is only present in the long isoform p75 (24–26). The mass spectrometry data analysis suggested the presence of the LEDGF-IBD peptides in the SPT5 BioID proximity labeling products (fig. S5E). To investigate the specific region within LEDGF responsible for inhibiting SPT5-PRD phosphorylation, we performed immunoprecipitation experiments using WT LEDGF and mutant LEDGF lacking either the PWWP domain or IBD and found that IBD in LEDGF mediated its interaction with SPT5 (Fig. 6, A and B).
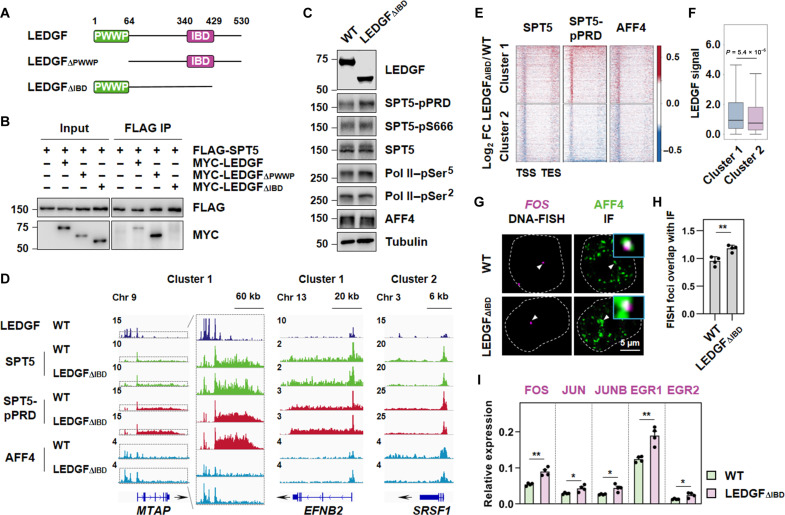
Fig. 6. LEDGF/p75 interacts with SPT5 PLD through its IBD and promotes transcriptional pausing at large subset of genes.
(A) Domain organization of LEDGF. (B) FLAG IPs showing the interaction of FLAG-SPT5 with MYC-LEDGF, MYC-LEDGFΔPWWP, and MYC-LEDGFΔIBD. (C) Western blot analysis of LEDGF, SPT5-pPRD, SPT5-pS666, SPT5, Pol II–pSer5, Pol II–pSer2, and AFF4 in WT and LEDGFΔIBD HCT 116 cells. α-Tubulin was used as a loading control. (D) Representative gene examples of LEDGF, SPT5, SPT5-pPRD, and AFF4 occupancies on the Cluster 1 or 2 genes in WT and LEDGFΔIBD HCT 116 cells. Y axis representing RPM. (E) Heatmaps showing log2 FC of SPT5, SPT5-pPRD, and AFF4 occupancy on scaled genes, ranked by decreasing occupancy distributed in two clusters (cluster 1 or 2: top 1000 genes) within both WT and LEDGFΔIBD HCT 116 cells. (F) LEDGF CUT&Tag signals of the two cluster genes at promoter sites in HCT 116 cells. (G) Confocal imaging of FOS DNA FISH with concurrent AFF4 IF in WT and LEDGFΔIBD HCT 116 cells after serum stimulation. Zoomed-in views of the merged regions are indicated by white arrows. (H) Fraction of overlap of FISH foci with IF puncta in (G). Two-tailed, unpaired Student’s t test was performed. **P < 0.01. (I) Reverse transcription quantitative PCR analysis showing RNA levels of FOS, JUN, JUNB, EGR1, and EGR2 in WT and LEDGFΔIBD HCT 116 cells after serum stimulation. Two-tailed, unpaired Student’s t test was performed. *P < 0.05 and **P < 0.01.
LEDGF/p75 promotes transcriptional pausing at large subset of genes
To investigate the impact of removing IBD from LEDGF/p75 on SPT5-PRD phosphorylation at the genome-wide level, we deleted IBD from LEDGF/p75 and generated LEDGF∆IBD cell line (fig. S7A). Immunoprecipitation assay confirmed that lack of IBD abolished the interaction between LEDGF/p75 and SPT5 (fig. S7B). Loss of IBD in LEDGF/p75 led to a profound increase in phosphorylation levels of SPT5 at PRD and also a substantial increase in nascent RNA synthesis (Fig. 6C and fig. S7, C and D).
In addition, genomic occupancy analysis revealed that phosphorylated SPT5 was notably increased in a large subset of genes (cluster 1), which preferentially enriched with high LEDGF levels at their promoters (Fig. 6, D to F, and fig. S7E). Low LEDGF-bound cluster 2 genes showed unchanged or even a slight decrease in SPT5 phosphorylation levels. Consistently, we also observed a specific increase of AFF4 occupancies at cluster 1 genes, but not Cluster 2, in LEDGF∆IBD cells (Fig. 6, D and E, and fig. S7E). Combined DNA FISH and IF analyses further demonstrated that the AFF4 condensates were substantially increased at the FOS locus in LEDGF∆IBD cells upon serum induction (Fig. 6, G and H). Consistently, the expression of FOS gene and other serum-inducible genes was notably up-regulated in LEDGF∆IBD cells upon serum treatment (Fig. 6I). Thus, our results strongly indicated that LEDGF could promote transcriptional pausing at large subset of genes through its IBD.
LEDGF-IBD prevents SPT5-PRD phosphorylation by SEC
To further investigate whether and how LEDGF-IBD is able to directly inhibit SEC-mediated phosphorylation of SPT5-PRD, we performed in vitro kinase assay using purified LEDGF-IBD (fig. S8A). The results indicated that phosphorylation of PRD-PLD by SEC was notably suppressed in the presence of LEDGF-IBD in vitro (Fig. 7A and fig. S8B). It has been reported that IBD-interacting domain, or IBD-binding motifs (IBMs), contains an FxGF motif on known LEDGF/p75-binding partners (37). The interaction of IBD-IBMs can be greatly enhanced by phosphorylation of IBMs (38). Sequence analysis of SPT5 PLD indicated that it may contain a degenerated version of IBM (fig. S8C). Further in vitro glutathione S-transferase (GST) pull-down assay also suggested weak direct interaction between LEDGF-IBD and SPT5-PLD or phosphorylation mimic PLD (fig. S8D).
