Abstract
The pathophysiology of neurodevelopmental disorders involves vulnerable neural populations, including striatal circuitry, and convergent molecular nodes, including chromatin regulation and synapse function. Despite this, how epigenetic regulation regulates striatal development is understudied. Recurrent de novo mutations in Zswim6 are associated with intellectual disability and autism. We demonstrate that ZSWIM6 localizes to the nucleus where it associates with repressive chromatin regulators. Disruption of Zswim6 in ventral telencephalic progenitors leads to increased chromatin accessibility and transcriptional dysregulation. Ablating Zswim6 in either striatal direct or indirect pathway spiny projection neurons resulted in similar cell-autonomous changes in excitatory but not inhibitory synaptic transmission. Specifically, Zswim6 disruption altered the desensitization properties of AMPA receptors, leading to enhanced synaptic recruitment of SPNs, explaining SPN-subtype specific effects on activity and behavioral sub-structure. Last, adult deletion of Zswim6 identified a continuing role in the maintenance of mature striatal synapses. Together, we describe a mechanistic role for Zswim6 in the epigenetic control of striatal synaptic development.
Chromatin regulator ZSWIM6 shapes striatal synapses, linking epigenetics to neurodevelopmental disorders.
INTRODUCTION
Increasing evidence suggests the pathophysiology of neurodevelopmental disorders (NDDs) involves dysfunction of vulnerable neural circuits along convergent molecular pathways (1, 2). As the input nucleus of the basal ganglia, the striatum is crucial for both motor and cognitive processes, mediating a range of attentional, decision-making, and motor control functions that are widely perturbed in neuropsychiatric disease (1, 3–5). Furthermore, mapping of autism and schizophrenia-associated gene candidates to cell type–specific transcriptomes throughout the brain consistently reveal striatal spiny projection neurons (SPNs) as vulnerable circuit elements (6, 7). Regarding molecular pathways of pathophysiology, NDD candidate genes are strongly enriched for both synaptic proteins and chromatin regulators. A substantial body of work has implicated synaptic adhesion molecules, postsynaptic scaffolding proteins, and glutamate receptor subunits in widespread excitatory synaptic dysfunction (8, 9), as well as the disruption of social/appetitive reward processing and motor control (1, 5, 10–13). While less studied, functionally diverse chromatin binding proteins affecting histone methylation, acetylation, ubiquitination, and adenosine 5′-triphosphate (ATP)–dependent chromatin remodeling have also been implicated in NDDs (14, 15). The degree to which dysfunction of chromatin regulation converges at the level of striatal synapse development and circuit function in NDD pathogenesis remains unclear.
Zswim6 (zinc finger SWIM domain containing protein 6), a gene of unknown function, has been implicated in schizophrenia and NDDs via genetic association studies (16–19). The presence of conserved SWIM and Sin3-like domains within ZSWIM6 suggest a possible role in the regulation of chromatin structure and gene expression (20, 21). Notably, two recurrent de novo point mutations that result in complex syndromic presentations involving intellectual disability (ID) and autistic features are each predicted to affect the Sin3-like domain—the R913Ter nonsense mutation introduces a premature stop codon upstream of the Sin3-like domain (16), while the R1163W missense mutation in the Sin3-domain is predicted to be functionally disruptive in this highly conserved region (17). In both human (Allen Brain Atlas) and mouse, Zswim6 is highly expressed in the developing telencephalon and adult striatum (22, 23). Detailed expression studies in mice show dynamic expression in the subventricular zone (SVZ) of the lateral ganglion eminence (LGE) followed by increasing postnatal expression in both direct and indirect pathway striatal SPNs, the principal striatal cell type (23). Consistent with a key role in brain development, constitutive knockout of Zswim6 in mice resulted in ~50% early postnatal lethality. Gross anatomical studies revealed a decrease in cortical and striatal volumes and reduced dendritic complexity and spine density of striatal SPNs. Last, Zswim6 knockout (KO) mice displayed multiple repetitive motor abnormalities and an increased sensitivity to subthreshold amphetamine challenge (22). Despite the converging evidence from human and animal studies suggesting a crucial role for Zswim6 in brain development, the cellular and circuit level functions of this protein remain a mystery (22).
Here, we demonstrate that Zswim6 is a previously unrecognized chromatin regulator that links gene regulation with SPN synapse and circuit function during striatal development. Tagged ZSWIM6 exhibited robust nuclear localization in neurons both in vitro and in vivo and interacted with chromatin and chromatin-associated proteins in heterologous cells. Assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) in postnatal day 8 (P8) conditional KO (cKO) mice revealed a broad increase in chromatin accessibility, consistent with a role for Zswim6 in the establishment of repressive chromatin structure. Single-nucleus RNA sequencing (snRNA-seq) in Zswim6 cKO mice revealed concomitant transcriptional dysregulation, with an overall bias toward up-regulation of transcripts in Zswim6 cKO mice, and functional enrichment for genes involved in synaptic function, protein translation, and axon development. Functionally, SPN subtype-specific cKOs resulted in dysregulated α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPAR)–dependent synaptic transmission and enhanced SPN recruitment by excitatory inputs. Behaviorally, SPN-specific cKO resulted in activity levels consistent with increased synaptic drive of SPNs and SPN subtype-specific impacts on locomotion and spontaneous behavioral substructure. Last, we show that adult deletion of Zswim6 results in similar synaptic phenotypes as those observed from late-embryonic deletion, reflecting a continued requirement for Zswim6 in adulthood. Together, these findings establish Zswim6 as a molecular link between chromatin regulation and synaptic function, which supports important aspects of striatal circuit development.
RESULTS
ZSWIM6 associates with chromatin modifiers and mediates a repressive chromatin state
As the conserved Sin3-like domain found at the C-terminal domain of ZSWIM6 is known to function in SIN3 to repress transcription in neurons via the RE1 silencing transcription factor (REST) complex (17, 24–26), we hypothesized that ZSWIM6 could function via chromatin regulation. To investigate the subcellular localization of ZSWIM6 protein, we used an adeno-associated virus (AAV) to express the full Zswim6 coding sequence with an N-terminal Flag tag under the elongation factor 1 alpha (EF1α) promoter (AAV-EF1α::Flag-Zswim6-pA). In cultured mouse cortical neurons [day in vitro (DIV) 12], the majority of ZSWIM6 protein (~75%) was found in the nucleus (fig. S1, A to C). We tested whether this localization was altered by bidirectional changes in neuronal activity by incubating cortical neuron cultures in media containing either tetrodotoxin (TTX) to decrease activity or picrotoxin (PTX) to indirectly increase activity via suppression of local inhibition (fig. S1A). We found that while TTX treatment did not alter ZSWIM6 localization, PTX exposure led to a small decrease in the amount of nuclear ZSWIM6 (fig. S1, B and C). While Zswim6 mRNA is expressed in a complex pattern throughout the developing telencephalon (23), the striatum shows a uniquely dynamic regulation, with early expression in the SVZ of the lateral ganglionic eminence, which gives rise to striatal GABAergic projection neurons (SPNs) and local circuit GABAergic interneurons. Following a decrease in Zswim6 expression during late embryogenesis, Zswim6 mRNA is increased in the postnatal striatum and maintained into adulthood (23). We used viral injection of AAV- EF1α::Flag-Zswim6 into adult striatum to confirm the predominantly nuclear localization of ZSWIM6 protein in striatal SPNs, similar to our result in cultured neurons (Fig. 1, A to D, and fig. S1D). Consistent with the robust nuclear localization of Flag-ZSWIM6, we noted a predicted nuclear localization sequence (NLS) near the N terminus of ZSWIM6 that is well-conserved across species (fig. S1E) (27). We generated a mutant version of our Flag-ZSWIM6 AAV in which the 10–amino acid putative NLS is removed (AAV- EF1α::Flag-∆NLS-Zswim6) and asked whether nuclear localization was disrupted. In cultured mouse cortical neurons, the nuclear enrichment of Flag-∆NLS-ZSWIM6 was markedly abolished, and in many neurons appeared enriched in the cytoplasm relative to the nucleus (Fig. 1, E and F). Thus, the putative NLS is necessary for enriched localization of ZSWIM6 protein to the nucleus of neurons.
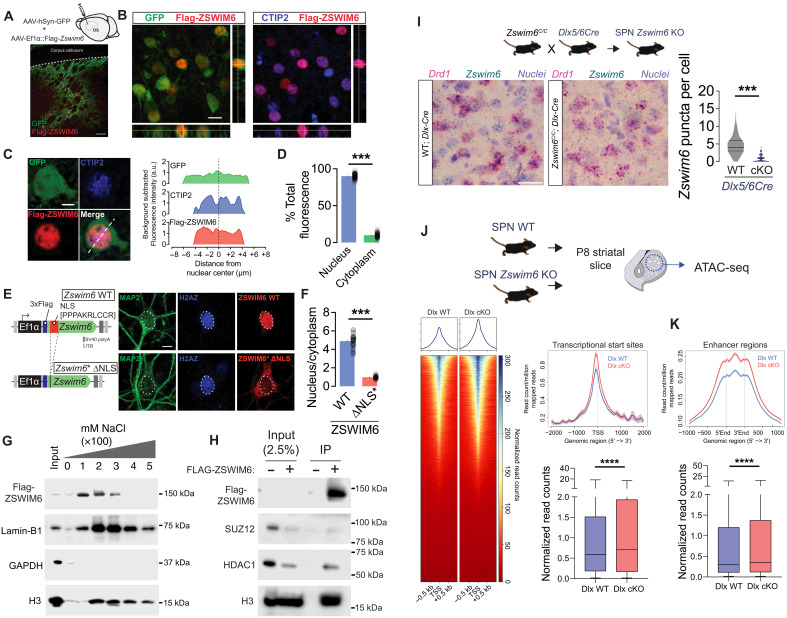
Fig. 1. ZSWIM6 localizes to the nucleus, interacts with chromatin, and regulates chromatin accessibility.
(A) Experimental schematic and image of AAV-hSyn-GFP and AAV-Ef1α-Flag-Zswim6 injection into dorsal striatum. Scale bar, 100 μm. (B) Confocal images showing Flag-ZSWIM6, cell-filling GFP, and nuclear CTIP2 in striatum. Orthogonal views confirm nuclear ZSWIM6 localization. Scale bar, 10 μm. (C) Individual labeled SPN demonstrating nuclear localization of Flag-ZSWIM6 (left) and corresponding line plot of fluorescent intensity (right) along the line indicated in the merged image. Scale bar, 4 μm. (D) Quantification of nuclear and cytoplasmic percentage of total Flag-ZSWIM6 in labeled SPN somas (means ± SEM, n = 38 cells from three animals, ***P < 1 × 10−12, paired t test). (E) Control and Zswim6*-ΔNLS AAV construct (left). DIV9 neurons transduced with Flag-ZSWIM6 and Flag-ZSWIM6*-ΔNLS AAV and stained for MAP2, H2AZ, and Flag. Scale bar, 5 μm. (F) Flag intensity (nucleus/cytoplasm) in control vs ΔNLS neurons (n = 29/28 cells, ***P < 1 × 10−12, paired t test). (G) Western blot following 100 to 300 mM salt extraction from HEK 293 T nuclear lysates with Flag-ZSWIM6 (n = 3). (H) Co-IP of SUZ12, HDAC1, and H3 with Flag-ZSWIM6 from HEK 293T nuclear lysates compared to untransfected control (n = 3). (I) Zswim6 SPN cKO breeding scheme (top). Images from BaseScope In Situ Hybridization (ISH) for Drd1 and Zswim6 mRNA in WT/cKO (left, scale bar: 20 μm) and quantification (right, n = 305/535 from one of two animal, ***P < 1 × 10−12, t test). (J) ATAC-seq schematic (top). Heatmaps (left) and summary graph (top, right) demonstrating normalized reads surrounding TSS in Dlx WT versus cKO P8 striatum. Box and whisker plots displaying fold change at TSS (****P < 1 × 10−12, t test). (K) Reads per million mapped reads within enhancer regions from Dlx WT versus cKO P8 striatum (top). Box and whisker plots displaying fold change within enhancer regions (****P < 1 × 10−12, t test). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To examine whether nuclear-localized ZSWIM6 protein associates with chromatin, we expressed Flag-tagged ZSWIM6 in heterologous cells [human embryonic kidney (HEK) 293T] and performed sequential salt extraction on isolated nuclei. ZSWIM6 protein was captured in eluates containing 100 to 300 mM NaCl, partially overlapping with early fractions in which histone H3 dissociates, indicating a loose association with chromatin (Fig. 1G). We next tested whether ZSWIM6 associates with endogenous chromatin-modifying proteins, focusing on known components of the polycomb repressive (SUZ12 and G9A), SIN3 (HDAC1), or SWI/SNF (BRG1) complexes (17, 28–30). We found that both SUZ12 and HDAC1 immunoprecipitated with Flag-tagged ZSWIM6 but were not detected in control immunoprecipitation (IPs) performed in mock-transfected cells (Fig. 1H). Histone H3 similarly precipitated with Flag-ZSWIM6, further confirming the association of ZSWIM6 with chromatin (Fig. 1H).
Given our data showing the association of ZSWIM6 with repressive chromatin regulators, we examined whether striatal deletion of Zswim6 altered global chromatin architecture. To do this, we crossed a Zswim6 conditional allele (Zswim6C/C) to the Dlx5/6-Cre line, which expresses Cre recombinase in all ventral GABAergic progenitors as early as embryonic day 12.5 (E12.5) (31, 32), verifying the effectiveness of recombination via in situ hybridization across the floxed exon (Fig. 1I). Isolated striatal tissue from P8 control Zswim6+/+; Dlx5/6-Cre [Dlx–wild type (WT)] and mutant Zswim6C/C; Dlx5/6-Cre (Dlx-cKO) mice were used for ATAC-seq, which uses Tn5 transposase activity to quantitatively probe general chromatin accessibility (33, 34). Strikingly, we found increased chromatin accessibility at transcriptional start sites (TSSs) of Dlx-cKO samples as compared to control (Fig. 1J and fig. S1F). Similarly, enhancer regions showed increased chromatin accessibility in Dlx-cKO samples, consistent with a broad loss of repressive chromatin upon deletion of Zswim6 (Fig. 1K). Together, our data suggest that ZSWIM6 protein largely localizes to the nucleus where it associates with specific chromatin regulatory proteins and its disruption in striatal progenitors leads to global opening of chromatin structure.
Zswim6 regulates transcription in both progenitors and developing striatal SPNs
Given the expression of Zswim6 in differentiating striatal progenitors (23) and its role in chromatin regulation, we hypothesized that loss of Zswim6 during embryonic development may result in transcriptional changes in both GABAergic progenitors and developing striatal neurons that normally express Zswim6. To test this, we used 10x snRNA-seq as it allowed us to detect transcriptional changes specific to individual striatal cell types. We isolated nuclei from P8 striatum as this time point marks the onset of excitatory synaptogenesis (35, 36) and provides a contemporaneous existence of both neural progenitors as well as developing and maturing SPNs (Fig. 2A) (37). After standard quality controls and the removal of putative doublets (see Materials and Methods), we obtained 45,732 nuclei from four Dlx-WT and cKO mice, with an average of 2760 unique molecular identifiers (median = 2523) and 1499 genes (median = 1418) detected per nucleus (fig. S2, A to C). We next performed an unbiased clustering analysis to identify cell types within the dataset (Fig. 2A). Using published data on adult and P9 striatum (37, 38), we were able to identify putative SPNs, neurogenic progenitors, and other known neuronal and glial cells type in both Dlx-WT and Dlx-cKO datasets (Fig. 2, A and B, and fig. S2D). We also detected two small clusters that did not correspond to known cell types within the striatum (fig. S2E). Both clusters were predominantly composed of Dlx-cKO nuclei, raising the possibility that they represent aberrant cell types in Dlx-cKO striatum (fig. S2F). Consistent with published work, we found highest levels of Zswim6 in clusters identified as neurogenic progenitors and “immature SPNs” (fig. S2D) (22, 23). Also, in agreement with previous work, Zswim6 was expressed in both dSPN and iSPN clusters, with a slight bias toward higher expression in iSPNs (fig. S2D) (23, 38). When comparing the relative proportion of cell types between genotypes, we noted that Dlx-cKO mutants had an ~43% increase in immature cells (neurogenic progenitors and immature SPNs, 10.4% Dlx-WT versus 14.9% Dlx-cKO; Fig. 2C).