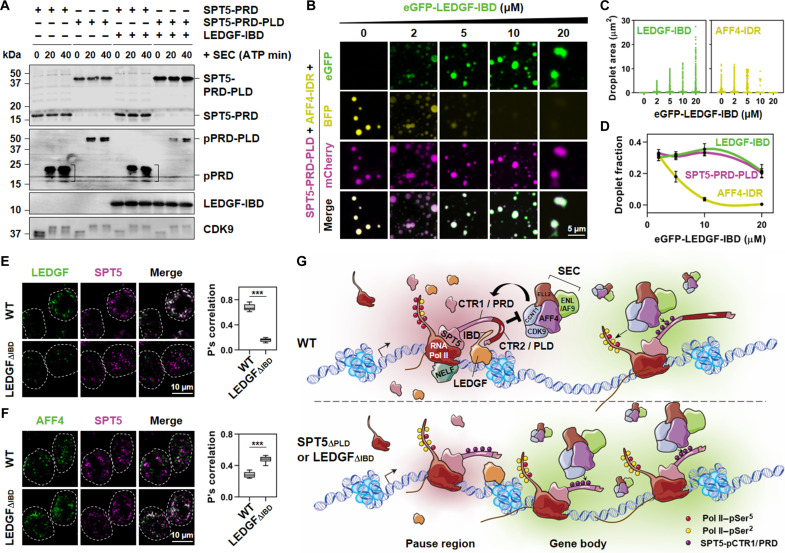
Fig. 7. LEDGF-IBD prevents SPT5-PRD phosphorylation by SEC.
(A) In vitro kinase assay of SPT5-PRD or SPT5-PRD-PLD, with or without LEDGF-IBD. (B) Fluorescence microscopy images showing co-phase–separated droplets formed in the presence of 10% PEG-8000 with 150 mM NaCl containing buffer and 2 μM mCherry-SPT5-PRD-PLD and BFP-AFF4-IDR along with increase concentration of eGFP-LEDGF-IBD (0, 2, 5, 10, and 20 μM). (C) Dot plot showing the droplet area in (B). Fields per condition n = 5. (D) Line plot showing the droplet fraction in (B). Error bars represent SDs. Fields per condition n = 5. (E) Confocal images (left) showing co-localization of SPT5 with LEDGF in nuclear puncta in WT and LEDGFΔIBD HCT 116 cells. Box plots (right) showing the mean values of the Pearson correlation coefficient of co-localization ratios of SPT5 with LEDGF. (F) Same as (E), but for SPT5 and AFF4. Results are representative of three biological replicates, each n > 20. Two-tailed, unpaired Student’s t test was performed. ***P < 0.001. (G) Models illustrating regulation of Pol II pausing and release by the interplay between SPT5 and LEDGF. See Discussion for detailed description.
Given the prion-like (28) nature of SPT5-PLD, characterized by ~10 repeats of an eight–amino acid sequence P-[T/S]-P-S-P-[Q/A]-[S/G]-Y, it can form condensates with other proteins via multivalent weak interactions. We found that LEDGF-IBD was able to phase separate and form heterotypic condensates with SPT5-PLD, but not with PRD (figs. S8E and S9, A and B). In vitro condensate formation assays also revealed that, when purified LEDGF-IBD and AFF4-IDR (the key domain for SEC condensate formation) (29) were incubated with either SPT5-PRD or SPT5-PLD, LEDGF-IBD only formed condensates with SPT5-PLD, while SPT5-PRD was excluded (figs. S8E and S9, A and B). Vice versa, AFF4-IDR only formed condensates with SPT5-PRD, and SPT5-PLD was excluded. We noticed that, while SPT5-PRD-PLD remained co-localized with AFF4-IDR in the presence of LEDGF-IBD, the condensate shape underwent changes (figs. S8E and S9, A and B). We thus further conducted dose-dependent experiments and observed that as the concentration of LEDGF-IBD increased, AFF4-IDR was progressively excluded from SPT5-PRD-PLD droplets (Fig. 7, B to D). This finding provides support that LEDGF-IBD might inhibit the association of AFF4 with SPT5 droplets. In addition, IF analyses demonstrated that in the absence of the IBD domain, the co-localization between SPT5 and LEDGF was attenuated, whereas the co-localization between SPT5 and AFF4 was enhanced (Fig. 7, E and F, and fig. S9C). These findings suggest that LEDGF-IBD may play a role in preventing SPT5 from co-localizing with SEC. Collectively, our data suggested that LEDGF-IBD might impede the binding of SEC to SPT5 via multivalent weak interaction-mediated phase separation, thereby preventing hyper-phosphorylation of SPT5.
DISCUSSION
The transcription regulator SPT5 plays a key role in both promoter proximal pausing and transcription elongation in metazoans (6, 14). Here, we demonstrate that PRD and PLD, the two adjacent domains within SPT5-CTR, are crucial for Pol II pausing and elongation, respectively. Mechanistically, IBD in LEDGF forms condensates with SPT5-PLD and prevents SEC-mediated phosphorylation of SPT5-PRD, leading to promoter-proximal pausing (Fig. 7G). Our study highlights that LEDGF and SEC act on PLD and PRD in SPT5 to coordinate transcriptional transition from pausing to elongation, a finding that notably expands our understanding of the complex regulatory network controlling gene expression.
Promoter proximal pausing plays a pivotal role in transcriptional regulation, ensuring the appropriate expression of genes (2, 3). This process is tightly controlled by a range of regulatory factors, including the well-established NELF and SPT5 (8). A recent study suggested that LEDGF binding at the promoter can help prevent the formation of R-loops (39). In our current study, we have identified LEDGF as a previously unknown factor involved in the regulation of promoter proximal pausing for a large subset of genes. However, the levels of SPT5 phosphorylation and AFF4 seem not markedly affected in another subset of genes, suggesting additional LEDGF-independent regulatory mechanism there exists. It has been shown that IBD in LEDGF is homologous to the structurally conserved transcription elongation factor SII (TFIIS) N-terminal domain, which specifically interacts with the intrinsically unstructured motifs of other transcription regulators (37). Earlier studies have established that LEDGF supports RNA Pol II in overcoming the nucleosome barrier during transcription elongation (40, 41). It is conceivable that LEDGF acts as a central hub to facilitate the transition from pausing to elongation by switching its IBD-interacting transcription regulatory partners.
The phosphorylation status of SPT5 plays a crucial role in determining the fate of the paused Pol II, affecting whether the paused Pol II can be released into the elongation stage or undergoes premature termination mediated by the Integrator-PP2A complex (INTAC) (35, 42–45). Our data indicate that SPT5, in addition to interacting with LEDGF, can also engage with the INTAC phosphatase PP2A (protein phosphatase 2A) catalytic subunit PPP2CA through its PLD domain. Notably, depletion of LEDGF results in reduced interaction between SPT5 and PPP2CA. Consequently, it is plausible to hypothesize that the disrupted interaction between SPT5 and PPP2CA in SPT5∆PLD and LEDGF depleted cells may hinder the dephosphorylation of SPT5-PRD, leading to an increase in its phosphorylation levels. However, it is important to acknowledge that PP2A has been observed to dephosphorylate not only SPT5-PRD but also S666 (20, 35). Notably, the removal of PLD had no impact on S666 phosphorylation, indicating that the specific regulation of PRD phosphorylation is unlikely to be solely attributed to PPP2CA. Therefore, further research is needed to elucidate how these factors collectively determine whether transcription progresses to release or terminates prematurely.