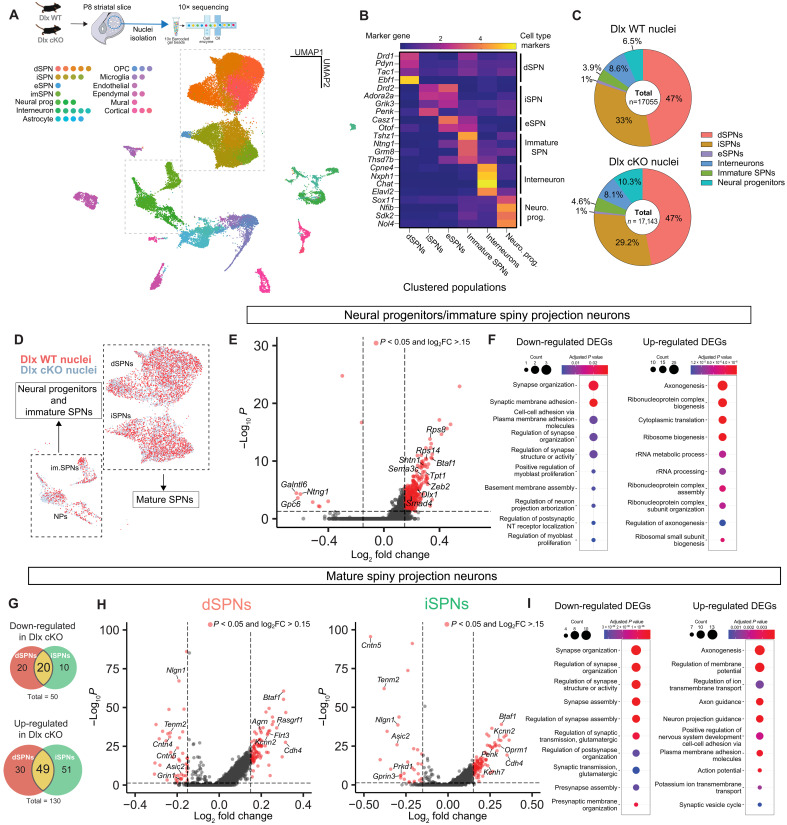
Fig. 2. Transcriptional dysregulation in P8 striatum of Dlx-cKO mice.
(A) snRNAseq workflow (top left). Uniform manifold approximation and projection (UMAP) plot of post-filtered nuclei (center-right, n = 4/4 Dlx WT/cKO animals, 44,460 nuclei. Nuclei belonging to two unidentified clusters in fig. S2E are not displayed). Cell types with cluster colors (left). (B) Heatmap showing normalized expression of select cell type markers across all striatal neurons and neural progenitors. (C) Numbers of indicated cell types in Dlx WT/cKO samples, displayed as percentage of all striatal neurons and neural progenitors. (D) Clusters of cells from UMAP plot shown in (A) that were selected for downstream analysis. Red and blue indicate Dlx WT/cKO nuclei, respectively. (E) Volcano plot of DEGs identified in Dlx WT/cKO neural progenitors/immature SPNs. (F) GO enrichment analysis illustrating enrichment of GO categories in down-regulated and up-regulated DEGs. (G) Venn diagrams illustrating the overlap of up- and down-regulated DEGs in mature iSPNs and dSPNs. (H) Volcano plot of DEGs identified in Dlx-WT/cKO dSPNs (left) and iSPN (right). (I) GO enrichment analysis illustrating enrichment of GO categories in down-regulated and up-regulated DEGs in mature SPNs. Dashed lines in all volcano plots indicate of an p value of <0.05, and log2 fold change (log2FC) of >0.15 or <−0.15. Genes that meet both criteria are colored red. Only genes with and false discovery rate–corrected P value of <0.05 were selected for further analysis.
We next performed a genotypic comparison of the transcriptional profiles of neural progenitors and immature SPNs as well as mature direct and indirect pathway SPNs (Fig. 2D). Consistent with increased chromatin accessibility seen in our ATAC-seq data, we found a strong bias in all cell groups toward an up-regulation of genes in Dlx-cKO samples compared to control (Fig. 2, E and H). This bias was particularly evident in immature cells (neurogenic progenitors and immature SPNs), with 279 up-regulated genes and only 9 down-regulated genes meeting criteria for statistical significance. Examination of the gene ontology groupings for up-regulated genes within immature cells revealed functional enrichment in transcripts relating to axonogenesis, ribosomal biogenesis, and protein translation (Fig. 2F and fig. S1G). We then compared the transcriptomes of control and Dlx-cKO mature SPNs, finding a similar but less prominent bias toward up-regulated genes in mutants (n = 50 down-regulated; n = 130 up-regulated; Fig. 2G). We found that over one-third of dysregulated genes were commonly shared between SPN pathways (40% for down-regulated; ~38% for up-regulated; Fig. 2G). When broadly looking at the Gene Ontology (GO) groupings of dysregulated genes, we found that down-regulated genes centered on synapse assembly and organization, while up-regulated gene networks included transcripts involved in axonogenesis and the regulation of membrane potential, particularly potassium channels (Fig. 2I). Together, these data suggest that Zswim6 functions in differentiating progenitors to repress transcriptional programs that maintain SVZ progenitors in a more immature proliferative state, allowing for the subsequent up-regulation of transcriptional programs involved in the development of SPNs, with a focus on synapse development.
Zswim6 plays a key role in the function of excitatory synapses onto striatal SPNs
Prior work on the whole brain constitutive knockout (KO) of Zswim6 uncovered a simplification of dendritic structure and a modest reduction in spines in the adult striatum (22). Given the broad synapse-related transcriptional changes observed in our Dlx-cKO mutants, we examined whether deletion of Zswim6 altered SPN morphology and synaptic connectivity in a subtype-specific manner. To do this, we crossed the Zswim6C/C with either the D1-Cre allele or the A2a-Cre allele to remove Zswim6 from dSPNs and iSPNs, respectively, creating experimental Zswim6C/C; D1-Cre (D1-cKO)/Zswim6C/C; A2a-Cre (A2A-cKO) mice or their respective littermate controls [Zswim6+/+; D1-Cre (D1-WT)/Zswim6+/+; A2a-Cre (A2A-WT)]. After confirming the SPN-specific deletion of Zswim6 via in situ hybridization (Fig. 3A), we used a dual-virus strategy involving co-injection of AAV5- EF1α::DIO-FLPo-myc and AAVDJ-hSyn::fDIO-mRuby into dorsal striatum of D1-cKO/A2A-cKO and their controls to create optimal sparse labeling for anatomical analyses (fig. S3A). In contrast with brain-wide Zswim6 KOs, we did not observe changes in the total dendritic length, number of primary dendrites, or spine density in either SPN-specific deletion (fig. S3, B to E). We did however note an increase in the number of filopodia-like protrusions per unit length in iSPNs, suggesting a bias toward more immature synapses (fig. S3E).
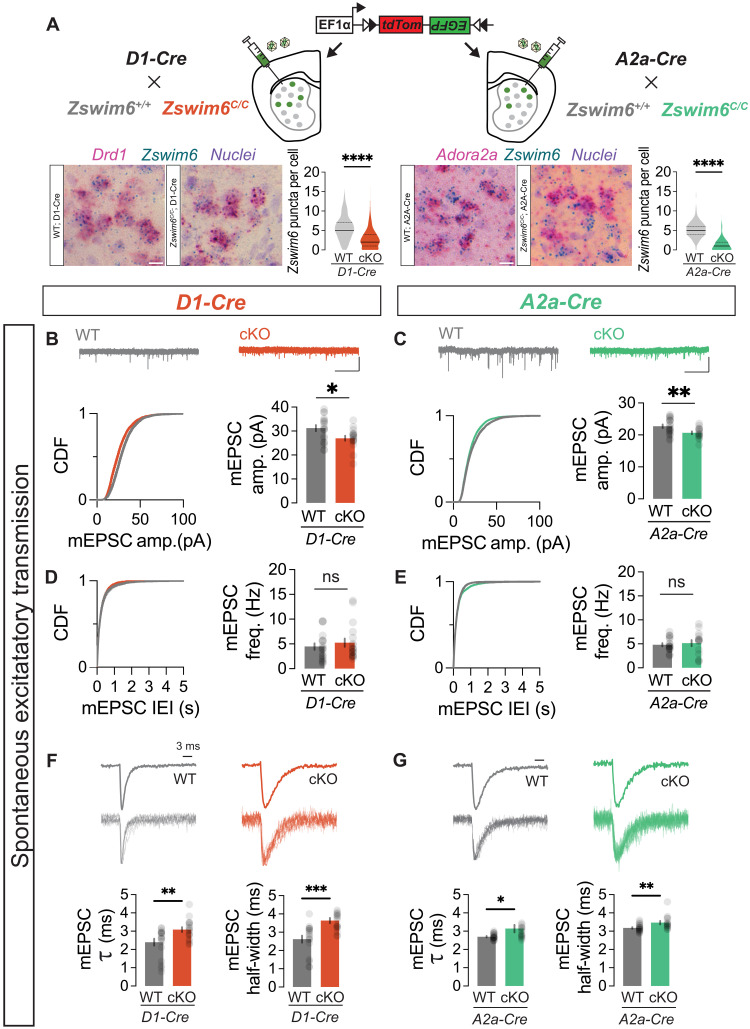
Fig. 3. Zswim6 cKO cell autonomously affects spontaneous excitatory synaptic transmission in SPNs.
(A) SPN subtype-specific cKO labeling strategy (top). BaseScope confirmation of Zswim6 mRNA reduction in D1 and A2A-cKO mice (bottom; scale bars, 10 μm) and quantification (D1-WT/cKO, n = 581/654, two of two animals, ***P < 1 × 10−12; A2A-WT/cKO, n = 627/572, 3 of 3 animals, ***P < 1 × 10−12; t tests). (B) mEPSC traces from D1-WT/cKO neurons. Scale bars, 1 s, 100 pA. Cumulative distribution (left) and average mEPSC amplitudes (right, n = 20/17 cells, five of six animals, *P = 0.0162, t test). (C) Similar to (B), but for A2a-WT/cKO data (n = 20/15 cells, four of four animals, **P = 0.0082, t test). (D) Cumulative distribution (left) and average mEPSC frequencies (right) from D1-WT/cKO neurons (n = 20/17 cells from five of six animals, P = 0.4028, t test). (E) Similar to (D), but for A2a-WT/cKO (n = 20/15 cells from four of four animals, P = 0.6191, t test). (F) Averaged and scaled superimposed traces of individual mEPSCs from D1-WT/cKO neurons. 𝜏 decay constant obtained from fitting to exponential function (left, n = 18/17 cells from five of six animals, **P = 0.0057, two-tailed unpaired t test) and half-width (right, ***P = 0.0002, t test). (G) Similar to (F), but for A2a-WT/cKO. Exponential function (left, n = 20/15 cells from four of four animals, *P = 0.0191, t test) and half-width (right, **P = 0.0061, t test). Data are presented as means ± SEM. Each shaded dot represents data from an individual experiment unless otherwise noted.
We then assessed spontaneous excitatory and inhibitory synaptic transmission in adult striatal circuits following cell-autonomous Zswim6 loss of function (LOF) in both SPN subtypes. We first injected all genotypes with AAV- EF1α::DO-tdTOM-DIO-EGFP to permit identification of WT and Zswim6 cKO SPN subtypes via green fluorescent protein (GFP) fluorescence (Fig. 3A). We measured miniature excitatory post synaptic currents (mEPSC) in the presence of TTX and PTX, finding that Zswim6 LOF resulted in decreased amplitude in both direct SPNs (dSPNs) and indirect SPNs (iSPNs) (Fig. 3, B and C). In contrast, mEPSC frequency was unchanged in both SPN subtypes (Fig. 3, D and E). We further analyzed the kinetics of averaged individual spontaneous events, finding an increased mEPSC decay constant (τ) in both SPN subtypes (Fig. 3, F and G), consistent with an elongated postsynaptic AMPAR response to glutamate. We next investigated spontaneous miniature inhibitory post synaptic currents (mIPSC) in the presence of TTX and 2,3-dihydroxy-6-nitro-7-sulphamoyl-benzo(F)quinoxaline (NBQX), observing no genotype specific differences for either amplitude, frequency, or decay kinetics in either SPN subtype (fig. S3, F to K).
To examine whether these changes in spontaneous excitatory synaptic transmission were recapitulated for action potential–dependent evoked excitatory transmission, we injected both experimental and control genotypes with our Cre-sensitive enhanced GFP (EGFP) virus at 6 to 8 weeks of age. Three weeks later, we prepared acute striatal slices. We then recorded from SPNs in voltage-clamp configuration while applying local electrical stimulation in the presence of PTX to selectively activate excitatory synapses. We first measured the ratio of AMPAR to N-methyl-d-aspartate receptor (NMDAR) currents, finding a reduction in this measure for both SPN subtypes (Fig. 4, A to C). While this could reflect decreases in AMPAR currents or increases in NMDAR currents, the observed reductions in mEPSC amplitudes are more consistent with either a reduction in synaptic AMPAR content or altered AMPAR channel properties, which are supported by the reduced mEPSC amplitude and altered kinetics, respectively (Fig. 3, B, C, F, G). To further test the possibility of NMDAR alterations, we measured the decay of the synaptic currents at +40 mV and found no change in D1-cKO or A2A-cKO SPNs, suggesting no persistence of the developmentally expressed Grin2b NMDAR subunit (fig. S4, A to D). Last, we tested short-term synaptic dynamics by measuring responses to pairs of stimuli at varying temporal intervals. Consistent with prior studies (39, 40), we found that paired-pulse ratios (PPRs) in the dorsal striatum of WT controls were depressing (PPR < 1) across interstimulus intervals (ISIs) in both SPN subtypes (Fig. 4, B and D). Unexpectedly, we found that SPN subtype–specific deletion increased synaptic facilitation, particularly at shorter ISIs (Fig. 4, B and D). Together, these data suggest that Zswim6 function is essential for glutamatergic but not GABAergic synaptic transmission onto both SPN subtypes, with Zswim6 disruption leading to reduced AMPAR content, prolonged AMPAR responses, and short-term excitatory synaptic facilitation.
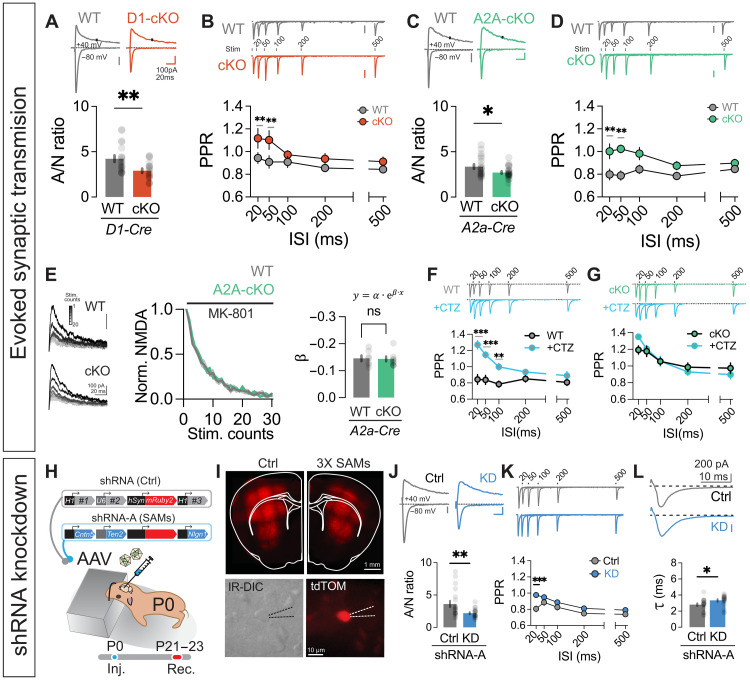
Fig. 4. Zswim6 cKO or knockdown of SAMs found to be down-regulated in Zswim6 cKO striatum disrupts evoked excitatory synaptic transmission.
(A) Evoked EPSC traces from D1-WT/cKO neurons at +40 mV and −80 mV for AMPA/NMDA ratio (top). Ratio comparison (bottom, n = 19/18cells, five of five animals, **P = 0.0042, t test). (B) Paired-pulse traces for D1-WT/cKO (top). Comparison of paired pulse ratio (bottom, n = 17/17 cells from five of five animals, repeated measure (RM)-two-way ANOVA, ISI F4,128 = 5.236, ***P = 0.0006, genotype F1,32 = 8.629, **P = 0.0061, interaction F4,128 = 2.173, P = 0.0757). (C) Similar to (A), but for A2A-WT/cKO (bottom, n = 20/24cells from five of five animals, *P = 0.0152, t test). (D) Similar to (B), but for A2A-WT/cKO (bottom, n = 16/19 cells from six of six animals, RM–two-way ANOVA, ISI F4,132 = 1.982, P = 0.1010, Genotype F1,33 = 10.10, **P = 0.0032, interaction F4,132 = 2.546, *P = 0.0424). (E) Evoked EPSCs at +40 mV in the presence of MK-801 for A2A-WT/cKO cells, indicating progressive blockade of NMDARs (left). Normalized NMDA currents versus stimulation counts (middle). Summary data form MK-801 blockade experiment comparing NMDAR decay constant (β) from A2A-WT/cKO neurons (right, n = 7/8 cells from two of two animals, P = 0.8870, t test). (F) A2A-WT paired-pulse traces ±CTZ (top). Ratio comparison (bottom, n = 9 cells from nine animals, RM–two-way ANOVA). (G) Similar to (F), but for A2A-cKO (bottom, n = 9 cells from five animals, RM–two-way ANOVA, ISI F4,64 = 28.05, ***P < 1 × 10−12, CTZ F1,16 = 0.04561, P = 0.8335, interaction 4,64 = 2.780, *P = 0.0341). (H) Knockdown strategy schematic using shRNA-expressing AAV. (I) Expression of NLS-mRuby2 in striatum (top) and images of individual neurons selected for recording (bottom). (J to L) Representative traces (top) and comparisons for (J) AMPA/NMDA ratio (bottom, n = 20/20 cells from four of four animals, **P = 0.0068, t test) or (K) PPR (n = 20/23 cells from four of four animals, RM–two-way ANOVA, ISI F4,164 = 21.33, ***P < 1 × 10−12, shRNA F1,41 = 5.049, *P = 0.0301, interaction F4,164 = 2.930, *P = 0.0225) or (L) τ (n = 20/20 cells from four of four animals, *P = 0.0195, t test) between control and SAMs shRNA. Significance from Šídák’s post hoc test for RM–two-way ANOVA.
Zswim6 haploinsufficiency phenocopies synaptic deficits in Zswim6 cKO mice
The large body of literature on human genetic disorders has increasingly highlighted the critical role of haploinsufficiency in disease pathogenesis (41, 42). Furthermore, de novo mutations in human Zswim6 which are associated with ID and autism-like phenotypes invariably are found in the context of a remaining wild-type Zswim6 allele (16). Thus, we asked whether conditional deletion of one copy of Zswim6 (Zswim6 cHet) is sufficient to cause synaptic dysregulation. Interestingly, in both Zswim6 cHets we observed reductions in AMPA/NMDA ratio and facilitation of PPRs nearly identical to those observed in Zswim6 cKO (fig. S4, E to H). Thus, SPN synaptic function is extremely sensitive to Zswim6 gene dosage. To assess the time course of our observed synaptic phenotype, we recorded from D1-cHets at an earlier time point (P18 to P22), which aligns with the end of rapid synaptic expansion in striatum (43). We did not find any differences in the AMPA/NMDA ratio or PPR D1-cKOs as compared to their respective controls (fig. S4, I to K). This suggests that Zswim6 gene disruption does not alter the initial induction or specification of synapses but rather disrupts their subsequent maintenance.
Zswim6 disruption alters AMPAR properties and SPN synaptic integration
While paired pulse facilitation is typically interpreted as indicating a decrease in presynaptic release probability, it can also result from postsynaptic mechanisms such as AMPAR desensitization (39). To disambiguate these possibilities, we used the use-dependent NMDAR antagonist MK-801 to probe alterations in release probability. Because MK-801 only binds NMDARs in the presence of bound glutamate, the decay rate of evoked NMDAR currents in the presence of MK-801 can be used to roughly approximate release probability (44–46). Hereafter, our electrophysiology analysis focused on iSPNs, as all prior data showed similar synaptic phenotypes in both SPN subtypes. We found no change in EPSC decay rate in the presence of MK-801 between A2A-cKO and controls (Fig. 4E). Together with the finding that mEPSC frequency is unaltered (Fig. 3E), it seems unlikely that Zswim6 deletion trans-synaptically alters release probability. Given that Zswim6 deletion prolonged spontaneous excitatory currents (Fig. 3, F and G), and prior work showing AMPARs at striatal synapse normally exhibit significant desensitization (39), we hypothesized that increased PPRs in Zswim6 cKO SPNs may result from impaired AMPAR desensitization. To test this, we recorded EPSCs in the presence of cyclothiazide (CTZ), an allosteric modulator of AMPARs that eliminates desensitization (39, 47, 48). Consistent with the substantial impact of AMPAR desensitization on the short-term synaptic dynamics within striatum (39), CTZ significantly increased PPR in control animals (Fig. 4F). In contrast, the elevated PPR in Zswim6 cKO SPNs was not further enhanced in the presence of CTZ (Fig. 4G), consistent with a baseline disruption of desensitization in A2A-cKOs.
Knockdown of synapse adhesions molecules down-regulated in Zswim6 cKO striatum partially phenocopies Zswim6-associated synaptic dysfunction
Given the preponderance of synaptic genes found to be dysregulated in Zswim6 cKO striatum (Fig. 2), we next asked whether knockdown of synapse-adhesion molecules (SAMs) could partially mediate the observed synaptic deficits seen across Zswim6 mutants. To this end, we generated AAVs to simultaneously express short hairpin RNAs (shRNAs) against three of the top down-regulated synaptic genes (Nlgn1, Cntn5, and Tenm2; 3× SAM shRNAs) and NLS-mRuby to identify transduced cells (Fig. 4H). We confirmed robust knockdown of Nlgn1 and Cntn5 in cultured neurons transduced with 3× SAMs shRNA compared to 3× control shRNA, although Tenm2 levels were not significantly reduced (fig. S4L). We injected 3X Control or SAMs shRNA AAVs into striatum of P0 to P1 WT mice and then assessed synaptic function by slice electrophysiology at P21 to P23. The knockdown of SAMs phenocopied the decrease in AMPA/NMDA ratio and partially phenocopied the increased PPR and τ observed in Zswim6 cKO animals (Fig. 4, J to L). In contrast, the knockdown of three nonsynaptic DEGs (Son, Rora, and Celf2) did not affect synaptic transmission (fig. S4, M to P). Thus, while an array of transcriptional dysregulation likely contributes to the aggregate Zswim6 cKO synaptic phenotypes, the transcriptional dysregulation of Nlgn1 and Cntn5 may play a contributing role.
Zswim6 disruption affects the dynamic recruitment of striatum in response to excitatory inputs
We next examined how the synaptic changes resulting from Zswim6 disruption could affect the dynamic recruitment of SPNs by excitatory inputs within striatum. We hypothesized that impaired desensitization would dominate during in vivo–like firing scenarios, effectively enhancing synaptic drive onto SPNs. To test this, we recorded iSPN spiking in current clamp in response to dynamic stimulus patterns in the absence of pharmacological blockers to best preserve local circuit responses (Fig. 5A). Consistent with strongly reduced desensitization in A2A-cKO iSPNs, we found that even with normalized synaptic input (Fig. 5C), iSPNs showed enhanced spiking fidelity as compared to controls across a range of instantaneous input frequencies (Fig. 5D), leading to increased total firing across trials (Fig. 5E). Given the potential impact of changes in excitability on this experiment, we examined rheobase, input resistance, and action potential threshold, finding no differences between genotypes (fig. S5, A to E). These data suggest that Zswim6 deletion leads to altered AMPAR function, including impaired receptor desensitization, which leads to enhanced excitatory synaptic recruitment of iSPNs.
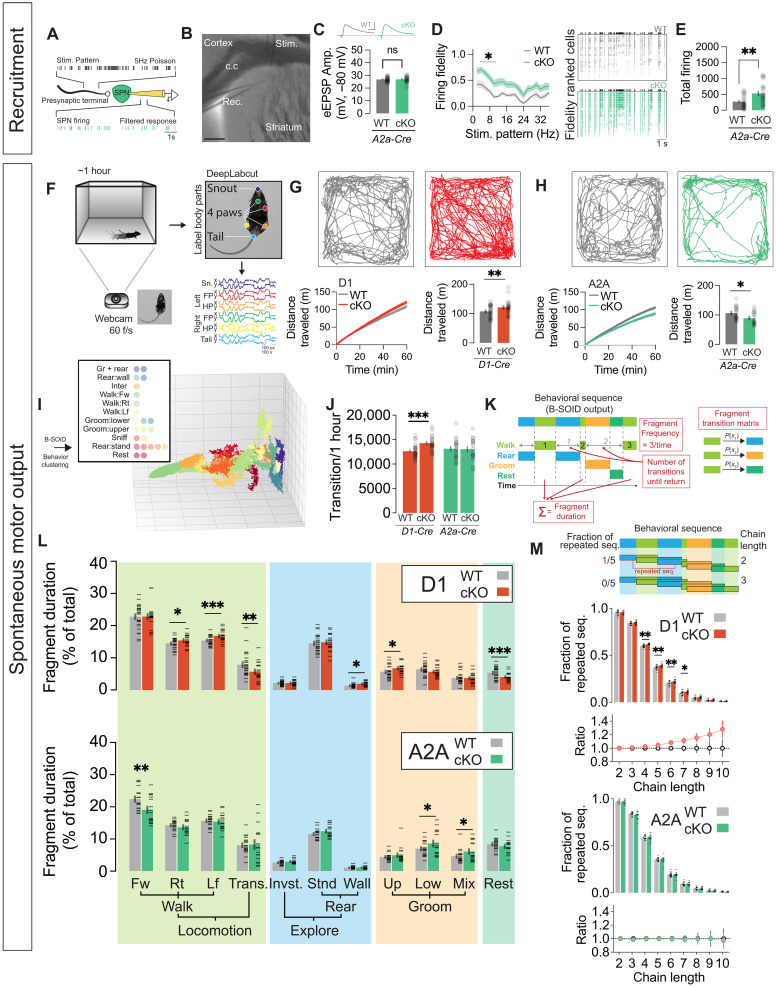
Fig. 5. Zswim6 cKO alters excitatory synaptic recruitment of SPNs and impacts spontaneous motor output.
(A) Cortico-striatal synaptic recruitment experiment design using 5-Hz Poisson stimulation. (B) Representative image showing locations of stimulation and recording pipettes. (C) Traces of evoked EPSP (top) and comparison of eEPSP amplitudes (bottom) from A2A-WT/cKO neurons that were used for subsequent assessment of synaptic recruitment (n = 19/20 cells from 5/4 animals, P = 0.8733, t test). (D) Firing fidelity across stimulation frequencies in A2A-WT/cKO neurons (left, n = 19/20 cells from 5/4 animals, RM–two-way ANOVA, Stim.Pattern F17,629 = 29.30, ***P < 1 × 10−12, genotype F1,37 = 8.208, **P = 0.0068, interaction F17,629 = 1.334, P = 0.1646. Raster plots (right) from all cells indicating SPN firing in relation to stimulation pattern (black). (E) Total firing of A2A-WT/cKO neurons during cortico-striatal recruitment (n = 19/20 cells from 5/4 animals, **P = 0.0050, t test). (F) Spontaneous motor behavior clustering strategy. (G) Tracking traces of D1-WT/cKO (top) for 14 min. Cumulative (bottom-left) and total distance traveled (bottom-right, n = 25/20 animals, **P = 0.0050, t test). (H) Similar to (G), but for A2A-WT/cKO (bottom-right, n = 19/17 animals, *P = 0.0222, t test). (I) B-SOiD clustering visualization. (J) Total number of behavioral segment transitions (D1-WT/cKO n = 25/20 animals, ***P = 0.0004, A2A-WT/cKO n = 19/17 animals, P = 0.7975, t test). (K) B-SOiD analysis illustration. (L) Percent duration of each behavioral cluster in D1- WT/cKO (top) and A2A-WT/cKO (bottom) animals (BCa Bootstrap test). (M) Ratio (top) and proportion of repeatedly identified sequences (bottom) plotted against different lengths of behavioral chains (BCa Bootstrap test). Data are presented as means ± SEM. Each shaded dot represents data from an individual experiment, unless otherwise noted.
SPN-specific disruption of Zswim6 differentially contributes to alterations in spontaneous motor output
Prior analyses of the constitutive Zswim6 KO mice showed increased rearing, rotational biases, stereotyped grooming patterns and an increased sensitivity to subthreshold doses of amphetamine (22). Here, we took advantage of our SPN subtype–specific Zswim6 cKO mice to understand the contribution of striatal Zswim6 function and their resulting synaptic phenotypes to the global generation of spontaneous motor behaviors in an unbiased manner. We recorded D1-cKO, A2A-cKO, and their respective controls through a plexiglass floor; used DeepLabCut to identify paws, nose, and tail; and used these data as input for the unsupervised behavioral classifier, B-SOID (Fig. 5, F and I) (49, 50). Before analyzing the statistics of individual motor states, we examined overall activity levels by tracking a central point between the hindlimbs and tail base. These overall activity measures were consistent with what would be expected if Zswim6 mutations caused enhanced glutamatergic drive onto specific SPN subtypes—D1-cKO mice had enhanced locomotion, while A2A-cKO had decreased locomotion as compared to their respective controls (Fig. 5, G and H).
We next took advantage of the detailed “motor state” information provided by our behavioral classifier to better understand how SPN-specific disruption of Zswim6 contributed to the frequency and patterns of spontaneous motor output. Unsupervised clustering of pose capture data produced 23 motor fragments, of which posthoc observer analysis revealed 11 main groups including locomotion, sniffing, upright standing, rearing, grooming, and rest (Fig. 5, I and L and see Materials and methods for each fragment abbreviation). Subdivisions within these cardinal poses frequently reported directional differences (e.g., walking left versus walking right) or distinct phases of a behavior (grooming face versus grooming body) (51–53). We first examined the total number of transitions between different motor fragments, finding that D1-cKOs had increased transitions as compared to their controls, while A2A-cKOs had a similar number of transitions as their littermate controls (Fig. 5J). We next examined the specific content of motor output (Fig. 5K)—the total percentage of time spent in each motor fragment (fragment duration, Fig. 5L), the number of visits to a given fragment (frequency, fig. S5F), and the number of other fragments visited before returning to a given fragment (transitions until return; fig. S5G). Consistent with our overall activity measures, D1-cKO mice exhibited more total time in “locomotion” and fewer in “rest” states, while A2A-cKO mice had fewer frames in forward locomotion but no changes in rest state occupancy (Fig. 5L). Both D1-cKO and A2A-cKO mice exhibited increases in time spent grooming (Fig. 5L). As most standard behavioral quantifications count the total number of observed behavioral events, we also quantified fragment frequency. Consistent with the increased number of total transitions (Fig. 5J), we found stronger effects for D1-cKOs than A2A-cKOs, with mutation-associated increases in visits to locomotion, “rearing,” and “grooming” motor fragments (fig. S5F). We also found that D1-cKO mice had fewer intervening behaviors before re-engaging in rearing and grooming (fig. S5G).
We carried out multiple analyses to examine whether SPN-specific Zswim6 disruption altered patterns of transition between motor fragments. First, we used transition matrices to quantify the likelihood of progressing between any two specific behavioral fragments (fig. S5H), assessing genotypic differences by dividing the cKO matrices and their respective controls. After thresholding ratios for reproducibility (P < 0.001) and effect size (> ~2.7× difference), we found no consistent fragment transitions in D1-cKOs. Unexpectedly, A2A-cKOs exhibited increased transitions between investigation and grooming, in both directions (fig. S5H). To quantify the extent of general behavioral repetitiveness (i.e., not linked to specific fragments), we used an adopted k-mer analysis that we call behavioral chain analysis (see Materials and Methods) (54). By quantifying the proportion of repeated sequences for increasing chain lengths (Fig. 5M, top), we found that D1-cKO mice exhibited a stronger propensity to repeat sequences of behavior over time. This observed effect was robust and consistent across various behavioral chain lengths, underscoring its reliability. This effect was not observed in the A2A-cKO mice (Fig. 5M). Together, these data suggest that while Zswim6 function in both SPN pathways uniquely contributes to alterations in motor output, Zswim6 disruption in dSPNs has broader behavioral impacts, increasing the total number of state transitions and increasing repetitiveness of behavior chains.