SPT5 PLD also contains multiple SP or TP consensus sequences that are likely to be phosphorylation sites. Previous in vitro kinase assay indicated that CTR2/PLD in SPT5 could be phosphorylated by CDK7 (46). In our Western blot analysis, SPT5 exhibits two distinct bands. The fainter upper band, which disappears after PLD removal or FP treatment (Fig. 3A), is plausible that the upper band represents the phosphorylated form of SPT5 at PLD. Here, we found that PLD inhibits the phosphorylation of PRD in SPT5 by SEC. Thus, it would be worthwhile to investigate in the future whether this intrinsic inhibition is achieved through competition between PLD and PRD for SEC or CDK7, and the potential impact of PLD phosphorylation on transcriptional pausing.
MATERIALS AND METHODS
Cell cultures
The HCT 116 and human embryonic kidney (HEK) 293 cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (HyClone) supplemented with 10% fetal bovine serum (FBS) (Ex-Cell Bio) and 1% penicillin-streptomycin (HyClone) and maintained at 37°C incubators with 5% CO2. For passaging, cells were washed with 1× phosphate-buffered saline (PBS; HyClone). Trypsin-EDTA (0.25%; Gibco) was used to detach cells from plates. Trypsin-EDTA was quenched by addition of FBS-containing medium. For serum stimulation, cells were first starved by washing three times with 1× PBS and culturing for 40 hours in serum-free medium and then either left untreated or treated with serum for indicated time before harvesting.
Generation of SPT5∆PRD and SPT5∆PLD HCT 116 cell lines
SPT5 single-guide RNA (sgRNA) oligos were cloned into lentiCRISPR v2. Lentiviral particle preparation was performed as described previously (29). HCT 116 cells were infected and selected with puromycin (2 μg/ml) for 48 hours in culture medium. The microhomologous recombination (47) precise integration into target chromosome (PITCh) sgRNA oligos were cloned into pX459 (see table S2 for oligo sequences). HCT 116 cells were transfected with pX459 containing PITCh sgRNA and homolog arm with repair fragment. The infected cells were maintained in the absence of puromycin until cell clones were ready to be picked. The clones were screened with polymerase chain reaction (PCR) and confirmed by thymine-adenine (TA) cloning plus DNA sequencing and Western blot.
Generation of BioID-tagged proximity labeling cell lines
pcDNA5/FRT FLAG-BioID-SPT5 or FLAG-BioID-SPT5∆PLD expression plasmid was transfected into HEK293 Flp-In TRex cells and selected with hygromycin (100 μg/ml). Expression of FLAG-BioID-SPT5 or FLAG-BioID-SPT5∆PLD proteins was induced with doxycycline (2 μg/ml) for 48 hours.
Generation of LEDGF-dTAG HCT 116 cell lines
To generate LEDGF-dTAG cells by the endogenous knock-in, PITCh plasmids (sgRNA cutting and microhomology-containing dTAG repair template plasmid) (47) were mixed with 1 × 106 HCT 116 cells followed by transfected with Lipofectamine 2000 (Invitrogen) (see table S2 for oligo sequences). After selected with puromycin (2 μg/ml) for 48 hours, cells were maintained in the absence of puromycin until cell clones were ready to be picked. The clones were screened with PCR and confirmed by TA cloning plus DNA sequencing and Western blot. For homogeneous knock-in clones, protein degradation efficiency was verified by dTAG-13 treatment for 3 hours followed by western blotting.
Generation of LEDGF∆IBD HCT 116 cell lines
To generate LEDGF∆IBD cells, HCT 116 cells were transfected with pX459 containing sgRNA and homolog arm with repair fragment. The transfected cells were maintained in the absence of puromycin until cell clones were ready to be picked. The clones were screened with PCR and confirmed by TA cloning plus DNA sequencing and Western blot.
Method details
Chromatin immunoprecipitation
ChIP was performed as previously described (29). First, ~2× 107 HCT 116 cells were cross-linked using 1% formaldehyde (Sigma-Aldrich) at room temperature for 10 min, and cross-linking was quenched by glycine (final concentration is 125 mM) for 5 min. Then, cells were rinsed twice with ice-cold 1× PBS, harvested by centrifugation, and resuspended in 10 ml of ChIP lysis buffer I [50 mM Hepes-KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Igepal, and 0.25% Triton X-100] in the presence of protease inhibitors cocktail (Sigma-Aldrich) for up to 10 min on ice. Next, the cells were suspended in ChIP lysis buffer II [10 mM tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and 1× protease inhibitor] for 10 min at room temperature. Fixed chromatin was sonicated into 200– to 500–base pair (bp) fragments (Sonicator, Diagenode) in ChIP lysis buffer III [10 mM tris-HCl (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, and 1× protease inhibitor]. Chromatin extracts were incubated with specific antibody (anti–Pol II, anti-SPT5, anti-SPT5-CTR1/PRD, anti-NELFE, and anti-AFF4) and protein A agarose beads at 4°C overnight. Immunoprecipitates were washed five times with radioimmunoprecipitation assay buffer [50 mM Hepes-KOH (pH 7.5), 500 mM LiCl, 1 mM EDTA, 1.0% NP-40, 0.7% and Na-deoxycholate] and twice with Tris and EDTA (TE) buffer. After the final wash, DNA was eluted with elution buffer [50 mM tris-HCl (pH 8.0), 10 mM EDTA, and 1.0% SDS] and reverse cross-linked at 65°C. Subsequently, the eluted DNA was treated with protease K and ribonuclease A and were purified by the FastPure Gel DNA Extraction Mini Kit (Vazyme). Last, ChIP-seq libraries were prepared using the VAHTS Universal DNA Library Prep Kit for Illumina V3 (Vazyme), and sequencing was performed using an Illumina NovaSeq 6000 platform.