Zswim6 continues to control excitatory synaptic function in adult striatal circuits
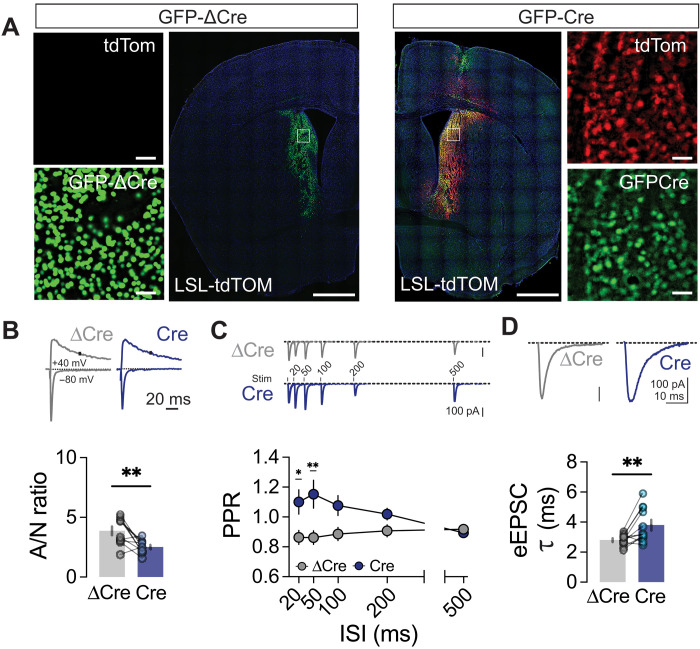
Embryonic deletion of Zswim6 leads to transcriptional dysregulation (Fig. 2) and synaptic dysfunction in mature SPNs (Figs. 3 and 4), suggesting that Zswim6 may control transcriptional programs involved in SPN differentiation and synaptic maturation. In contrast to other brain regions, Zswim6 is also highly expressed in adult SPNs (22, 23). To test whether Zswim6 has similar or divergent functions at this stage, we injected adult (3-month-old) Zswim6C/C animals with either AAV-DJ-hSyn::GFP-Cre or a truncated inactive Cre recombinase control (AAV-DJ-hSyn::GFP-ΔCre; 1 injection per hemisphere of each animal) to conditionally ablate Zswim6 in adult striatum. We confirmed the efficacy of this strategy by injecting both viruses into Ai14 Cre reporter mice, which express tdTomato in the presence of Cre recombinase (Fig. 6A). While nuclear GFP was noted in both hemispheres, only the side with active Cre recombinase had tdTomato+ cells. Three weeks after our injections into Zswim6C/C mice, we assessed evoked excitatory synaptic transmission in acute striatal slices. Similar to earlier SPN-specific Zswim6 deletions, Zswim6C/C neurons expressing GFP-Cre exhibited decreased AMPA/NMDA ratios (Fig. 6B) and increased PPR compared to neurons expressing GFP-ΔCre (Fig. 6C). Furthermore, we found elongated decay kinetics of glutamatergic synaptic responses at −80 mV, consistent with previously observed changes to AMPAR desensitization (Fig. 6D). We found similar changes to AMPA/NMDA ratios, PPRs, and evoked EPSC kinetics in sparse viral injections of Cre, providing further evidence that these synaptic phenotypes are caused by cell-autonomous mechanisms (fig. S6). Thus, Zswim6 is not only crucial for SPN synaptic development but also required for maintenance of synaptic function in adulthood.
Fig. 6. Zswim6 is required in adulthood to maintain excitatory synaptic transmission.
(A) Representative images of GFP-ΔCre (left) and GFP-Cre AAV injection sites in adult Ai14 reporter mice. Cre-mediated expression of tdTomato reporter occurs only in the presence of functional Cre. Scale bars, 1000 and 50 μm for insets. (B) Representative traces of evoked EPSCs from ΔCre and Cre expressing neurons in Zswim6C/C mice, obtained with cells clamped +40 mV and −80 mV to determine AMPA/NMDA ratio (top). Comparison of AMPA/NMDA ratio (bottom, n = 10/10 cells from three animals, **P = 0.0088, t test). (C) Representative traces of from ΔCre and Cre expressing neurons in Zswim6C/C mice, obtained during paired-pulse protocol (top). Comparison of paired pulse ratio (bottom, n = 11/11cells from three animals, RM–two-way ANOVA, ISI F4,80 = 1.343, p = 0.2613, genotype F1,20 = 9.148, **P = 0.0067, interaction F4,80 = 3.497, *P = 0.0110). (D) Representative traces showing EPSC decay from ΔCre and Cre expressing neurons from Zswim6C/C mice (top). Comparison of eEPSC decay constant (𝜏) between ΔCre and Cre expressing neurons in Zswim6C/C mice (bottom, **P = 0.0086, t test). Data are presented as means ± SEM. Each shaded dot represents data from an individual experiment, unless otherwise noted. Significance from Šídák’s post hoc test for RM–two-way ANOVA.
DISCUSSION
Emerging behavioral and genetic analyses point toward striatal dysfunction as a central pathway in NDD pathogenesis. At the behavioral level, striatal circuits have been directly implicated in a range of motor and cognitive control processes that are broadly disrupted in neuropsychiatric disorder (1, 11, 13, 55, 56). At the genetic level, NDD candidate genes are frequently enriched in major striatal cell types (1). Despite this, the genetic and molecular mechanisms of striatal circuit development remain comparatively understudied compared to other brain regions. Here, we identify Zswim6 as a previously undiscovered epigenetic regulator that controls SPN synaptic development and function. Tagged ZSWIM6 protein exhibits robust nuclear localization and interacts with HDAC1 and SUZ12 in heterologous cells. Consistent with the hypothesis that Zswim6 is a chromatin regulator, conditional deletion of Zswim6 in SPN progenitors caused an increase in chromatin accessibility and transcriptional dysregulation. In agreement with the preponderance of dysregulated synaptic genes in Zswim6 cKO SPNs, specific deletion of Zswim6 in either dSPNs or iSPNs produced cell-autonomous alterations in excitatory synaptic transmission including a dramatic reduction in AMPAR desensitization. In the face of dynamic patterns of excitation, impaired AMPAR desensitization enhances SPN recruitment, suggesting a mechanism by which Zswim6 mutation could alter striatal processing and behavior. Unbiased behavioral analysis revealed distinct phenotypes resulting from SPN-subtype specific Zswim6 deletion that are broadly consistent with opposing roles of these SPN subtypes in motor behavior. Last, the deletion of Zswim6 in adulthood recapitulated the synaptic phenotypes observed following late embryonic deletion, suggested a continued requirement for Zswim6 in adulthood to maintain striatal circuit function.
ZSWIM6 modifies chromatin accessibility
Prior structural modeling of the C-terminal of ZSWIM6 revealed similarity to the four paired amphipathic helix (PAH) domains of the transcriptional coregulators SIN3A and SIN3B, two mammalian SIN3 isoforms (17). SIN3 is an evolutionarily conserved master scaffold protein and transcriptional corepressor that forms a complex with HDAC1/HDAC2 (20, 29, 57). Here, we found that tagged ZSWIM6 interacted with HDAC1, suggesting that it functions in part by recruiting core repressive chromatin modifiers. Whether ZSWIM6 directly interacts with HDAC1 or is part of larger protein complex containing HDAC1 remains unclear. Given that the HDAC/SIN3A interaction depends on a region of SIN3A adjacent to the PAH3 domain that is not present in ZSWIM6 (24), it is likely that ZSWIM6 interacts with as-yet unknown factors to form a larger repressive complex incorporating HDAC1. We also found that ZSWIM6 interacts with SUZ12, a component of the PRC2. Similar to SIN3/HDAC complexes, the PRC2 complex mediates closed repressive chromatin states, although it does so via histone H3 methylation at lysine 27 (H3K27) as opposed to histone deacetylation associated with HDAC1 activity (28, 58).
While our data suggest two mechanisms by which Zswim6 may exert epigenetic control, it is also possible that Zswim6 modulates chromatin through other mechanisms such as histone ubiquitination (59). Analysis of ZSWIM6 primary amino acid sequence reveals BC box and Cul2 box domains, key sites for ubiquitin ligase function (17, 22). While ZSWIM8 and its Caenorhabditis elegans homolog, EBAX-1, were both found to function as part of the E3 ligase complex (60), ZSWIM8 exhibits far less sequence homology to ZSWIM4 to ZSWIM6 and exhibits predominantly cytoplasmic localization in heterologous cells (60). Unbiased methods such as mass spectrometry will be necessary to fully characterize the mechanisms of epigenetic regulation by ZSWIM6.
Transcriptional dysregulation following Zswim6 disruption
Our biochemical and ATAC-seq data suggest a role for ZSWIM6 in establishing repressive chromatin architecture, which is typically associated with transcriptional down-regulation. Consistent with this, snRNA-seq revealed a bias toward gene up-regulation in P8 SPNs and immature neurons/progenitors in Dlx-cKO mice (Fig. 2). In progenitors/immature SPNs, up-regulated differentially expressed genes (DEGs) were enriched in GO terms exclusively associated with axonogenesis, protein translation, and ribosomal assembly. In mature SPNs, up-regulated DEGs related to axonogenesis, ion transport, and neuronal excitability. In contrast with immature cells, mature SPNs also exhibited multiple down-regulated DEGs centered on synaptic function. One possible explanation for this finding is that the deletion of Zswim6 has a primary effect of chromatin derepression, and the subsequent perturbed developmental state of these striatal progenitors interferes with downstream transcriptional networks essential for synaptic development. Genetic disruption of the PRC2 repressive complex was found to not only reduce H3K27me3-mediated transcriptional repression in SPNs but also has a secondary effect of down-regulating many SPN-specific genes (61). Alternatively, it is possible that ZSWIM6 also recruits factors that promote transcription of certain genes, a type of complex bifunctionality that has been observed for SIN3 histone deacetylase complexes (20). A more comprehensive analysis of ZSWIM6 interaction networks and chromatin immunoprecipitation sequencing experiments should be informative in this regard.
Zswim6 regulates SPN synaptic function
We found that late-embryonic deletion of Zswim6 in either iSPNs or dSPNs resulted in a dysregulation of spontaneous excitatory transmission, manifest as a reduction in mEPSC amplitude but not frequency and slowed decay kinetics of averaged mEPSC events. We also examined action potential-evoked excitatory synaptic transmission, finding that the ratio of AMPAR to NMDAR-mediated currents was reduced in both SPN knockouts and the decay of spontaneous responses at −80 mV was slower. Together, these data are most consistent with a reduction in the number or properties of synaptic AMPARs. We did not observe changes in measures of spontaneous inhibitory transmission, revealing a specificity not immediately evident from the observed transcriptional dysregulation. We also noted that loss of Zswim6 in both SPN subtypes lead to an increase in the paired pulse ratio, which is frequently associated with alterations in presynaptic release probability. In this case, we observed a similar decay rate of NMDAR currents in the presence of the use dependent antagonist MK-801, strongly suggesting equivalent release probabilities between A2A-WT and A2A-cKO iSPNs.
Because AMPAR desensitization can also affect PPR, and glutamatergic synapses in striatum are known to exhibit strong desensitization (39), we next asked whether a loss of desensitization in Zswim6 cKO SPNs may be responsible for increasing PPR. We found that blocking desensitization with CTZ substantially increased PPR in WT SPNs, while this effect was occluded in Zswim6 cKO SPNs. The simplest interpretation of these experiments is that AMPAR responses in SPNs, which are normally strongly desensitizing, lose this property in Zswim6 cKO SPNs. This hypothesis is further supported by the prolonged decay kinetics observed in the spontaneous excitatory responses. Candidate mechanisms underlying the observed changes in AMPAR-mediated transmission include reduced synaptic AMPAR localization, a change in AMPAR subunit composition, or changes in AMPAR-interacting proteins such as transmembrane AMPA receptor regulatory proteins (TARPs) (62, 63). Additional electrophysiology analyses have revealed two other important aspects of Zswim6 contribution to synaptic function—(i) The gene demonstrates haploinsufficiency, as D1-cHets and A2a-cHets both exhibited phenotypes of similar effect size as those in their respective cKO genotypes (fig. S4, E to H), and (ii) the impact of Zswim6 disruption are less relevant to the initial formation of synapses but rather affect synapse maintenance, as evidenced by the absence of phenotypes at P18 to P22 that are readily observed at 3 months in D1-cHets (fig. S4, I to K). These phenotypes are consistent with that of synaptic adhesion molecules such as Neuroligin and Neurexin, wherein gene disruption does not affect the initial formation or specification of synapses (64, 65).
While our P8 snRNA-seq data revealed the dysregulation of many synaptic genes, these did not include AMPAR subunits, AMPAR auxiliary proteins, or other obvious candidates for direct modulation of AMPARs. In our first attempts to link the specific impacts of Zswim6-associated transcriptional dysregulation with our observed physiological phenotypes, we “screened” the top down-regulated genes shared by SPN subtype using multi-shRNA constructs. While we believe that the Zswim6 synaptic phenotype is likely the result of small transcriptional changes across many synapse-associated molecules, here we attempted to probe a small number of potential candidate contributors. Knockdown of Cntn5 and Nlgn1 partially phenocopied the synaptic phenotype seen across Zswim6 mutants, while simultaneous knockdown of nonsynaptic targets (Son, Rora, and Celf2) had no effects on striatal circuits. These data provide initial evidence that the synaptic phenotypes could be downstream of transcriptional dysregulation of synaptic adhesion molecules, for which disruptions are known to result in mislocalization of synaptic proteins including glutamate receptors (66–69). Last, the deletion of Zswim6 in adult striatum phenocopied the dysregulated synaptic transmission we observed following embryonic Zswim6 deletion, indicating that Zswim6 is required for synaptic function in adulthood. This may reflect a continued requirement for Zswim6 in maintaining mature SPN identity. Consistent with this idea, the disruption of the repressive PRC2 complex (containing the Zswim6-interacting protein SUZ12) was shown to induce ectopic expression of non-SPN genes and down-regulation of SPN-specific genes (61). Thus, continued repression of non-SPN genes appears to be necessary for functional SPN maintenance.
Zswim6-associated alterations in circuit output and behavior
The input-output transformations occurring at the neural circuit level integrate the effects of dynamic patterns of synaptic excitation and inhibition together with cellular excitability. To best model the effects of Zswim6 mutations on striatal circuit processing, we used a previously established naturalistic stimulus protocol in slice (70), finding that Zswim6 cKO iSPNs were recruited more efficiently than WT iSPNs. Because we did not observe evidence of altered intrinsic excitability in Zswim6 cKO iSPNs, we suggest that reductions in AMPAR desensitization outweigh reductions in total synaptic AMPAR current when SPNs are integrating dynamic excitatory input. Together, we believe that changes in the short-term filtering properties of excitatory synapses onto SPNs effectively produce an enhancement of synaptic drive in mice with disrupted Zswim6 function. These results are similar to work at retinogeniculate synapses, wherein loss of CKAMP44, an AMPAR auxiliary subunit, leads to reduced synaptic AMPAR content and reduced receptor desensitization. In an analogous manner to what we observe in Zswim6 cKOs, the net effect of CKAMP44 LOF is to increase ON- and OFF-firing rate increases within thalamus (71).
The established roles of striatal SPN pathways in motor output allowed us to make several basic predictions about behavioral effects of Zswim6 mutation–associated changes in circuit output. Consistent with models of bidirectional spiny neuron control of motor behavior and our observed enhancements of excitatory drive, we found that D1-cKOs exhibited increased distance traveled, while A2A-cKOs had decreased distance traveled in open field exploration. Our use of a machine classifier on pose-tracking data provided further insight into the effects of Zswim6 on substructure of spontaneously generated motor states. At the broadest level, we found that D1-cKOs had an increased number of transitions between differing motor states, while A2A-cKO did not alter total transition numbers. Furthermore, we noted the D1-cKOs spent more time walking, turning, and rearing, variations that could largely be explained by increased selection frequency of these fragments. In a competitive model of basal ganglia function, in which different dSPNs subpopulations represent alternative actions, a broad increase of excitatory drive onto dSPNs might be expected to increase transitions between motor states as multiple possible fragments are brought closer to selection threshold. We also found evidence for repeated motor sequences from both SPN subtype disruptions, with D1-cKOs showing more general repetition of motor sequences and A2A-cKOs showing biasing toward a specific fragment transition. Further testing of these ideas will require simultaneous in vivo recordings during spontaneous motor exploration. Nevertheless, our data demonstrate how similar synaptic phenotypes across SPN subtypes can generate the full range of motor abnormalities observed in Zswim6 mutants.
Multi-hit NDD models and convergence at the synapse
NDDs exhibit complex multi-genic etiologies, wherein the disruption of multiple loci jointly contributes to pathophysiology. Studies of the genetic contributions to neurodevelopmental disease have revealed strong signals for genes encoding synaptic proteins and epigenetic regulators (8, 9, 14, 15). Despite the puzzling nature of these divergent signals, these data have been taken to mean that it is the combined or synergistic effects of mutations that sufficiently perturb neural circuit function to affect behavior. Our work on Zswim6 lends support to the idea that epigenetic dysfunction may disproportionately impair synaptic function as a central pathogenesis mechanism. Furthermore, this pathophysiological process is seen with loss of just one Zswim6 allele, highlighting the sensitivity of synapses to proper chromatin regulation. Another key part of this chromatin synapse model is that epigenetic dysregulation can affect multiple synaptic molecules simultaneously, a key concept given the highly redundant nature of the synaptic adhesion landscape. Simultaneous disruption of epigenetic and synaptic molecules will provide more direct tests of this convergent pathogenesis model.