Cleavage under targets and tagmentation
CUT&Tag was performed using the Hyperactive Universal CUT&Tag Assay Kit for Illumina Pro (Vazyme) according to the manufacturer’s protocol. For each CUT&Tag reaction, 5× 105 HCT 116 cells were harvested using StemPro Accutase (Gibco) to prevent overdigestion. After centrifugation, cells were washed twice with Wash Buffer. Concanavalin A–coated magnetic beads (ConA Beads Pro, Vazyme) were pre-activated and resuspended in an equal volume of Binding Buffer. Subsequently, 10 μl of activated ConA Beads Pro were added to cells, followed by a 10-min incubation under gentle rotation. The bead-bound cells were magnetized to remove excess liquid and then resuspended in 50 μl of Antibody Buffer. Next, 1 μg of LEDGF (Bethyl) antibody was added, and the mixture was rotated 2 hours at room temperature. After incubation, the bead-bound cells were subjected to successive incubations with goat anti-rabbit immunoglobulin G (IgG; diluted 1:100) in 50 μl of Dig-wash Buffer for 1 hour at room temperature. Subsequently, the cells were washed four times with Dig-wash Buffer to remove unbound antibodies. The pA/G-Tnp Pro was prepared in Dig-300 Buffer to a final concentration of 0.04 μM. The bead-bound cells were resuspended in 100 μl of pA/G-Tnp Pro mix and incubated at room temperature for 1 hour before supernatant removal. Following thorough washing, the tagmentation reaction was initiated in 40 μl of Dig-300 Buffer with 10 μl of 5× TTBL at 37°C for 1 hour. To halt the reaction, 2 μl of 10% SDS was added, and appropriate amount of DNA Spike-in was added. After a 10-min incubation at 55°C, DNA purification was performed using DNA Extract Beads Pro (Vazyme). The purified DNA was then subjected to amplification using TruePrep Index Kit V2 for Illumina (Vazyme) with a universal i5 primer and a uniquely barcoded i7 primer. The exact number of PCR cycles was determined by quantitative PCR before amplification, with 11 cycles typically sufficient to yield an adequate library for sequencing. Following library size selection with VAHTS DNA Clean Beads (Vazyme) to achieve library sizes ranging from 200 to 700 bp, sequencing was performed using an Illumina NovaSeq 6000 platform.
BioID proteomics
The BioID procedures below were carried out as described previously (48). When cells are 80% confluent, replace cell culture medium and add biotin (Sigma-Aldrich) to a final concentration of 50 μM and incubate for 16 to 18 hours. Approximately 2 × 107 cells for each sample were collected, washed with PBS twice in plate, and resuspended in lysis buffer [8 M urea, 50 mM tris-HCl (pH 7.4), 1 mM dithiothreitol (DTT)] containing protease inhibitor cocktail (Sigma-Aldrich). After added with Pierce Universal Nuclease (Vazyme), cells were incubated for 10 min at room temperature and collected by centrifugation at 4°C and then lysed by sonication. After ultracentrifuge, the lysate was precleared by Gelatin Sepharose Beads (Smart-Lifesciences) and then mixed with streptavidin-conjugated Sepharose Beads (Sigma-Aldrich). After incubation at 4°C for 4 hours, the beads were spun down and washed four more times with wash buffer [8 M urea, 50 mM tris-HCl (pH 7.4)] before resuspending beads by 50 mM ammonium bicarbonate containing 1 mM biotin. Last, liquid chromatography–mass spectrometry analysis was performed.
Recombinant protein purification
cDNAs encoding SPT5-PRD, SPT5-PRD-PLD, and LEDGF-IBD were inserted to the pET16b vectors; cDNAs encoding SPT5-PRD-PLD, SPT5-PRD, and SPT5-PLD were inserted to the modified pET16b-mCherry (monomeric Cherry) vectors; cDNAs encoding LEDGF-IBD or AFF4-IDR were separately inserted to the modified pET16b-eGFP (enhanced green fluorescent protein) or blue fluorescent protein (BFP) vectors. All expression constructs were sequenced to ensure the sequence accuracy. The fluorescence-tagged fusion proteins were expressed using the Escherichia coli BL21 expression system. Briefly, recombinant plasmids were transformed into BL21. Mid-log phase of the transformed BL21 cells were induced by 1 mM isopropyl-β-d-thiogalactopyranoside for 4 hours to express the proteins of interest. Cells were harvested by centrifugation and stored at −80°C until needed. To purify the recombinant proteins, cell pellets from 500 ml of culture were resuspended in 40 ml of lysis buffer [50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 10 mM imidazole, and 0.05% Tween 20] in the presence of phenylmethanesulfonyl fluoride (PMSF) and homogenized using a high-pressure homogenizer for 5 cycles at 10,000 MPa. The crude lysate was cleared by centrifugation at 20,000g for 1 hour at 4°C. Ni–nitrilotriacetic acid (NTA) agarose (1 ml; QIAGEN) was pre-equilibrated with lysis buffer and then added to the cleared lysate. After overnight incubation at 4°C, the lysate agarose slurry was washed five times with 10 ml of wash buffer [50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 30 mM imidazole, and 0.05% Tween 20]. Protein was eluted with 5 mL of elution buffer [50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 250 mM imidazole, and 0.05% Tween 20]. The purified recombinant proteins were analyzed by Coomassie-stained SDS–polyacrylamide gel electrophoresis (PAGE). Recombinant proteins were concentrated and desalted using Amicon Ultra centrifugal filters (Merck Millipore).
cDNAs encoding LEDGF-IBD were inserted to the pGEX-5X-1 vectors. To purify the recombinant proteins, cell pellets from 500 ml of culture were resuspended in 40 ml of lysis buffer [50 mM tris-HCl (pH 8.0) and 150 mM NaCl] in the presence of PMSF and homogenized using a high-pressure homogenizer for five cycles at 10,000 MPa. The crude lysate was cleared by centrifugation at 20,000g for 1 hour at 4°C. GST agarose (1 ml; Smart-Lifesciences) was pre-equilibrated with lysis buffer and then added to the cleared lysate. After overnight incubation at 4°C, the lysate agarose slurry was washed once with 10 ml of high-salt buffer [50 mM tris-HCl (pH 8.0) and 300 mM NaCl] and twice with no salt buffer [50 mM tris-HCl (pH 8.0)]. Protein was eluted with 5 ml of elution buffer [50 mM NaH2PO4 (pH 8.0) and 10 mM glutathione]. The purified recombinant proteins were analyzed by Coomassie-stained SDS-PAGE. Recombinant proteins were concentrated and desalted using Amicon Ultra centrifugal filters (Merck Millipore).
In vitro droplet assay
The purified recombinant proteins were concentrated dialyzed against droplet formation buffer [50 mM tris-HCl (pH 7.5), 10% glycerol, and 1 mM DTT] and desalted using Amicon Ultra centrifugal filters (Merck Millipore). Indicated concentrations of recombinant BFP-AFF4-IDR, mCherry-SPT5-PRD-PLD, mCherry-SPT5-PRD, mCherry-SPT5-PLD, or eGFP-LEDGF-IBD proteins were then added to droplet formation buffer containing indicated concentration of salt in the presence of 10% crowding agent PEG-8000. The protein solution was immediately loaded onto a coverslip and imaged with Zeiss microscope with a 20× objective.