MATERIALS AND METHODS
Animal
Animal experiment procedures were approved by the University of Pennsylvania Institutional Animal Care and Use Committee, and all experiments were conducted in accordance with the National Institutes of Health Guidelines for the Use of Animals (protocol: 805643). This study follows the ARRIVE guidelines for reporting animal research. The ARRIVE checklist has been completed. For all electrophysiology and behavioral experiments, animals were generated by intercrossing a Zswim6 conditional allele (wherein exon 3 is surrounded by loxP sites) with either A2a-Cre mice from the Jackson Laboratory (B6.FVB(Cg)-Tg(Adora2a-cre)KG139Gsat/Mmcd) or D1-Cre mice from the Jackson Laboratory (B6;129-Tg(Drd1-cre)120Mxu/Mmjax). iSPN or dSPN cKO mice and their littermate controls were obtained by intercrossing Zswim6C/+; A2a-Cre or Zswim6C/+; D1-Cre animals, respectively. For ATAC-seq and snRNA-seq experiments, Dlx5/6-Cre mice from the Jackson Laboratory [Tg(dlx5a-cre)1Mekk/J, stock no. 008199] were intercrossed with the Zswim6 conditional allele. Following this initial cross, experimental animals were obtained from the following breedings: Zswim6C/C; Dlx5/6-Cre × Zswim6C/C (for Dlx-cKO mice) and Zswim6+/+; Dlx5/6-Cre × Zswim6+/+ (for Dlx-WT mice). For the primary culture and neonatal injection, we used timed pregnant mice at E16.5, which were obtained from Charles River Laboratories. All were grouped with littermates on a 12:12 light-dark cycle and provided ad libitum food and water.
Cloning of plasmids
The Zswim6 coding sequence was cloned from Zswim6 cDNA isolated from E13.5 mouse brain and then subcloned into pKMyc, a mammalian expression vector (Addgene, catalog no. 19400). Because of the highly repetitive, guanine-cytosine–rich (GC-rich) nature of the N-terminal coding sequence, a codon-optimized version of the sequence corresponding to amino acids 1 to 731 was synthesized (GenScript) and used to replace the endogenous cDNA sequence. The Flag-Zswim6 expression plasmid was created by inserting a 3X Flag sequence directly upstream of the Zswim6 coding sequence using the Xba I and Age I restriction sites. To create the Ef1α-Flag-Zswim6 AAV construct, an AAV backbone (gift from W. Xu, Südhof laboratory) was first modified by removing the existing multiple cloning site (MCS), β-globin intron, human growth hormone polyadenylate [poly(A)] and sequences and replacing them with a linker containing a new MCS. Flag-Zswim6 was then subcloned into this AAV backbone using Xba I and Eco RI sites and then replacing the existing cytomegalovirus promoter with the Ef1α core promoter using Mlu I and Xba I sites. Last, the SV40 poly(A) sequence was added downstream of Flag-Zswim6 using Xho I and Hpa I sites. The ∆NLS variant of the Ef1α-Flag-Zswim6 AAV construct was made by removing the N-terminal sequence of Zswim6 that includes the NLS by cutting with Bam HI and Bsp EI, the latter of which cuts within the Zswim6 coding sequence (CDS). This sequence was replaced with a sequence (synthesized by IDT) missing the 30 nucleotides corresponding to the NLS but identical to WT Zswim6 in the flanking regions. pAAV-mDlx-CreGFP was made from the existing pAAV-mDlx-NLS-mRuby2 backbone (Addgene, #99130) by removing NLS-mRuby using Spe I and Asc I sites and replacing it with CreGFP fusion protein sequence PCRed from pAAV-hSyn-CreGFP (72). To generate Ef1α-FLEX-Flag-Zswim6, the Ef1α-Flag-Zswim6 was converted into a FLEX construct in three cloning steps that avoided polymerase chain reactioning (PCRing) Zswim6 CDS due to its highly repetitive, GC-rich nature near the N terminus. First pairs of loxP sites were PCRed from pAAV-Ef1α-DIO-ChIEF-2A-mVenus (construct previously cloned by M.V.F) and cloned into Ef1α-FLEX-Flag-Zswim6 so as to flank Flag-Zswim6 on 5′ and 3′ sides. By adding strategically positioned Xba I and Eco RI sites to the loxP PCR products after the insertion of the two pairs of loxP sites, the resulting backbone contained Eco RI and Xba I, which were flanked by the double loxP sites and in the opposite 5′-3′ order as the original plasmid. Thus, Flag-Zswim6 could then be cut from Ef1α-FLEX-Flag-Zswim6 using Xba I and Eco RI sites (which flank Flag-Zswim6 on the 5′ and 3′ sides, respectively) and ligated into the modified, loxP site-containing plasmid in reverse orientation.
3X shRNA plasmid backbones were generated by PCRing the H1 and tandem H1-U6 promoters from the L315 lentiviral vector (gift from Südhof laboratory) and subcloned into an AAV backbone (gift from W. Xu, Südhof laboratory). The hSyn promoter was PCRed from pAAV-hSyn-EGFP (Addgene, #50465) and then subcloned into the H1-U6-H1 backbone. Last, NLS-mRuby2 was PCRed from the pAAV-mDlx-NLS-mRuby2 backbone (Addgene, #99130) and subcloned to generate pAAV-H1-U6-hSyn-NLS-mRuby-H1. Each polymerase III promoter had adjacent cloning sites for shRNAs, which were sequentially cloned into the 3X shRNA backbone. Sequences for control and knockdown shRNAs (table S1) were obtained from published work (73–77), with the exception of Rora shRNA, which we designed using the shRNA design tool freely available from VectorBuilder. Control shRNA sequences were specifically selected from publications, which did not detect synaptic phenotypes when expressing their control shRNAs compared to untransfected neurons (78–80).
Primary neuronal culture
Primary cortical neurons were prepared from E16.5 CD1 mouse embryos (Charles River Laboratories). Timed pregnant females were deeply anesthetized with isoflurane and then decapitated. Cortices were dissected in “dissecting solution” (136 mM NaCl, 5.4 mM KCl, 0.17 mM Na2HPO4, 0.22 mM KHPO4, 0.25 mM Hepes, and 43.8 mM sucrose in ddH2O) and then incubated in papain solution [papain (10 U/ml), 0.5 mM l-cysteine, and 0.25 mM EDTA, Worthington Biochemical, cataog no. LS003126, in dissecting solution] for 5 min at 37°C. Cortices were washed 1X in dissecting solution and then washed 3× in trypsin inhibitor solution containing 1% bovine serum albumin (BSA) and 1% trypsin inhibitor (Sigma-Aldrich, catalog no. T6522) in dissection solution. Trypsin inhibitor solution was removed and replaced by Neurobasal media (Thermo Fisher Scientific, catalog no. 21103049). A single-cell suspension was then made by triturating four to five times with a fire-polished Pasteur pipette. Before plating, glass coverslips (Bellco Glass) were coated with poly-d-lysine (MP Biomedicals) and Laminin (Corning). Neurons were plated on coverslips in 24-well plates (150,000 neurons per well) in Neurobasal supplemented with B27 (Thermo Fisher Scientific), GlutaMAX (Thermo Fisher Scientific), and penicillin-streptomycin (Invitrogen). Medium was replaced with fresh supplemented Neurobasal media (kept in incubator to maintain temperature and pH) 1 to 2 hours after plating. To prevent glial overgrowth, AraC was added at DIV5 at a final concentration of 0.5 μM. For Flag-Zswim6 expression experiments, 2 μl of AAV-EF1α-Flag-Zswim6 virus was added to each well on DIV7. For experiments comparing localization of Flag-ZSWIM6 and Flag-∆NLS-ZSWIM6, 2 μl of each virus in separate wells was added at DIV5.
For cortico-striatal cocultures used for shRNA validation, primary neurons from E16.5 embryos were isolated using the same protocol, except that dorsal striatum/LGE was isolated a dissociated separately from cortex. Total neurons (300,000; 65% striatal and 35% cortical) were plated and maintained in 12-well plates under the same conditions as described above for cortical cultures, except that AraC was omitted due to apparent toxicity to striatal neurons, which we observed in pilot experiments. Viruses were added at DIV3 to DIV6, and neurons were harvested for quantitative PCR (qPCR) experiments 6 to 7 days later.
Stereotaxic surgery
Intracranial surgery was conducted on a stereotaxic surgery frame (Kopf Instrument, Model 1900) under isoflurane anesthesia (1.5 to 2% + oxygen 1 liter/min). Animal body temperature was maintained at 30°C during surgery using a feedback thermocontroller (Harvard Apparatus, #50722F). Skin was cleaned with Nair hair remover followed by application of betadine to disinfect the area. Before surgery, bupivacaine (2 mg/kg) was administered subcutaneously, and the mouse was given a single dose of meloxicam (5 mg/kg). Skin was carefully opened along the anterior-posterior (AP) midline, bregma was set to zero. A craniotomy was performed with an arm-attached drill above the target site. Virus or tracer was loaded into mineral oil (Sigma-Aldrich, M3516)–filled glass pipette (WPI, TW100F-3) and delivered at rate of 30 nl/min using a micro-infusion pump (Harvard Apparatus, #70-3007). At least 5 min after infusion, the pipette was slowly withdrawn (1 mm/min) from the brain, and the skin was sutured. Animals were monitored up to 1 hour following regaining consciousness, then transferred to the home cage, and monitored after 24, 48, and 72 hours. Injection coordinates for the dorsal striatum were: AP, +0.65 mm; medio-lateral (ML), ±1.6 mm; dorso-ventral (DV), −2.8 mm.
For the viral injection procedure on neonatal mice, each pup was placed on ice to induce anesthesia without direct contact, preventing cold-related injuries. Once immobilized, the pup was transferred to a three-dimensional printed stereotaxic frame (81), which secured their posture, and surrounded with ice to maintain anesthesia. The virus was loaded into a glass pipette using a Nanoject II (Drummond Instrument, no. 3-000-204) system. A small incision was made on the skull at the designated injection site using a sharp syringe, and then the injection pipette was positioned at this site. The virus was then injected into three different DV positions; each site received 100 nl of virus. The injection coordinates for the pups were AP, +0 mm; ML, ±1.4 mm; and DV positions at −2.2, −1.82, and −1.4 mm.
For conditional gene deletion in SPNs, male mice aged 5 to 6 weeks were stereotactically injected with purified AAV.dj/Syn-GFP-Cre and AAV.dj/Syn-GFP-ΔCre (control) into the right and left striatal hemispheres, respectively. To achieve sparse labeling of SPNs, viral solutions were diluted 1:10 in sterile phosphate-buffered saline (PBS) before injection. Electrophysiological recordings were performed 3 weeks after injection to allow sufficient viral expression and gene deletion.
Chromatin sequential salt extraction
Salt extractions were performed as described (82). Briefly, a 2X radioimmunoprecipitation assay (RIPA) solution was made [100 mM tris (pH 8.0), 2% NP-40, and 0.5% sodium deoxycholate] and mixed with varying concentrations of a 5 M NaCl solution to generate RIPA containing 0, 100, 200, 300, 400, and 500 mM NaCl. Pelleted cells were resuspended in modified buffer A [25 mM Hepes (pH 7.6), 25 mM KCl, 5 mM MgCl2, 0.05 mM EDTA, 0.1% NP-40, and 10% glycerol] with protease inhibitors and rotated at 4°C for 5 min. Nuclei were pelleted by centrifugation at 6000g for 5 min at 4°C. Supernatant was removed and saved, and 200 μl of RIPA with 0 mM NaCl and protease inhibitors was added to the sample. Samples were mixed by pipetting 10 times and incubated on ice for 3 min before centrifuging for 3 min at 6500g at 4°C. Supernatant was saved, and RIPA steps were repeated for all NaCl concentrations. Samples were boiled and sonicated before analyzing by Western blot.
Flag-ZSWIM6 co-IP for chromatin-associated proteins
Chromatin immunoprecipitation was performed using a protocol adapted from Lee et al. (83). Frozen pellets of ~25 M HEK cells (American Type Culture Collection, CRL-11268) (untransfected and transfected with Flag-Zswim6) were resuspended in 3 ml of lysis buffer 1 [50 mM Hepes-KOH (pH 7.5 to 8), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1X cOmplete protease inhibitor, and 1X PhosSTOP phosphatase inhibitor], rotated 10 min at 4C, and pelleted by centrifuging for 5 min at 1350g, 4°C. Pellets were resuspended in 3 ml of lysis buffer 2 [10 mM tris-HCl (pH 8), 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 1 mM PMSF, 1X cOmplete protease inhibitor, and 1X PhosSTOP phosphatase inhibitor], rotated 10 min at RT, and pelleted as described above. Pellets were then resuspended in 3 ml of lysis buffer 3 [10 mM tris-HCl (pH 8), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, 1% Triton X-100, 1 mM DTT, 1 mM PMSF, 1X cOmplete protease inhibitor, and 1X PhosSTOP phosphatase inhibitor]. Samples were sonicated 3X, 3 s each, using a tip sonicator. Next, 0.1× volume of 100% Triton-X was then added to each sample. Samples were centrifuged for 10 min at 18,000g at 4°C, and the supernatant was aliquoted for input (5%) and IPs (95%). For Flag immunoprecipitation, 200 μl of anti-Flag M2 beads (Millipore, M8823) were added to IP samples and rotated overnight at 4°C. Flag-ZSWIM6 binding partners were eluted by competition with 3X Flag peptide (150 μg/ml; Millipore, F4799). Input and IP samples were boiled 10 min with loading buffer containing 2.5% β-mercaptoethanol. Candidate binding partners were analyzed by Western blotting using the following antibodies: anti-Flag (1:1000; Sigma-Aldrich, F1804), anti-SUZ12 (1:1000; Cell Signaling Technology, 3737), anti-HDAC1 (1:1000; Abcam, ab19845), and anti-histone H3 (1:10,000; Abcam, ab1791).
cDNA synthesis and RT-qPCR
After allowing shRNA viruses to incubate on corticostriatal cultures for 6 to 7 days, total RNA was extracted using the QIAGEN RNeasy Mini Kit. RNA concentration was determined using a Nanodrop, and then first-strand cDNA synthesis was carried out using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). cDNA synthesis was carried out using 200 ng of total RNA with random hexamer primers. qPCR was performed using Sybr Premix Ex Taq II (Takara), with 5 ng of cDNA per reaction, on an Applied Biosystems StepOne Plus instrument. All qPCR primers (table S2) flanked exon-exon junctions to avoid amplification of genomic DNA, and the specificity was confirmed by single peaks in the melt curves for all primers used. To calculate normalized expression of target genes, Ct values for each gene in each sample were normalized to the average Ct values of ActB and Hprt in that sample.
ATAC sequencing
Nuclei for ATAC-seq were first isolated from striatal tissue. Briefly, the tissue was homogenized in homogenization buffer [0.32 M sucrose, 5 mM CaCl2, 10 mM tris-Cl (pH 8), 3 mM MgAc2, 0.1% Triton X-100, and 0.1 mM EDTA] and was gently pipetted on top of a sucrose cushion [1.8 M sucrose, 10 mM tris-Cl (pH 8), and 15 mM MgAc2] in an ultracentrifuge tube. Nuclei were pelleted in an ultracentrifuge for 90 min at 4°C at 100,000g. The homogenization buffer and sucrose cushion were removed, and 100,000 nuclei per sample were collected for ATAC. Nuclei were resuspended in cold lysis buffer [100 μl per 100,000 cells; 10 mM tris-Cl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 0.1% (v/v) NP-40, 0.1% (v/v) Tween 20, and 0.01% (v/v) digitonin] and washed in wash buffer [10 mM tris-Cl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, and 0.1% (v/v) Tween 20]. Transposition was performed with Tagment DNA TDE1 (Illumina, 15027865). Transposition reactions were cleaned with AMPure XP beads (Beckman, A63880), and libraries were generated by PCR with NEBNext High-Fidelity 2× PCR Master Mix (NEB, M0541). Library size was confirmed on a Bioanalyzer before sequencing on the NextSeq 550 platform (40–base pair read length, paired end). For ATAC-seq analysis, alignments were performed with Bowtie2 (2.1.0) (84) using the mm10 genome with the pipeline at https://github.com/shenlab-sinai/chip-seq_preprocess. Reads were mapped using NGS plot. Three biological replicates per condition were sequenced, and each replicate yielded greater than 60 million reads. Replicates were merged to generate average profile plots.