In vitro kinase assay
To purify the SEC kinase, ~2 × 107 cells expressed FLAG-AFF4 for each assay were collected, washed with PBS once, and lysed in high-salt lysis buffer [20 mM Hepes (pH 7.4), 10% glycerol, 0.35 M NaCl, 1 mM MgCl2, 0.5% Triton X-100, and 1 mM DTT] containing proteinase inhibitors (Sigma-Aldrich). After incubation at 4°C for 30 min, the lysate was cleared twice by centrifugation at 4°C. The balance buffer [20 mM Hepes (pH 7.4), 1 mM MgCl2, and 10 mM KCl] was added to the resulting supernatant to make the final NaCl concentration of 300 mM. The lysate was then mixed with ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich). After incubation at 4°C for 4 hours, the beads were spun down and washed three times with wash buffer [10 mM Hepes (pH 7.4), 1 mM MgCl2, 300 mM NaCl, 10 mM KCl, and 0.2% Triton X-100] before eluting by elution buffer [10 mM Hepes (pH 7.9), 0.1 M NaCl, 1.5 mM MgCl2, and 0.05% Triton X-100] containing FLAG peptide (200 μg/ml; Sigma-Aldrich). Kinase assays were done as previously described (49). Commonly, 5 μg of recombinant SPT5-PRD or SPT5-PRD-PLD and 5 μg of LEDGF-IBD were incubated with SEC in the presence or absence of 1 mM adenosine 5′-triphosphate for indicated times at 30°C in kinase buffer [50 mM tris-HCl (pH 7.5), 2 mM DTT, and 5 mM MgCl2]. Reactions were terminated by boiling in SDS gel sample buffer and analyzed by Western blot using the anti–SPT5-CTR or anti–SPT5-pPRD antibodies.
Immunoprecipitation
Cells (5 × 106) were harvested for each immunoprecipitation and incubated with ice-cold high-salt lysis buffer [20 mM Hepes (pH 7.9), 25% glycerol, 420 mM NaCl, 1.55 mM MgCl2, 0.2 mM EDTA, and 1× protease inhibitor] with gentle rotation for 30 min at 4°C. After centrifugation, the balance buffer [20 mM Hepes (pH 7.4), 1 mM MgCl2, and 10 mM KCl] was added to the supernatant to make the final NaCl concentration 300 mM. The lysate was then incubated with antibodies and protein A beads overnight at 4°C. The beads were washed three times with wash buffer [10 mM Hepes (pH 7.9), 150 mM NaCl, 1.5 mM MgCl2, 10 mM KCl, and 0.02% Triton X-100] before boiling in SDS loading buffer.
Pull-down assay
Indicated concentrations of recombinant GST and GST-LEDGF-IBD proteins were resuspended in lysis buffer [50 mM tris-HCl (pH 8.0) and 150 mM NaCl] containing proteinase inhibitors (Sigma-Aldrich) and incubated with GST beads for 2 hours at 4°C. Then, the beads were washed once with high-salt buffer [50 mM tris-HCl (pH 8.0) and 300 mM NaCl] and twice with no salt buffer [50 mM tris-HCl (pH 8.0)]. After incubating with SPT5-PRD, SPT5-PRD-PLD, or SPT5-PRD-PLD (PLD-S/T-E) overnight at 4°C, the beads were washed as above. SDS loading buffer was mixed with the beads for Western blotting.
Western blot
The Western blotting method was conducted as previously described (29). Briefly, the lysates of the cells were subjected to SDS-PAGE gels and then transferred onto polyvinylidene fluoride membrane. Thereafter, the membrane was incubated with the anti–Pol II (1:5000), anti–Pol II–pSer5 (1:5000), anti–Pol II–pSer2 (1:5000), anti-SPT5 (1:5000), anti–SPT5-pPRD (1:5000), anti–SPT5-pS666 (1:5000), anti-NELFE (1:5000), anti-CDK9 (1:5000), anti-AFF4 (1:5000), anti-AFF1 (1:3000), anti-PAF1 (1:2000), anti-LEDGF (1:5000), anti-PPP2CA (1:5000), anti-TCERG1 (1:4000), anti-BRD4 (1:3000), anti-FLAG (1:5000), anti-MYC (1:5000), anti-HIS (1:5000), anti-streptavidin (1:2000), or anti–α-tubulin (1:1000) antibody that was diluted in TBST and incubated overnight at 4°C. Subsequently, horseradish peroxidase–conjugated secondary antibodies (Sigma-Aldrich) were used at a dilution of 1:5000. ECL substrate (Merck Millipore) was applied to the membrane for imaging using autoradiography.
Generation of LEDGF antibody
LEDGF Rabbit Polyclonal was generated in house. Human LEDGF (328 to 530 amino acids) was expressed as His-tag fusion proteins in PET-16b, purified on NTA-agarose according to QIAGEN’s protocol and sent to HUA BIO Co. Ltd. for immunization into rabbits.
Immunofluorescence
Cells grown on coverslips were fixed with 4% paraformaldehyde (PFA) in PBS for 10 min at room temperature. After three washes in PBS for 5 min, cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. Following rinses with PBS, cells were incubated in blocking buffer (2% bovine serum albumin and 0.3% Triton X-100) for at least 1 hour at room temperature and then in diluted primary antibodies (1:1000 anti-SPT5, 1:200 anti-AFF4, 1:200 anti-LEDGF, 1:200 anti-NELFE, and 1:800 anti–Pol II–pSer5) in blocking buffer overnight at 4°C. After three washes in PBS, coverslips were incubated at room temperature with secondary antibodies (1:2000 goat anti-rabbit IgG Alexa Fluor 488, 1:2000 goat anti-rabbit IgG Alexa Fluor 647, and 1:2000 goat anti-mouse IgG Alexa Fluor 488) in the dark for 1 hour, followed by three washes with PBS. Coverslips were mounted on slides using VECTASHIELD Mounting Medium with 4′,6-diamidino-2-phenylindole (DAPI). Three-dimensional images were acquired at Zeiss LSM 700 confocal microscope with 63×, 1.4 numerical aperture (NA) oil immersion objective lens using Zen Light Edition acquisition software and charge-coupled device (CCD) camera. Images were post-processed using Zen Light Edition and Fiji Is Just ImageJ (FIJI) (50).
DNA FISH combined with IF
Immunostaining was performed as above described. After immunostaining, cells were postfixed with 4% PFA in PBS for 10 min at room temperature and washed three times with PBS. Cells were then dehydrated by 70, 85, and then 100% ethanol at room temperature. FOS DNA FISH probe (1 μl) was diluted in 7 μl of FISH Hybridization Buffer and 2 μl of water and added on slides. Cell attached coverslips were placed on the top of slides (cell-side toward the hybridization mixture). Coverslips were sealed with rubber cement. Genomic DNA and probes were denatured by incubating slides at 78°C for 5 min and then hybridized at 37°C overnight in the dark. The coverslip was detached from the slide and washed in pre-warmed Wash Solution 1 [0.3% Igepal (Sigma-Aldrich, CA-630) or NP-40/0.4× saline-sodium citrate buffer (SSC)] at 73°C for 2 min and in Wash Solution 2 (0.1% Igepal or NP-40/2× SSC) for 1 min at room temperature. Coverslips were mounted using VECTASHIELD Mounting Medium with DAPI. Images were acquired at Zeiss LSM 700 confocal microscope with 63×, 1.4 NA oil immersion objective lens using Zen Light Edition acquisition software and CCD camera. Images were post-processed using Zen Light Edition and FIJI. The FOS DNA FISH probe was designed to hybridize to the FOS gene and generated by Empire Genomics. Design Region: CHR14: 75745480-75748937, Locus: 14q24.3.
Nascent RNA synthesis assay
Nascent RNA was examined using the Click-iT RNA Alexa Fluor 488 kit (Invitrogen) according to the manufacturer’s protocol (51). Briefly, 1 mM EU was added and incubated under specific cell culture conditions for 30 min. Proceed immediately to cell fixation by 3.7% formaldehyde in PBS for 15 min, then permeabilize with 0.5% Triton X-100 in PBS for 15 min, and incubate with Click-iT reaction cocktail for 30 min at room temperature. Wash cells once with 1 ml of Click-iT reaction rinse buffer. Coverslips were mounted using VECTASHIELD Mounting Medium with DAPI. Images were acquired at Zeiss LSM 700 confocal microscope with 63×, 1.4 NA oil immersion objective lens using Zen Light Edition acquisition software and CCD camera. Images were post-processed using Zen Light Edition and FIJI.
Lentivirus-mediated RNA interference
Human LEDGF, PPP2CA, TCERG1, BCOR, SCAF11, and BRD4 (52) shRNA oligos (see table S2 for oligo sequences) were cloned into the pLKO.1 vector. Nontargeting shRNA construct (SHC002) was purchased from Sigma-Aldrich. Lentiviral particle preparation and infection were performed as described previously (49). Briefly, around 60% confluent HEK293T cells in 15-cm tissue culture plate were co-transfected with 8 μg of the shRNA construct or nontargeting control shRNA, 6 μg of psPAX2 packaging plasmids, and 2 μg of pMD2.G envelope plasmids using Lipofectamine 2000. The media were replaced with fresh culture media after 16 hours of transfection. The lentiviral supernatants were collected 48 and 72 hours after transfection and filtered through the 0.45-μm filters. Cells were infected with the filtered lentiviral supernatants containing polybrene (8 μg/ml; Sigma-Aldrich). Twenty-four hours after infection, cells were subjected to selection with puromycin (2 μg/ml) for an additional 48 hours.
Prediction of PLD
Protein disorder propensity plot for SPT5 was prepared using the PLAAC (prion-like amino acid composition) webtool (http://plaac.wi.mit.edu/) with background set to 0%.
Prediction of phosphorylation sites
Protein posttranslation modification was predicted using the PhosphoSitePlus webtool (www.phosphosite.org/).
ChIP-seq analysis
Adaptors and low-quality reads removal were performed using Trim Galore v0.6.7 (www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with the parameter “-q 20.” Paired-reads were aligned to the human h19 genomes using Bowtie v2.4.5 with default parameters (53). PCR duplicates were removed by Picard 2.27.4 (https://broadinstitute.github.io/picard/). Normalized bigwig and log2 fold change of ChIP-seq signal were generated using deepTools v3.5.1 bamCoverage and bigwigCompare (54). Heatmaps and metaplots of ChIP-seq signal were generated with deepTools v3.5.1 computeMatrix, plotHeatmap, and plotProfile. We selected transcripts annotated in the NCBI RefSeq database from UCSC Table Browser. Next, we chose transcripts that promoters overlap with peaks called in untreated HCT 116 cells. Transcripts that are <2 kb long and <2 kb from the nearest genes were filtered out.
CUT&Tag analysis
Raw CUT&Tag adaptors and low-quality reads were removed as described in ChIP-seq analysis, and the paired-reads were aligned to the human h19 genomes using Bowtie v2.4.5 with the options “--local --very-sensitive --no-mixed --no-discordant -I 10 -X 700” (53). PCR duplicates were filtered using Picard 2.27.4. Normalized bigwig and log2 fold change were generated as described for ChIP-seq above.
Transcriptional pause index calculation
The pause index was defined as the ratio of average signal in promoter region and average signal in body region. The promoter referred to the region containing 100 bp upstream to 300 bp downstream of the transcription start sites (TSS). The gene body referred to the region from 300 bp to transcript end sites (TES).
Peak calling and visualization
Peaks were called using MACS 2.2.7.1 with a P value cutoff of 1 × 10−7, and peaks region were annotated with ChIPseeker R package v1.30.3 (55).
Live cell cluster quantification
FIJI was used to identify clusters and characterize their numbers, sizes, and shapes. All images were equally thresholded, and clusters areas were identified using the “Analyze Particles” function of FIJI. Clusters identified in >20 cells were quantified. The mean intensity within the clusters (C-in) and in the bulk (C-out) were calculated for each channel. The violin plots show the distributions of mean clusters of each cell. For co-localization analysis, the phase-separated puncta were analyzed using “Colocalization_Finder” plugin on FIJI (https://imagej.nih.gov/ij/plugins/colocalization-finder.html). Results shown are representative of at least three biological replicates.
In vitro droplet quantification
FIJI was used to identify droplets and characterize their numbers, sizes, and shapes. All images were equally thresholded, and droplets areas were identified using the “Analyze Particles” function of FIJI. Hundreds of droplets identified in typically six independent fields of view were quantified. Results shown are representative of at least two biological replicates. To determine the condensed fraction, we first calculated the sum total of intensities in all droplets within a specific field (I-in). In addition, we calculated the sum total intensity in the bulk dilute phase outside the droplets (I-out) for each channel. The condensed fraction was then computed as the (I-in)/[(I-in) + (I-out)]. For calculating the partition ratio, we obtained the average intensity of each droplet (C-in) as well as the average intensity of the bulk dilute phase outside the droplet (C-out) for each channel. The partition coefficient was then computed as the ratio of (C-in)/(C-out).
Statistical analyses
Wilcoxon test was used for ChIP-seq and CUT&Tag, and others were used for paired t test in this study.