Nuclei extraction
P8 male and female mice were deeply anesthetized with isoflurane and then decapitated. Brains were quickly removed and transferring to sucrose cutting solution (26 mM NaHCO3, 1 mM NaH2PO4, 2.5 mM KCl, 228 mM sucrose, 11 mM glucose, 7 mM MgSO4, and 0.5 mM CaCl2). Sections (400 μm) were cut on a vibratome (VT1200s, Leica) and then transferred to artificial cerebrospinal fluid (ACSF; 119 mM NaCl, 26.2 mM NaHCO3, 1 mM NaH2PO4, 2.5 mM KCl, 11 mM glucose, 1.3 mM MgSO4, and 2.5 mM CaCl2). Sucrose and ACSF solutions were both bubbled with a 95% O2 and 5% CO2 mixture at all times. With the slices in ACSF, striatum was then dissected from each slice (four slices per animal), then snap-frozen in liquid nitrogen, and stored at −80. Nuclei extraction was carried out using a protocol optimized by the Macosko laboratory (dx.doi.org/10.17504/protocols.io.7xchpiw). Fluorescence-activated cell sorter (FACs) capture was omitted, as no fluorophores were present and minimal nuclei clumping was observed.
10X snRNA-seq and initial data processing
Nuclei capture, 10X sequencing, read alignment, and initial quality control steps were carried out by the Next Generation Sequencing Core (NGSC) at the Perelman School of Medicine (University of Pennsylvania). Initial data processing was carried out using the CellRanger v6.1.2 pipeline. Briefly, raw base call (BCL) files were demultiplexed using the mkfastq command, generating FASTQ files. Extracted and quality control passed FASTQ reads were aligned to the GRCm38/mm10 genome, and single-cell nucleus count data were generated using the count command.
snRNA-seq analysis was done using the Seurat analysis pipeline in R (Seurat v4.3.0) (85). First, nuclei containing <200 features per nucleus or >4000 features per nucleus or greater than 1% mitochondrial RNA reads were discarded. Next, data in each biological replicate (four of four Dlx WT/cKO samples) was processed individually according to the Seurat analysis pipeline, identifying principal components (PCs) in each sample. Jackstraw analysis was used to determine statistically significant PCs. Each sample was found to have at least 40 statistically significant PCs, so 40 PCs were used for all downstream steps.
DoubletFinder (v2.0.3) (86) was then used to identify and remove doublets. The DoubletFinder pipeline was used to identify optimal pK values for each sample, and a doublet rate of 8% was assumed on the basis of the number of nuclei loaded for 10X capture. Following identification of putative doublets, data from all samples were merged into a single Seurat object, and data were renormalized and clustered. Before discarding putative doublets, the DoubletFinder pipeline was run again on the merged dataset, resulting in identification of additional putative doublets (n = 1232) that generally clustered with previously identified doublets. Thus, all putative doublets (n = 6704 of 52,436 nuclei) were removed, and an additional cluster that was composed of ~40% doublets and localized in between other identifiable clusters was removed. Last, the remaining singlets (n = 45,732) were renormalized, scaled, and clustered for downstream analysis.
Cluster identification and DEG analysis
Cell types represented in clusters were determined by identifying top DEGs for each cluster and manually cross referencing these against marker genes in published neonatal (P9) and adult (P60) datasets (37, 38). Of the 36 clusters, only two were not easily identifiable and were both were composed largely of Dlx cKO nuclei (fig. S2, E and F). For differential gene expression (DEG) analysis, we separately analyzed clusters corresponding to dSPNs, iSPNs, and a group composed of neurogenic progenitors and immature SPNs (immature cells). DEGs were identified with the FindMarkers Seurat function using the Wilcoxon rank sum test. Criteria for inclusion as a DEG were an average log2 fold change of 0.15 and a false discovery rate–adjusted P value < 0.05. In addition, DEGs corresponding to mitochondrial genes or X/Y chromosome genes were discarded. Functional enrichment was determined using the ClusterProfiler R package (87).
Immunohistochemistry
Mice were deeply anesthetized with pentobarbital then transcardially perfused with 1X PBS followed by 4% paraformaldehyde (PFA) in PBS. Brains were post-fixed overnight at 4°C in 4% PFA, after which PFA was removed and replaced with PBS. A vibratome was used to cut 50-μm sections, which were then blocked for 1 hour at room temperature in PBS containing 0.5% Triton X-100, 10% FBS, and 1% BSA. Sections were incubated in the same blocking buffer with the appropriate primary antibodies overnight at 4°C, washed 3X in PBS, then incubated with secondary antibodies for 2 hours at room temperature, and washed 3X in PBS before mounting (see the Antibodies section). Sections were mounted on slides and coverslipped with VectaMount mounting media (Vector Laboratories).
Immunocytochemistry
Neuronal cultures were fixed on DIV9 to DIV12 after addition of AAV-Ef1α-Flag-Zswim6 or AAV- Ef1α-Flag-∆NLS-Zswim6 virus at DIV5 to DIV7. Neuronal medium was removed and replaced with 4% PFA + 2% sucrose in 1X PBS and incubated for 8 min at room temperature. Coverslips were then washed 3× in PBS and then blocked for 1 hour at room temperature in PBS containing 1% ovalbumin (Sigma-Aldrich), 0.2% gelatin from cold water fish (Sigma-Aldrich), 0.01% saponin (Sigma-Aldrich), and 0.1% Triton X-100 (Sigma Aldrich). Coverslips were then incubated in the same blocking solution with appropriate primary antibodies overnight at 4°C, washed 3X in PBS, then incubated with appropriate secondary antibodies for 2 hours at room temperature (see the Antibodies section) and washed 3X in PBS before mounting on slides using Aqua-Mount mounting media (Sigma-Aldrich).
Dendrite and spine morphology
Mice were euthanized 2 weeks following injection of AAV.DJ/Syn-fDIO-mRuby + AAV5/ef1α-DIO-FLPo-Myc to achieve sparse labeling of dSPNs or iSPNs in D1-Cre or A2a-Cre mice, respectively. Mice were deeply anesthetized with pentobarbital then transcardially perfused with 1X PBS followed by 4% PFA in PBS. Brains were post-fixed overnight at 4°C in 4% PFA, after which PFA was removed and replaced with PBS. A vibratome was used to cut 100-μm sections, which were immediately mounted and coverslipped with VectaMount (Vector Laboratories).
BaseScope in situ hybridization
BaseScope [Advanced Cell Diagnostics Inc. (ACD)] was used to detect Zswim6, Adora2a, and Drd1 mRNA transcripts. BaseScope is similar to RNAscope but leverages chromogenic dyes in place of fluorescent dyes to enable higher sensitivity detection of short RNA or DNA sequences. We designed a Zswim6 BaseScope probe consisting of two ZZ pairs targeting the E3-E4 and E4-E5 exon-exon junctions. Tissue for the BaseScope assay was obtained from 4- to 6-month-old WT and Zswim6 cKO mice crossed to each Cre line used in the study. Mice were deeply anesthetized with pentobarbital then transcardially perfused with 1X ribonuclease-free PBS followed by 4% PFA in PBS. Brains were post-fixed overnight at 4°C in 4% PFA, after which PFA was removed and washed with 1X PBS. PBS was then removed and replaced with 30% (w/v) sucrose/PBS. Brains were then stored at 4°C for at least 48 hours before cryosectioning. Free-floating cryosections (20 μm) were obtained using a cryostat microtome. The BaseScope duplex assay was then carried out according to the ACD protocol, the only modification being that H2O2 pretreatment was carried out before mounting sections on slides in wells containing 0.3% H2O2 (to avoid bubbles forming under sections on slides). Sections were then imaged on a Leica DM6 microscope.
Spine and dendrite morphology
Brain sections were imaged on a Leica SP5 confocal using a 40× oil immersion objective. Images were acquired in resonance scanning mode (8000 Hz) as z-stacks, with images at 0.5-μm intervals. Images for analysis of dendrites were acquired using 1.7× zoom to capture the entire dendritic arbor, while images for spine analysis for captured with 2.5× zoom to obtain higher resolution. When selecting cells for dendrite morphology analysis, care was taken to select cells farther away from the viral injection site, where the density of labeled cells was low and entire dendritic arbors of individual cells could be visualized. For analysis of dendrite morphology, raw z-stacks were imported into ImageJ and analyzed by manually tracing the entire dendritic arbor of each neuron through the entire z-stack. All analysis was done with the experimenter blinded to genotype. For spine number and morphology quantification, raw z-stacks were imported into ImageJ, then a gaussian blur (sigma = 0.7) was applied. Spines and filopodia were manually counted by an experimenter blinded to genotype.
In vivo Flag-ZSWIM6 localization
Images were acquired as z-stacks (0.5-μm intervals) in conventional scan mode (400 Hz) on a Leica SP5 confocal using a 40X oil immersion objective. Raw image stacks were pre-processed with the “remove outliers” ImageJ function (threshold = 50, radius = 2.0), and then a gaussian blur was applied (sigma = 0.6). Neurons with clear Flag-ZSWIM6, GFP, and CTIP2 fluorescent signals were selected for analysis. To prevent misidentifying out-of-focus Flag-ZSWIM6 signal as being localized to the nucleus, a maximum projection of only two optical sections (1 μm) was made, corresponding to the approximate center of the nucleus in the axial plane. Regions of interest (ROIs) corresponding to the entire cell body (GFP outline) and the nucleus (CTIP2 outline) were manually drawn, and Flag- ZSWIM6 fluorescent intensity was measured in each region. The total intensity of cytoplasmic Flag-ZSWIM6 was calculated by subtracting the intensity within the nucleus (CTIP2 outline) from the intensity in the entire cell body (GFP outline).
In vitro Flag-ZSWIM6 localization
Images were acquired as z-stacks (0.5-μm intervals) in resonant scan mode (8000 Hz) on a Leica SP5 confocal using a 40× oil immersion objective. A Gaussian blur was applied (sigma = 0.5), and then maximum projections of each channel were used for analysis using custom ImageJ macros. Briefly, MAP2 and H2AZ channels were manually thresholded to generate masks and ROIs for the somatodendritic compartment and nucleus, respectively. Flag-ZSWIM6 intensity was measured within the soma/dendrite mask and separately measured within the nucleus. Total fluorescent intensity within the soma/dendrites outside of the nucleus was calculated by subtracting the total intensity within nucleus from total intensity in the soma/dendrite mask (which included the nucleus). A corresponding calculation was done for soma/dendrite area, allowing for calculation of mean fluorescence intensity in the soma/dendrites excluding the nucleus. For initial experiments characterizing Flag-ZSWIM6 localization (fig. S1), the soma/dendrite mask included the soma and proximal dendrites. For comparison of WT Flag-Zswim6 and Flag-∆NLS-Zswim6 localization (Fig. 1), we noted that the mislocalized ∆NLS variant primarily accumulated outside of the nucleus but within the soma. To best capture this phenotype in comparison to WT Flag-Zswim6, we restricted our analysis to the cell soma and nucleus only.
BaseScope analysis
Using a 20× objective, two to three fields of view were obtained for each animal. The number of Zswim6 puncta per SPN (identified by hematoxylin stain and Adora2a or Drd1 mRNA for iSPNs and dSPNs, respectively) was manually quantified by an experimenter blinded to genotype.
Primary antibodies
Mouse anti-Flag M2 [immunohistochemistry (IHC): 1:500, immunocytochemistry (ICC): 1:1000, Western: 1:000; Sigma-Aldrich, F1804], rat anti-Ctip2 (IHC:1:500; Abcam, ab18465), rabbit anti-Flag (ICC: 1:500; Millipore, F7425), rabbit anti-H2A.Z (ICC: 1:500; Abcam, 4174), chicken anti-MAP2 (ICC: 1:5000; Novus Biologicals NB300-213), rat anti-SUZ12 (Western: 1:1000; Cell Signaling Technology, 3737), rabbit anti-HDAC1 (Western: 1:1000; Abcam, ab19845), rabbit anti-histone H3 (Western: 1:10,000; Abcam, ab1791), mouse anti-GAPDH (Western: 1:3000; Abcam, ab8245), and rabbit anti-Lamin B1(Western: 1:10,000; Abcam, ab16048).
Secondary antibodies
All secondary antibodies for ICC and IHC were diluted 1:500. The following antibodies were used: goat anti-mouse Cy3 (the Jackson Laboratory), goat anti-rat Cy5 (Invitrogen), donkey anti-rabbit Alexa-488 (Invitrogen), goat anti-Chicken Alexa-488 (the Jackson Laboratory), goat anti-rabbit Alexa-555 (Invitrogen), and goat anti-mouse Alexa-647 (Invitrogen). For Western secondary antibodies, all secondary antibodies for Western blotting were diluted 1:5000. The following antibodies were used: goat anti-rabbit horseradish peroxidase (HRP) (Abcam, ab6721) and sheep anti-mouse HRP (Cytiva, NA931).
Electrophysiology
Male and female adult mice (2 to 3 months old) were used for all acute slice electrophysiology experiments except for P0 shRNA experiments (recorded at P21 to P23; Fig. 4, H to L) and the “developmental analysis” (recorded at P18 to P22 in fig. S4, I to K). Before recording, dual-color virus (AAV-DJ-EF1α::DO/DIO-tdTomato/eGFP) was injected into dorsal striatum to visualize SPN subtypes. Sample size was determined on the basis of the experimental design. For paired conditions, we aimed to record from >10 neurons, while for unpaired conditions, we targeted >17 neurons per group to ensure adequate statistical power and account for variability in electrophysiological responses. All mice were allowed to recover for 3 to 4 weeks before recording. Mice were deeply anesthetized and trans-cardially perfused with ice-cold ACSF containing 124 mM NaCl, 2.5 mM KCl, 1.2 mM HaH2PO4, 24 mM NaHCO3, 5 mM Hepes, 13 mM glucose, 1.3 mM MgSO4, and 2.5 mM CaCl2. After perfusion, the brain was quickly removed, submerged, and coronally sectioned on a vibratome (VT1200s, Leica) at 250-μm thickness in ice-cold ACSF. Slices were transferred to NMDG-based recovery solution at 32°C of the following composition 92 mM NMDG, 2.5 mM KCl, 1.2 mM NaH2PO4, 30 mM NaHCO3, 20 mM Hepes, 25 mM glucose, 5 mM sodium ascorbate, 2 mM thiourea, 3 mM sodium pyruvate, 10 mM MgSO4, and 0.5 mM CaCl2. After 12- to 15-min recovery, slices were transferred to room temperature ACSF chamber (20° to 22°C) and left for at least 1 hour before recording. Following recovery, slices were placed in a recording chamber, fully submerged at a flow rate of 1.4 to 1.6 ml/min and maintained at 29° to 30°C in oxygenated (95% O2 and 5% CO2) ACSF.
In voltage-clamp recordings, recording pipettes were fabricated by pulling borosilicate glass (World Precision Instruments, TW150-3). These pipettes exhibited a tip resistance ranging from 3 to 5 megohm when filled with an internal solution comprising the following concentrations: 130 mM CsMeSO4, 5 mM CsCl, 10 mM Hepes, 2.5 mM MgCl, 0.6 mM EGTA, 1 mM QX-314, 10 mM Na-phosphocreatine, 4 mM NaATP, and 0.3 mM Na–guanosine 5′-triphosphate (GTP), with a pH adjusted to 7.3 to 7.4 using CsOH. Striatal neurons were identified under visual control using infrared differential interference contrast (IR-DIC) optics (Olympus, BX51). Visual identification of D1-SPNs and D2-SPNs was based on expression of GFP/tdTomato (Chroma filter cube, #49002 and #49005). To record miniature excitatory postsynaptic currents (mEPSCs), the cell was voltage-clamped at −80 mV, and spontaneous events were recorded for a minimum of 10 min. This recording was performed in the presence of PTX (100 μM; Sigma-Aldrich P1675) and TTX citrate (1 μM; Cayman Chemical company 14964) to block inhibitory and action potential-dependent synaptic activity, respectively. Data for analysis were collected from an 8- to 10-min time window following the establishment of whole-cell configuration. For each recording session, a minimum of 200 events were analyzed to generate cumulative distribution functions using the built-in function in Mini Analysis (Synaptosoft).
The decay constant () of mEPSCs was determined by fitting the 10 to 90% decay phase of the trace. mEPSC events were aligned on the basis of 10 to 90% rise phase from 0 to peak. The fitting was performed using the following equation
| (1) |
To measure the AMPA/NMDA ratio, cellular responses were recorded by clamping voltage at two different membrane potentials in presence of PTX. The first potential was set at −80 mV to measure inward AMPA currents, while the second potential was set at +40 mV to measure outward AMPA/NMDA currents. The NMDA current was isolated by averaging the response during a specific time window of 50 to 55 ms following stimulation. The decay constants (, ) of NMDA current were determined by fitting a window of the trace ranging from 50 to 1500 ms. The fitting was performed using the following equation
| (2) |
To measure paired-pulse ratio, the AMPA current was recorded using a protocol consisting of two consecutive electrical stimulations with varying interstimulation intervals in presence of PTX. The intervals used in this study were set at 20, 50, 100, 200, and 500 ms. Each set of sweeps was separated by a 15-s interval to mitigate potential plasticity effects resulting from the previous stimulus. To obtain use-dependent NMDA decay, slice was incubated in the ACSF containing MK-801 (50 μM; Tocris 0914) and NBQX (10 μM; Abcam, ab120046), PTX. After a 10-min incubation period, electrical stimulation was delivered with a consistent intensity. Each set of sweeps was separated by a 20-s interval for data acquisition.