Acknowledgments
We are grateful to the Lin and Luo laboratory members for helpful discussion of this study. We are grateful to F. X. Chen for the SPT5-pPRD and SPT5-pS666 antibodies and K. Chen for the DNA Clean Beads.
Funding: This work was funded by grants from the National Natural Science Foundation of China (32030017 and 32350004 to C.L.; 32470627 to Z.L.; and 32300453 to C.G.), the National Key R&D Program of China (2018YFA0800100 to C.L.), and Shenzhen Science and Technology Program (JCYJ20220530160417038 to C.L. and JCYJ20220530160416037 to Z.L.), as well as the fellowship of China National Postdoctoral Program for Innovative Talents (BX20230069 to C.G.), the China Postdoctoral Science Foundation (2023M730574 to C.G.), and the Jiangsu Funding Program for Excellent Postdoctoral Talent (2023ZB707 to C.G.).
Author contributions: Conceptualization: C.G., Z.L., and C.L. Investigation: C.G., H.F., S.Sh., Y.Z., X.D., B.D., and J.W. Formal analysis: C.G., S.Si., and Z.L. Software: Z.L. Visualization: Z.L. Resources: Z.G., Z.L., and C.L. Data curation: S.Si. Funding acquisition: C.G., Z.L., and C.L. Validation: C.G., B.D., H.Y., Z.L., and C.L. Writing—original draft: C.G. and Z.L. Writing—review and editing: J.W., Z.L., and C.L. Supervision: Z.G., Z.L., and C.L. Project administration: C.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. ChIP-seq and CUT&Tag sequencing data have been deposited in NCBI Gene expression Omnibus under accession number GSE250380.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 and S2
References
REFERENCES AND NOTES
- 1.Chen F. X., Smith E. R., Shilatifard A., Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell. Bio. 19, 464–478 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Core L., Adelman K., Promoter-proximal pausing of RNA polymerase II: A nexus of gene regulation. Gene Dev. 33, 960–982 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dollinger R., Gilmour D. S., Regulation of promoter proximal pausing of RNA polymerase II in metazoans. J. Mol. Biol. 433, 166897 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo C. H., Luo Z. J., Lin C. Q., Dynamic regulation of promoter-proximal RNA polymerase II pausing. Chin. Sci. B. Chin. 65, 4084–4094 (2020). [Google Scholar]
- 5.Aoi Y., Shilatifard A., Transcriptional elongation control in developmental gene expression, aging, and disease. Mol. Cell 83, 3972–3999 (2023). [DOI] [PubMed] [Google Scholar]
- 6.Decker T. M., Mechanisms of transcription elongation factor DSIF (Spt4-Spt5). J. Mol. Biol. 433, 166657 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Hartzog G. A., Fu J. H., The Spt4-Spt5 complex: A multi-faceted regulator of transcription elongation. Bba-Gene Regul. Mech. 1829, 105–115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi Y., Shibata H., Handa H., Transcription elongation factors DSIF and NELF: Promoter-proximal pausing and beyond. Bba-Gene Regul. Mech. 1829, 98–104 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Cheng B., Price D. H., Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J. Biol. Chem. 282, 21901–21912 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Kim J. B., Sharp P. A., Positive transcription elongation factor b phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276, 12317–12323 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Yamada T., Yamaguchi Y., Inukai N., Okamoto S., Mura T., Handa H., P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21, 227–237 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Swanson M. S., Malone E. A., Winston F., Spt5, an essential gene important for normal transcription in Saccharomyces-cerevisiae, encodes an acidic nuclear-protein with a carboxy terminal repeat. Mol. Cell. Biol. 11, 3009–3019 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wada T., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G. A., Winston F., Buratowski S., Handa H., DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Gene Dev. 12, 343–356 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song A. X., Chen F. X., The pleiotropic roles of SPT5 in transcription. Transcr. Austin. 13, 53–69 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werner F., A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J. Mol. Biol. 417, 13–27 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu Y. J., Gilmour D. S., Identification of regions in the Spt5 subunit of DRB sensitivity-inducing factor (DSIF) that are involved in promoter-proximal pausing. J. Biol. Chem. 292, 5555–5570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernecky C., Plitzko J. M., Cramer P., Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp. Nat. Struct. Mol. Biol. 24, 809–815 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Ehara H., Yokoyama T., Shigematsu H., Yokoyama S., Shirouzu M., Sekine S. I., Structure of the complete elongation complex of RNA polymerase II with basal factors. Science 357, 921–924 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Fong N., Sheridan R. M., Ramachandran S., Bentley D. L., The pausing zone and control of RNA polymerase II elongation by Spt5: Implications for the pause-release model. Mol. Cell 82, 3632–3645.e4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu S. B., Peng L., Xu C., Wang Z., Song A., Chen F. X., SPT5 stabilizes RNA polymerase II, orchestrates transcription cycles, and maintains the enhancer landscape. Mol. Cell 81, 4425–4439.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Sansó M., Levin R. S., Lipp J. J., Wang V. Y. F., Greifenberg A. K., Quezada E. M., Ali A., Ghosh A., Larochelle S., Rana T. M., Geyer M., Tong L., Shokat K. M., Fisher R. P., P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Gene Dev. 30, 117–131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Z. J., Lin C. Q., Shilatifard A., The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 13, 543–547 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Guo C. H., Zhang Y., Shuai S., Sigbessia A., Hao S., Xie P., Jiang X., Luo Z., Lin C., The super elongation complex (SEC) mediates phase transition of SPT5 during transcriptional pause release. EMBO Rep. 24, e55699 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge H., Si Y. Z., Roeder R. G., Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 17, 6723–6729 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh D. P., Kimura A., Chylack L. T., Shinohara T., Lens epithelium-derived growth factor (LEDGF/p75) and p52 are derived from a single gene by alternative splicing. Gene 242, 265–273 (2000). [DOI] [PubMed] [Google Scholar]
- 26.van Nuland R., van Schaik F. M. A., Simonis M., van Heesch S., Cuppen E., Boelens R., Timmers H. T. M., van Ingen H., Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain. Epigenetics Chromatin 6, 12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu A., Sanchez T. W., Casiano C. A., DFS70/LEDGFp75: An enigmatic autoantigen at the interface between autoimmunity, AIDS and cancer. Front. Immunol. 6, 1–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakravarty A. K., Jarosz D. F., More than just a phase: Prions at the crossroads of epigenetic inheritance and evolutionary change. J. Mol. Biol. 430, 4607–4618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo C. H., Che Z., Yue J., Xie P., Hao S., Xie W., Luo Z., Lin C., ENL initiates multivalent phase separation of the super elongation complex (SEC) in controlling rapid transcriptional activation. Sci. Adv. 6, eaay4858 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roux K. J., Kim D. I., Raida M., Burke B., A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Appel L. M., Franke V., Benedum J., Grishkovskaya I., Strobl X., Polyansky A., Ammann G., Platzer S., Neudolt A., Wunder A., Walch L., Kaiser S., Zagrovic B., Djinovic-Carugo K., Akalin A., Slade D., The SPOC domain is a phosphoserine binding module that bridges transcription machinery with co- and post-transcriptional regulators. Nat. Commun. 14, 166 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ardehali M. B., Damle M., Perea-Resa C., Blower M. D., Kingston R. E., Elongin A associates with actively transcribed genes and modulates enhancer RNA levels with limited impact on transcription elongation rate. J. Biol. Chem. 296, 100202 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebmeier C. C., Erickson B., Allen B. L., Allen M. A., Kim H., Fong N., Jacobsen J. R., Liang K., Shilatifard A., Dowell R. D., Old W. M., Bentley D. L., Taatjes D. J., Human TFIIH Kinase CDK7 regulates transcription-associated chromatin modifications. Cell Rep. 20, 1173–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhattacharya S., Levy M. J., Zhang N., Li H., Florens L., Washburn M. P., Workman J. L., The methyltransferase SETD2 couples transcription and splicing by engaging mRNA processing factors through its SHI domain. Nat. Commun. 12, 1443 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang K. L., Jee D., Stein C. B., Elrod N. D., Henriques T., Mascibroda L. G., Baillat D., Russell W. K., Adelman K., Wagner E. J., Integrator recruits protein phosphatase 2A to prevent pause release and facilitate transcription termination. Mol. Cell 80, 345–358.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nabet B., Roberts J. M., Buckley D. L., Paulk J., Dastjerdi S., Yang A., Leggett A. L., Erb M. A., Lawlor M. A., Souza A., Scott T. G., Vittori S., Perry J. A., Qi J., Winter G. E., Wong K. K., Gray N. S., Bradner J. E., The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 14, 431–441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cermakova K., Demeulemeester J., Lux V., Nedomova M., Goldman S. R., Smith E. A., Srb P., Hexnerova R., Fabry M., Madlikova M., Horejsi M., de Rijck J., Debyser Z., Adelman K., Hodges H. C., Veverka V., A ubiquitous disordered protein interaction module orchestrates transcription elongation. Science 374, 1113–1121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma S., Čermáková K., de Rijck J., Demeulemeester J., Fábry M., el Ashkar S., van Belle S., Lepšík M., Tesina P., Duchoslav V., Novák P., Hubálek M., Srb P., Christ F., Řezáčová P., Hodges H. C., Debyser Z., Veverka V., Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 115, E7053–E7062 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jayakumar S., Patel M., Boulet F., Aziz H., Brooke G. N., Tummala H., Pradeepa M. M., PSIP1/LEDGF reduces R-loops at transcription sites to maintain genome integrity. Nat. Commun. 15, 361 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koutná E., Lux V., Kouba T., Škerlová J., Nováček J., Srb P., Hexnerová R., Šváchová H., Kukačka Z., Novák P., Fábry M., Poepsel S., Veverka V., Multivalency of nucleosome recognition by LEDGF. Nucleic Acids Res. 51, 10011–10025 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LeRoy G., Oksuz O., Descostes N., Aoi Y., Ganai R. A., Kara H. O., Yu J. R., Lee C. H., Stafford J., Shilatifard A., Reinberg D., LEDGF and HDGF2 relieve the nucleosome-induced barrier to transcription in differentiated cells. Sci. Adv. 5, eaay3068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng H., Qi Y., Hu S., Cao X., Xu C., Yin Z., Chen X., Li Y., Liu W., Li J., Wang J., Wei G., Liang K., Chen F. X., Xu Y., Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase. Science 370, eabb5872 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Xu C. L., Li C., Chen J., Xiong Y., Qiao Z., Fan P., Li C., Ma S., Liu J., Song A., Tao B., Xu T., Xu W., Chi Y., Xue J., Wang P., Ye D., Gu H., Zhang P., Wang Q., Xiao R., Cheng J., Zheng H., Yu X., Zhang Z., Wu J., Liang K., Liu Y.-J., Lu H., Chen F. X., R-loop-dependent promoter-proximal termination ensures genome stability. Nature 621, 610–619 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu S. B., Peng L., Song A., Ji Y.-X., Cheng J., Wang M., Chen F. X., INTAC endonuclease and phosphatase modules differentially regulate transcription by RNA polymerase II. Mol. Cell 83, 1588–1604.e5 (2023). [DOI] [PubMed] [Google Scholar]
- 45.Fujiwara R., Zhai S. N., Liang D., Shah A. P., Tracey M., Ma X. K., Fields C. J., Mendoza-Figueroa M. S., Meline M. C., Tatomer D. C., Yang L., Wilusz J. E., IntS6 and the integrator phosphatase module tune the efficiency of select premature transcription termination events. Mol. Cell 83, 4445–4460.e7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larochelle S., Batliner J., Gamble M. J., Barboza N. M., Kraybill B. C., Blethrow J. D., Shokat K. M., Fisher R. P., Dichotomous but stringent substrate selection by the dual-function Cdk7 complex revealed by chemical genetics. Nat. Struct. Mol. Biol. 13, 55–62 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Sakuma T., Nakade S., Sakane Y., Suzuki K. T., Yamamoto T., MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protoc. 11, 118–133 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Sears R. M., May D. G., Roux K. J., BioID as a tool for protein-proximity labeling in living cells. Methods Mol. Biol. 2012, 299–313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin C., Smith E. R., Takahashi H., Lai K. C., Martin-Brown S., Florens L., Washburn M. P., Conaway J. W., Conaway R. C., Shilatifard A., AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 37, 429–437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jao C. Y., Salic A., Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U.S.A. 105, 15779–15784 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Francisco J. C., Dai Q., Luo Z., Wang Y., Chong R. H. H., Tan Y. J., Xie W., Lee G. H., Lin C., Transcriptional elongation control of hepatitis B virus covalently closed circular DNA transcription by super elongation complex and BRD4. Mol. Cell. Biol. 37, e00040-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramirez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu G. C., Wang L. G., He Q. Y., ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Lancaster A. K., Nutter-Upham A., Lindquist S., King O. D., PLAAC: A web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30, 2501–2502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Che Z. Z., Liu X., Dai Q., Fang K., Guo C., Yue J., Fang H., Xie P., Luo Z., Lin C., Distinct roles of two SEC scaffold proteins, AFF1 and AFF4, in regulating RNA polymerase II transcription elongation. J. Mol. Cell Biol. 15, mjad049 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 and S2
References