To record mIPSCs, an ACSF solution containing NBQX, AP5 (50 μM; Cayman Chemical Company, 14539), and PTX was used. The recording pipette was filled with an internal solution consisting of 135 mM CsCl, 10 mM Hepes, 0.6 mM EGTA, 2.5 mM MgCl2, 4 mM NaATP, 0.3 mM NaGTP, 10 mM 2Na-phosphocreatine, 0.1 mM spermine, and 1 mM QX-314, with the pH adjusted to 7.3 to 7.4 using CsOH. The cells were held at a membrane potential of −70 mV for the recordings. The decay constants (, ) of mIPSC were determined by fitting the 10 to 90% decay phase of the trace. The fitting was performed using the following equation
| (3) |
For current-clamp recordings, recording pipette filled with internal solution containing 140 mM k-gluconate, 5 mM KCl, 0.2 mM EGTA, 2 mM MgCl2, 10 mM Hepes, 4 mM MgATP, 0.3 mM NaGTP, and 10 mM 2Na-phosphocreatine (pH adjusted to 7.3 to 7.4 using KOH). To assess the passive membrane properties, cells were maintained at their resting membrane potential, and a ramp protocol was applied. The ramp protocol consisted of a gradual increase in current injection over a 500 ms period, from 0 to 500 pA. The rheobase was inferred by identifying the time point at which the first action potential occurred in response to the ramp protocol. To assess synaptic recruitment using near-natural stimulus pattern, we used stimulus protocol based on the in vivo firing pattern (70). To establish the stimulation intensity before delivering the stimulus pattern, the stimulation intensity was adjusted to evoke a response of approximately 25 mV (Fig. 5C). Before administering the in vivo pattern stimulus, the membrane potential was maintained at −60 mV using current injection, and the protocol was executed for a duration of 5 min.
Recordings were performed using a MultiClamp 700B (Molecular Devices) and Igor7-based Recording Artist (WaveMetrics; recording artist addon, developed by R. C. Gerkin, github: https://github.com/rgerkin/recording-artist), filtered at 2.8 kHz, and digitized at 10 kHz. Axon terminals were stimulated with brief (0.2 ms) pulses using isoflex isolator. Input and series resistance were monitored continuously, and experiments were discarded if either parameter changed by >20%. Data were analyzed using mini-analysis (Synaptosoft) and Igor7. Data analysis was conducted using coded files, with group identities revealed only after analysis completion. Key findings were confirmed across three separate cohorts of animals tested over a 12-month period.
Video recording in open field
The open field setup was constructed using transparent acrylic material, with dimensions of 38.1 cm by 30.48 cm by 25.4 cm. The open field box was enclosed within a custom-built cabinet equipped with two fans to reduce ambient noise levels and provide ventilation. A recording webcam (Brio, Logitech) was positioned approximately 60 cm below the open field box to capture movement. Each recording session was 1 hour. The frame rate was set at a fixed value of 60 frames per second (fps).
Behavioral analysis
To extract the coordinates for each body part, we employed the DeepLabCut (DLC) (49) algorithm (version 2.2.0.6) for training a model. In each iteration, we manually labeled 10 body parts, including the snout, left forepaw, right forepaw, torso, left hindpaw, right hindpaw, pelvis, tail base, tail-mid, and tail tip. In every iteration, body parts were manually identified by visual inspection from at least 75 frames extracted by the DLC using a k-means clustering algorithm. Human observers labeled the body parts based on their visual observations. A total of 10 reference videos were used to train the model, and the training process consisted of a maximum of 1,030,000 iterations. The model was trained using the resnet_50 network and the default augmentation method. After training the model, the reference videos that were labeled by the DLC model were further inspected through human observation. A total of five training iterations were conducted, with inspections carried out between each iteration. To handle low confidence labeled body parts with a likelihood below 0.9, a correction process was performed using the built-in fillmissing function in MATLAB. The fillmissing function used the shape-preserving piecewise cubic spline interpolation method to interpolate low confidence frames.
To track animal movement, three lower limb body parts, namely, the left hindpaw, right hindpaw, and tail base, were averaged to approximate a central location, which was then used for conducting basic open field locomotion analysis. For quantification of the diversity of spontaneously generated motor output, the estimated coordinates obtained from DLC were transferred into the B-SOiD (50) algorithm to perform unbiased behavior clustering. To train the B-SOiD model effectively and capture genotypic differences in behavior, 10 reference videos were carefully selected. These reference videos encompassed a range of genotypes, including four videos from D1-WT, two videos from D1-cKO, two videos from A2A-WT, and two videos from A2A-cKO. By incorporating samples from each genotype, the training process aimed to encompass the behavioral variations specific to each genotype, facilitating subsequent analysis and comparison of genotypic differences in behavior. The total duration of the videos amounted to 240 min, with a frame rate of 60 fps. Following the recommendation of the B-SOiD creators, we used six body parts for the analysis. These body parts included the snout, left forepaw, right forepaw, left hindpaw, right hindpaw, and tail base. To reduce the dimensionality of the behavioral instance features, a nonlinear transformation was applied. After transformation of a total of 134,990 behavioral instances in the reduced eight-dimensional feature space, the instances were subjected to clustering. The clustering process was performed with a range of 0.4 to 3.0% of the minimum cluster size, resulting in the identification of 23 distinct clusters representing different behavior patterns. To generate the model, a random forest classifier was used. The model performance of the classifier was evaluated on 20% of the data, and it achieved an accuracy of 94%. Video snippets from each of the 23 clusters were extracted from all 10 reference videos for further visual inspection. This process resulted in the generation of 100 video snippets for each cluster, approximately 2300 video snippets in total (23 clusters × 10 snippets/cluster × 10 videos). To identify and categorize specific behaviors represented by each cluster, a human experimenter examined each snippet of the data. By visually inspecting the behavior patterns within each cluster, the experimenter determined the corresponding behavior cluster. In addition, each cluster was inspected using a transition matrix to identify natural behavior sequences. If a specific sequence of behaviors, such as initiating rearing, rearing, and terminating rearing, occurred consistently and sequentially within a cluster, then they were combined into the same “fragment.” This process aimed to capture and group together behavior sequences that commonly occurred together, allowing for a more coherent and accurate representation of the animal’s natural behavior patterns. These include forward walk (FW), left turn (Lf), right turn (Rt), transition from walk (Trans.), Investigation (Invst.), rearing with straight up (Stand), wall rearing (Wall), upper body grooming (Upper), lower body grooming (Lower), both types of grooming (Mix), and rest. The analysis of % duration, frequency, and the number of clusters until return was conducted.
For the analysis of behavior chains, the entire list of behavior fragments was divided into segments ranging from 2 to 10 elements with moving window. Within each combination of behavior fragments, individual fragments were analyzed to determine the number of times they appeared. Behavior chains that were counted only once within the session were defined as “nonrepeated sequences,” while all other behavior chains were categorized as “repeated sequences.” The “fraction of repeated sequences” was quantified by taking the ratio of the number of unique repeated sequences to the total count of behavior sequences identified. Sample size for behavioral experiments was determined on the basis of previous studies in the field and power analysis. We aimed for a minimum of 15 animals per group to detect biologically significant differences with adequate statistical power, accounting for potential variability in behavioral responses. Data analysis was conducted using coded files, with group identities revealed only after analysis completion.
Acknowledgments
We acknowledge the NGSC at the Perelman School of Medicine, University of Pennsylvania for performing single-nucleus sequencing and initial data processing. We also acknowledge the excellent technical support of A. Jean-Louis, A. Cowell, and W. Tu.
Funding: This work was supported by the National Institute of Mental Health grant R01 MH118369 (to M.V.F.), the Tourette Association of America Young Investigator Award (to M.V.F.), Eagles Autism Foundation grant (to M.V.F.), the Foundation for OCD Research (to M.V.F.), the Penn Autism Spectrum Program of Excellence (to M.V.F. and E.K), the National Institute of Mental Health grants 1DP2MH129985 (to E.K) the Klingenstein-Simons Fellowship from the Esther A. & Joseph Klingenstein Fund (to E.K.), and the Simons Foundation, the Alfred P. Sloan Foundation Research Fellowship FG-2020-13529 (to E.K.)
Author contributions: Conceptualization: K.C., N.T.H., D.J.T., S.A.A., E.K., and M.V.F. Methodology: K.C., N.T.H., D.J.T., S.A.A., E.K., and M.V.F. Software: N.T.H. Validation: K.C., N.T.H., F.D., I.B., and M.V.F. Formal analysis: K.C., N.T.H., E.R.F., S.L., F.D., I.B., and M.V.F. Investigation: K.C., N.T.H., E.R.F., S.L., J.G., F.D., I.B., D.J.T., M.V.F. Resources: K.C., N.T.H., S.L., F.D., D.J.T., S.A.A., M.V.F. Data curation: K.C., N.T.H., and M.V.F. Writing—original draft: K.C., N.T.H., and M.V.F. Writing—review and editing: K.C., N.T.H., D.J.T., S.A.A., E.K., and M.V.F. Visualization: K.C., N.T.H., F.D., and M.V.F. Supervision: K.C., N.T.H., S.A.A., E.K., and M.V.F. Project administration: K.C., N.T.H., E.K., and M.V.F. Funding acquisition: E.K. and M.V.F.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data necessary to evaluate the conclusions of this study are presented in the paper or the Supplementary Materials. Additional datasets generated and analyzed during this study have been archived and are publicly available at Zenodo (10.5281/zenodo.13861327). ATAC-seq and snRNA-seq data are available from NCBI GEO: GSE282824 (ATAC-seq) and GSE282825 (snRNA-seq). Data processing pipeline used for ATAC-seq can be found at https://github.com/shenlab-sinai/chip-seq_preprocess. Data processing pipeline for snRNA-seq can be found on the M.V.F. laboratory Github (https://github.com/Fuccillo-Lab).
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 and S2
REFERENCES AND NOTES
- 1.Fuccillo M. V., Striatal circuits as a common node for autism pathophysiology. Front. Neurosci. 10, 27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Souza L., Channakkar A. S., Muralidharan B., Chromatin remodelling complexes in cerebral cortex development and neurodevelopmental disorders. Neurochem. Int. 147, 105055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ring H. A., Serra-Mestres J., Neuropsychiatry of the basal ganglia. J. Neurol. Neurosurg. Psychiatry 72, 12–21 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCutcheon R. A., Abi-Dargham A., Howes O. D., Schizophrenia, dopamine and the striatum: From biology to symptoms. Trends Neurosci. 42, 205–220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balleine B. W., Delgado M. R., Hikosaka O., The role of the dorsal striatum in reward and decision-making. J. Neurosci. 27, 8161–8165 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang J., Gilman S. R., Chiang A. H., Sanders S. J., Vitkup D., Genotype to phenotype relationships in autism spectrum disorders. Nat. Neurosci. 18, 191–198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skene N. G., Bryois J., Bakken T. E., Breen G., Crowley J. J., Gaspar H. A., Giusti-Rodriguez P., Hodge R. D., Miller J. A., Munoz-Manchado A. B., O'Donovan M. C., Owen M. J., Pardinas A. F., Ryge J., Walters J. T. R., Linnarsson S., Lein E. S., Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, Sullivan P. F., Hjerling-Leffler J., Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 50, 825–833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Rubeis S., He X., Goldberg A. P., Poultney C. S., Samocha K., Cicek A. E., Kou Y., Liu L., Fromer M., Walker S., Singh T., Klei L., Kosmicki J., Shih-Chen F., Aleksic B., Biscaldi M., Bolton P. F., Brownfeld J. M., Cai J., Campbell N. G., Carracedo A., Chahrour M. H., Chiocchetti A. G., Coon H., Crawford E. L., Curran S. R., Dawson G., Duketis E., Fernandez B. A., Gallagher L., Geller E., Guter S. J., Hill R. S., Ionita-Laza J., Gonzalez P. J., Kilpinen H., Klauck S. M., Kolevzon A., Lee I., Lei I., Lei J., Lehtimaki T., Lin C. F., Ma'ayan A., Marshall C. R., McInnes A. L., Neale B., Owen M. J., Ozaki N., Parellada M., Parr J. R., Purcell S., Puura K., Rajagopalan D., Rehnstrom K., Reichenberg A., Sabo A., Sachse M., Sanders S. J., Schafer C., Schulte-Ruther M., Skuse D., Stevens C., Szatmari P., Tammimies K., Valladares O., Voran A., Li-San W., Weiss L. A., Willsey A. J., Yu T. W., Yuen R. K., The DDD Study, Homozygosity Mapping Collaborative for Autism, UK10K Consortium, The Autism Sequencing Consortium, Cook E. H., Freitag C. M., Gill M., Hultman C. M., Lehner T., Palotie A., Schellenberg G. D., Sklar P., State M. W., Sutcliffe J. S., Walsh C. A., Scherer S. W., Zwick M. E., Barett J. C., Cutler D. J., Roeder K., Devlin B., Daly M. J., Buxbaum J. D., Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moretto E., Murru L., Martano G., Sassone J., Passafaro M., Glutamatergic synapses in neurodevelopmental disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry 84, 328–342 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Alabi O. O., Davatolhagh M. F., Robinson M., Fortunato M. P., Vargas Cifuentes L., Kable J. W., Fuccillo M. V., Disruption of Nrxn1α within excitatory forebrain circuits drives value-based dysfunction. eLife 9, e54838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothwell P. E., Fuccillo M. V., Maxeiner S., Hayton S. J., Gokce O., Lim B. K., Fowler S. C., Malenka R. C., Sudhof T. C., Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell 158, 198–212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor S. C., Ferri S. L., Grewal M., Smernoff Z., Bucan M., Weiner J. A., Abel T., Brodkin E. S., The role of synaptic cell adhesion molecules and associated scaffolding proteins in social affiliative behaviors. Biol. Psychiatry 88, 442–451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peixoto R. T., Chantranupong L., Hakim R., Levasseur J., Wang W., Merchant T., Gorman K., Budnik B., Sabatini B. L., Abnormal striatal development underlies the early onset of behavioral deficits in Shank3B(−/−) mice. Cell Rep. 29, 2016–2027.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabriele M., Lopez Tobon A., D'Agostino G., Testa G., The chromatin basis of neurodevelopmental disorders: Rethinking dysfunction along the molecular and temporal axes. Prog. Neuropsychopharmacol. Biol. Psychiatry 84, 306–327 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Ciptasari U., van Bokhoven H., The phenomenal epigenome in neurodevelopmental disorders. Hum. Mol. Genet. 29, R42–R50 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer E. E., Kumar R., Gordon C. T., Shaw M., Hubert L., Carroll R., Rio M., Murray L., Leffler M., Dudding-Byth T., Oufadem M., Lalani S. R., Lewis A. M., Xia F., Tam A., Webster R., Brammah S., Filippini F., Pollard J., Spies J., Minoche A. E., Cowley M. J., Risen S., Powell-Hamilton N. N., Tusi J. E., Immken L., Nagakura H., Bole-Feysot C., Nitschke P., Garrigue A., de Saint Basile G., Kivuva E., DDD Study, Scott R. H., Rendon A., Munnich A., Newman W., Kerr B., Besmond C., Rosenfeld J. A., Amiel J., Field M., Gecz J., A recurrent de novo nonsense variant in ZSWIM6 results in severe intellectual disability without frontonasal or limb malformations. Am. J. Hum. Genet. 101, 995–1005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith J. D., Hing A. V., Clarke C. M., Johnson N. M., Perez F. A., Park S. S., Horst J. A., Mecham B., Maves L., Nickerson D. A., University of Washington Center for Mendelian Genomics, Cunningham M. L., Exome sequencing identifies a recurrent de novo ZSWIM6 mutation associated with acromelic frontonasal dysostosis. Am. J. Hum. Genet. 95, 235–240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanagishita T., Eto K., Yamamoto-Shimojima K., Segawa O., Nagata M., Ishihara Y., Miyashita Y., Asano Y., Sakata Y., Nagata S., Yamamoto T., A recurrent de novo ZSWIM6 variant in a Japanese patient with severe neurodevelopmental delay and frequent vomiting. Hum. Genome Var. 8, 16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ripke S., O'Dushlaine C., Chambert K., Moran J. L., Kahler A. K., Akterin S., Bergen S. E., Collins A. L., Crowley J. J., Fromer M., Kim Y., Lee S. H., Magnusson P. K., Sanchez N., Stahl E. A., Williams S., Wray N. R., Xia K., Bettella F., Borglum A. D., Bulik-Sullivan B. K., Cormican P., Craddock N., de Leeuw C., Durmishi N., Gill M., Golimbet V., Hamshere M. L., Holmans P., Hougaard D. M., Kendler K. S., Lin K., Morris D. W., Mors O., Mortensen P. B., Neale B. M., O'Neill F. A., Owen M. J., Milovancevic M. P., Posthuma D., Powell J., Richards A. L., Riley B. P., Ruderfer D., Rujescu D., Sigurdsson E., Silagadze T., Smit A. B., Stefansson H., Steinberg S., Suvisaari J., Tosato S., Verhage M., Walters J. T., Multicenter Genetic Studies of Schizophrenia Consortium, Levinson D. F., Gejman P. V., Kendler K. S., Laurent C., Mowry B. J., O'Donovan M. C., Owen M. J., Pulver A. E., Riley B. P., Schwab S. G., Wildenauer D. B., Dudbridge F., Holmans P., Shi J., Albus M., Alexander M., Campion D., Cohen D., Dikeos D., Duan J., Eichhammer P., Godard S., Hansen M., Lerer F. B., Liang K. Y., Maier W., Mallet J., Nertney D. A., Nestadt G., Norton N., O'Neill F. A., Papadimitriou G. N., Ribble R., Sanders A. R., Silverman J. M., Walsh D., Williams N. M., Wormley B., Psychosis Endophenotypes International Consortium, Arranz M. J., Bakker S., Bender S., Bramon E., Collier D., Crespo-Facorro B., Hall J., Iyegbe C., Jablensky A., Kahn R. S., Kalaydjieva L., Lawrie S., Lewis C. M., Lin K., Linszen D. H., Mata I., McIntosh A., Murray R. M., Ophoff R. A., Powell J., Rujescu D., Van Os J., Walshe M., Weisbrod M., Wiersma D., Wellcome Trust Case Control Consortium 2, Donnelly P., Barroso I., Blackwell J. M., Bramon E., Brown M. A., Casas J. P., Corvin A. P., Deloukas P., Duncanson A., Jankowski J., Markus H. S., Mathew C. G., Palmer C. N., Plomin R., Rautanen A., Sawcer S. J., Trembath R. C., Viswanathan A. C., Wood N. W., Spencer C. C., Band G., Bellenguez C., Freeman C., Hellenthal G., Giannoulatou E., Pirinen M., Pearson R. D., Strange A., Su Z., Vukcevic D., Donnelly P., Langford C., Hunt S. E., Edkins S., Gwilliam R., Blackburn H., Bumpstead S. J., Dronov S., Gillman M., Gray E., Hammond N., Jayakumar A., McCann O. T., Liddle J., Potter S. C., Ravindrarajah R., Ricketts M., Tashakkori-Ghanbaria A., Waller M. J., Weston P., Widaa S., Whittaker P., Barroso I., Deloukas P., Mathew C. G., Blackwell J. M., Brown M. A., Corvin A. P., McCarthy M. I., Spencer C. C., Bramon E., Corvin A. P., O'Donovan M. C., Stefansson K., Scolnick E., Purcell S., McCarroll S. A., Sklar P., Hultman C. M., Sullivan P. F., Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 45, 1150–1159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams G. E., Chandru A., Cowley S. M., Co-repressor, co-activator and general transcription factor: The many faces of the Sin3 histone deacetylase (HDAC) complex. Biochem. J. 475, 3921–3932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarova K. S., Aravind L., Koonin E. V., SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem. Sci. 27, 384–386 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Tischfield D. J., Saraswat D. K., Furash A., Fowler S. C., Fuccillo M. V., Anderson S. A., Loss of the neurodevelopmental gene Zswim6 alters striatal morphology and motor regulation. Neurobiol. Dis. 103, 174–183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang C. C., Kuo H. Y., Chen S. Y., Lin W. T., Lu K. M., Saito T., Liu F. C., Developmental characterization of schizophrenia-associated gene zswim6 in mouse forebrain. Front. Neuroanat. 15, 669631 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laherty C. D., Yang W. M., Sun J. M., Davie J. R., Seto E., Eisenman R. N., Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89, 349–356 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Chen Z. F., Paquette A. J., Anderson D. J., NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 20, 136–142 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Schoenherr C. J., Anderson D. J., The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science 267, 1360–1363 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Kosugi S., Hasebe M., Tomita M., Yanagawa H., Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl Acad. Sci. U.S.A. 106, 10171–10176 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margueron R., Reinberg D., The polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grzenda A., Lomberk G., Zhang J. S., Urrutia R., Sin3: Master scaffold and transcriptional corepressor. Biochim. Biophys. Acta 1789, 443–450 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang L., Nogales E., Ciferri C., Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog. Biophys. Mol. Biol. 102, 122–128 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monory K., Massa F., Egertova M., Eder M., Blaudzun H., Westenbroek R., Kelsch W., Jacob W., Marsch R., Ekker M., Long J., Rubenstein J. L., Goebbels S., Nave K. A., During M., Klugmann M., Wolfel B., Dodt H. U., Zieglgansberger W., Wotjak C. T., Mackie K., Elphick M. R., Marsicano G., Lutz B., The endocannabinoid system controls key epileptogenic circuits in the hippocampus. Neuron 51, 455–466 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou L., Bar I., Achouri Y., Campbell K., De Backer O., Hebert J. M., Jones K., Kessaris N., de Rouvroit C. L., O'Leary D., Richardson W. D., Goffinet A. M., Tissir F., Early forebrain wiring: Genetic dissection using conditional Celsr3 mutant mice. Science 320, 946–949 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kee J., Thudium S., Renner D. M., Glastad K., Palozola K., Zhang Z., Li Y., Lan Y., Cesare J., Poleshko A., Kiseleva A. A., Truitt R., Cardenas-Diaz F. L., Zhang X., Xie X., Kotton D. N., Alysandratos K. D., Epstein J. A., Shi P. Y., Yang W., Morrisey E., Garcia B. A., Berger S. L., Weiss S. R., Korb E., SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry. Nature 610, 381–388 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tepper J. M., Sharpe N. A., Koos T. Z., Trent F., Postnatal development of the rat neostriatum: Electrophysiological, light- and electron-microscopic studies. Dev. Neurosci. 20, 125–145 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Kuo H. Y., Liu F. C., Synaptic Wiring of Corticostriatal Circuits in Basal Ganglia: Insights into the Pathogenesis of Neuropsychiatric Disorders. eNeuro 6, ENEURO.0076.19.2019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson A. G., Kulkarni A., Harper M., Konopka G., Single-cell analysis of Foxp1-driven mechanisms essential for striatal development. Cell Rep. 30, 3051–3066.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saunders A., Macosko E. Z., Wysoker A., Goldman M., Krienen F. M., de Rivera H., Bien E., Baum M., Bortolin L., Wang S., Goeva A., Nemesh J., Kamitaki N., Brumbaugh S., Kulp D., McCarroll S. A., Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carter A. G., Soler-Llavina G. J., Sabatini B. L., Timing and location of synaptic inputs determine modes of subthreshold integration in striatal medium spiny neurons. J. Neurosci. 27, 8967–8977 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding J., Peterson J. D., Surmeier D. J., Corticostriatal and thalamostriatal synapses have distinctive properties. J. Neurosci. 28, 6483–6492 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice A. M., McLysaght A., Dosage-sensitive genes in evolution and disease. BMC Biol. 15, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basilicata M. F., Keller Valsecchi C. I., The good, the bad, and the ugly: Evolutionary and pathological aspects of gene dosage alterations. PLOS Genetics 17, e1009906 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peixoto R. T., Wang W., Croney D. M., Kozorovitskiy Y., Sabatini B. L., Early hyperactivity and precocious maturation of corticostriatal circuits in Shank3B−/− mice. Nat. Neurosci. 19, 716–724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenmund C., Clements J. D., Westbrook G. L., Nonuniform probability of glutamate release at a hippocampal synapse. Science 262, 754–757 (1993). [DOI] [PubMed] [Google Scholar]
- 45.Jahr C. E., High probability opening of NMDA receptor channels by L-glutamate. Science 255, 470–472 (1992). [DOI] [PubMed] [Google Scholar]
- 46.Hessler N. A., Shirke A. M., Malinow R., The probability of transmitter release at a mammalian central synapse. Nature 366, 569–572 (1993). [DOI] [PubMed] [Google Scholar]
- 47.Partin K. M., Patneau D. K., Mayer M. L., Cyclothiazide differentially modulates desensitization of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor splice variants. Mol. Pharmacol. 46, 129–138 (1994). [PubMed] [Google Scholar]
- 48.Yamada K. A., Tang C. M., Benzothiadiazides inhibit rapid glutamate receptor desensitization and enhance glutamatergic synaptic currents. J. Neurosci. 13, 3904–3915 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathis A., Mamidanna P., Cury K. M., Abe T., Murthy V. N., Mathis M. W., Bethge M., DeepLabCut: Markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Hsu A. I., Yttri E. A., B-SOiD, an open-source unsupervised algorithm for identification and fast prediction of behaviors. Nat. Commun. 12, 5188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berridge K. C., Fentress J. C., Disruption of natural grooming chains after striatopallidal lesions. Psychobiology 15, 336–342 (1987). [Google Scholar]
- 52.Berridge K. C., Whishaw I. Q., Cortex, striatum and cerebellum: Control of serial order in a grooming sequence. Exp. Brain Res. 90, 275–290 (1992). [DOI] [PubMed] [Google Scholar]
- 53.Sjobom J., Tamte M., Halje P., Brys I., Petersson P., Cortical and striatal circuits together encode transitions in natural behavior. Sci. Adv. 6, eabc1173 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mapleson D., Garcia Accinelli G., Kettleborough G., Wright J., Clavijo B. J., KAT: A K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics 33, 574–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grissom N. M., McKee S. E., Schoch H., Bowman N., Havekes R., O'Brien W. T., Mahrt E., Siegel S., Commons K., Portfors C., Nickl-Jockschat T., Reyes T. M., Abel T., Male-specific deficits in natural reward learning in a mouse model of neurodevelopmental disorders. Mol. Psychiatry 23, 544–555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thabault M., Turpin V., Maisterrena A., Jaber M., Egloff M., Galvan L., Cerebellar and striatal implications in autism spectrum disorders: From clinical observations to animal models. Int. J. Mol. Sci. 23, 2294 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Halder D., Lee C. H., Hyun J. Y., Chang G. E., Cheong E., Shin I., Suppression of Sin3A activity promotes differentiation of pluripotent cells into functional neurons. Sci. Rep. 7, 44818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G., Genome regulation by polycomb and trithorax proteins. Cell 128, 735–745 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Vaughan R. M., Kupai A., Rothbart S. B., Chromatin regulation through ubiquitin and ubiquitin-like histone modifications. Trends Biochem. Sci. 46, 258–269 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z., Hou Y., Guo X., van der Voet M., Boxem M., Dixon J. E., Chisholm A. D., Jin Y., The EBAX-type Cullin-RING E3 ligase and Hsp90 guard the protein quality of the SAX-3/Robo receptor in developing neurons. Neuron 79, 903–916 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.von Schimmelmann M., Feinberg P. A., Sullivan J. M., Ku S. M., Badimon A., Duff M. K., Wang Z., Lachmann A., Dewell S., Ma'ayan A., Han M. H., Tarakhovsky A., Schaefer A., Polycomb repressive complex 2 (PRC2) silences genes responsible for neurodegeneration. Nat. Neurosci. 19, 1321–1330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bissen D., Foss F., Acker-Palmer A., AMPA receptors and their minions: Auxiliary proteins in AMPA receptor trafficking. Cell Mol. Life Sci. 76, 2133–2169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Henley J. M., Wilkinson K. A., Synaptic AMPA receptor composition in development, plasticity and disease. Nat. Rev. Neurosci. 17, 337–350 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Chubykin A. A., Atasoy D., Etherton M. R., Brose N., Kavalali E. T., Gibson J. R., Südhof T. C., Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron 54, 919–931 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soler-Llavina G. J., Fuccillo M. V., Ko J., Südhof T. C., Malenka R. C., The neurexin ligands, neuroligins and leucine-rich repeat transmembrane proteins, perform convergent and divergent synaptic functions in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 16502–16509 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanamura K., Washburn H. R., Sheffler-Collins S. I., Xia N. L., Henderson N., Tillu D. V., Hassler S., Spellman D. S., Zhang G., Neubert T. A., Price T. J., Dalva M. B., Extracellular phosphorylation of a receptor tyrosine kinase controls synaptic localization of NMDA receptors and regulates pathological pain. PLOS Biol. 15, e2002457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hruska M., Henderson N. T., Xia N. L., Le Marchand S. J., Dalva M. B., Anchoring and synaptic stability of PSD-95 is driven by ephrin-B3. Nat. Neurosci. 18, 1594–1605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han Y., Cao R., Qin L., Chen L. Y., Tang A. H., Sudhof T. C., Zhang B., Neuroligin-3 confines AMPA receptors into nanoclusters, thereby controlling synaptic strength at the calyx of Held synapses. Sci. Adv. 8, eabo4173 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Letellier M., Sziber Z., Chamma I., Saphy C., Papasideri I., Tessier B., Sainlos M., Czondor K., Thoumine O., A unique intracellular tyrosine in neuroligin-1 regulates AMPA receptor recruitment during synapse differentiation and potentiation. Nat. Commun. 9, 3979 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davatolhagh M. F., Fuccillo M. V., Neurexin1 differentially regulates synaptic efficacy within striatal circuits. Cell Rep. 34, 108773 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X., Aslam M., Gollisch T., Allen K., von Engelhardt J., CKAMP44 modulates integration of visual inputs in the lateral geniculate nucleus. Nat. Commun. 9, 261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aoto J., Martinelli D. C., Malenka R. C., Tabuchi K., Sudhof T. C., Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell 154, 75–88 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng Y. R., Tran N. M., Krishnaswamy A., Kostadinov D., Martersteck E. M., Sanes J. R., Satb1 regulates contactin 5 to pattern dendrites of a mammalian retinal ganglion cell. Neuron 95, 869–883.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ueda M., Matsuki T., Fukada M., Eda S., Toya A., Iio A., Tabata H., Nakayama A., Knockdown of Son, a mouse homologue of the ZTTK syndrome gene, causes neuronal migration defects and dendritic spine abnormalities. Mol. Brain 13, 80 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacPherson M. J., Erickson S. L., Kopp D., Wen P., Aghanoori M. R., Kedia S., Burns K. M. L., Vitobello A., Tran Mau-Them F., Thomas Q., Gold N. B., Brucker W., Amlie-Wolf L., Gripp K. W., Bodamer O., Faivre L., Muona M., Menzies L., Baptista J., Guegan K., Male A., Wei X. C., He G., Long Q., Innes A. M., Yang G., Nucleocytoplasmic transport of the RNA-binding protein CELF2 regulates neural stem cell fates. Cell Rep. 35, 109226 (2021). [DOI] [PubMed] [Google Scholar]
- 76.Chih B., Engelman H., Scheiffele P., Control of excitatory and inhibitory synapse formation by neuroligins. Science 307, 1324–1328 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Ichinose S., Susuki Y., Hosoi N., Kaneko R., Ebihara M., Hirai H., Iwasaki H., Interaction between Teneurin-2 and microtubules via EB proteins provides a platform for GABAA receptor exocytosis. eLife 12, e83276 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brouwer M., Farzana F., Koopmans F., Chen N., Brunner J. W., Oldani S., Li K. W., van Weering J. R., Smit A. B., Toonen R. F., Verhage M., SALM1 controls synapse development by promoting F-actin/PIP2-dependent Neurexin clustering. EMBO J 38, e101289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hasegawa Y., Mao W., Saha S., Gunner G., Kolpakova J., Martin G. E., Futai K., Luciferase shRNA presents off-target effects on voltage-gated ion channels in mouse hippocampal pyramidal neurons. eNeuro 4, ENEURO.0186.17.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grinman E., Nakahata Y., Avchalumov Y., Espadas I., Swarnkar S., Yasuda R., Puthanveettil S. V., Activity-regulated synaptic targeting of lncRNA ADEPTR mediates structural plasticity by localizing Sptn1 and AnkB in dendrites. Sci. Adv. 7, eabf0605 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Olivetti P. R., Lacefield C. O., Kellendonk C., A device for stereotaxic viral delivery into the brains of neonatal mice. BioTechniques 69, 307–312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Porter E. G., Connelly K. E., Dykhuizen E. C., Sequential salt extractions for the analysis of bulk chromatin binding properties of chromatin modifying complexes. J. Vis. Exp. 55369 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee T. I., Johnstone S. E., Young R. A., Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc 1, 729–748 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McGinnis C. S., Murrow L. M., Gartner Z. J., DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., Fu X., Liu S., Bo X., G., Yu, clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 and S2