Abstract
Background
Systemic lupus erythematosus (SLE) is a complex and incurable autoimmune disease, so several drug remission for SLE symptoms have been developed and used at present. However, treatment varies by patient and disease activity, and existing medications for SLE were far from satisfactory. Novel drug targets to be found for SLE therapy are still needed.
Methods
Mendelian randomization (MR), an observational study way, was performed to explore potential drug targets for SLE using protein quantitative trait loci (pQTL) from recently published genome-wide association studies (GWAS) of cerebrospinal fluid (CSF) and plasma proteins, which obtained genetic instruments for 154 CSF proteins of 971 participants, and 734 plasma proteins of 23591 participants. Bidirectional Mendelian randomization analysis, colocalization analysis, and phenotype scanning were performed to find key proteins for SLE. In addition, external data verification was implemented to further consolidate the Mendelian randomization findings. Candidate proteins as targets to find drugs and discuss the druggability. Finally, Network pharmacology and molecular docking methods were used to verify the effects of Voclosporin and Cyclosporine on SLE targets. Protein-protein interaction (PPI) and core target analysis of candidate drugs and SLE overlapping targets were performed to identify potential hub targets and interactions. The affinity between drug targets and SLE targets was confirmed by molecular docking.
Results
In the preliminary analysis, we identified four key proteins as possible drug targets in CSF and plasma proteins, included ICAM-1(P = 4.62E-05, OR = 0.90(0.86, 0.95)), sICAM-1(P = 4.62E-05, OR = 0.49(0.35, 0.69)), FCG2B (P = 7.63E-11, OR = 0.57(0.48, 0.67)), PPP3CA; PPP3R1 (P = 5.47E-07, OR = 0.66(0.57, 0.78)). Among them, ICAM1 was detected in both CSF and plasma proteins. By excluding reverse causality, confounding factors, and linkage disequilibrium (LD), we identified PPP3CA; PPP3R1 as novel drug targets for SLE, including Voclosporin and Cyclosporine. Finally, the Drugbank database shows that novel drugs contain 33 targets for treating SLE. PPI suggested that SIRT1, ACE, PTGS2, and BACE1 were pivotal targets for SLE treatment. In addition, the molecular docking showed that the bioactive molecules of Voclosporin and Cyclosporine had a good affinity with the target of SLE.
Conclusions
Our integrative analysis suggested that levels of circulating PPP3CA; PPP3R1 had causal effects on SLE risk and served as potential treatment targets. Moreover, this study provides new evidence for Voclosporin as an SLE treatment through Mendelian randomization and Network pharmacology, and warrants further clinical investigation.
1. Introduction
Systemic lupus erythematosus (SLE) is a chronic immune system disorder with abnormalities of immune cells, manifesting as the dysregulated immune system, characterized by overactivated B and T cells attacking normal cells and tissues, which affects multiple organs and systems [1]. According to published epidemiological surveys, the incidence and prevalence of SLE are high in North America (241 per 100,000 people) [2]. The prevalence of SLE is estimated to increase annually. In addition, as a systemic disorder, SLE frequently affects the blood, kidneys, respiratory, nervous, and gastrointestinal systems [3], and the specific etiology and the pathophysiology of SLE have remained elusive. Notably, the clinical presentation of SLE is varied and unpredictable leading to delayed diagnosis and difficult treatment. Previous studies have shown that early detection and medication regimens have substantially improved SLE survival outcomes [4]. Clinical medications generally treat SLE including glucocorticoids, antimalarial agents, nonsteroidal anti-inflammatory drugs (NSAIDs), immunosuppressive agents, and B cell-targeting biologics [1]. Nonetheless, treatment varies by patient and disease activity, and novel drug targets for SLE therapy are still needed.
Proteins are engaged in complex biological regulation in living organisms and generally serve as drug targets for diseases. Because of the blood-brain barrier, circulating proteins are divided into plasma proteins and cerebrospinal fluid (CSF) proteins. On the one hand, plasma proteins play crucial roles in a range of biological processes and represent major druggable targets [5]. On the other hand, it is generally acknowledged that CSF is produced in the brain, and CSF proteins are considered ideal biomarkers for observing disease states in the brain [6]. So plasma proteins and CSF proteins are separately adopted in this study to improve the result accuracy of the drug targets of SLE. pQTL can be divided into two categories: cis-pQTL and trans-pQTL. In contrast to cis-pQTL, which are located near the target protein, trans-pQTL are typically found on another chromosome. These trans-pQTL do not directly affect the gene or gene domain of the target protein. Previous studies have shown that selection of cis-pQTL for IV can reduce the effects of false positives and pleiotropy [7]. Hence, cis-pQTL are considered to have more direct biological effects on proteins.
Mendelian randomization (MR) is a statistical method that utilizes genetic variation as an instrumental variable (IV) to draw the causal relationship between exposure and outcome [8]. Compared with traditional observational studies, MR analysis can reduce both confounding variables and reverse causation because alleles are random allocation during meiosis, and provide better evidence of causal inference [9]. It’s worth noting that IV fulfilled three assumptions: a, IV was strongly associated with exposure and independent of each other (P<5×10−8, LD <0.001); b, IV was not associated with the outcome; c, IV was not associated with confounders [10] (S1 Fig). MR analysis has been extensively applied in recent years for the development and repurposing of drug targets [11]. In fact, many of experimental clinical drugs have failed to achieve efficacy in clinical trials. However, if a protein drug target linked with the disease is confirmed by genetic association, its market approval will go up [12].
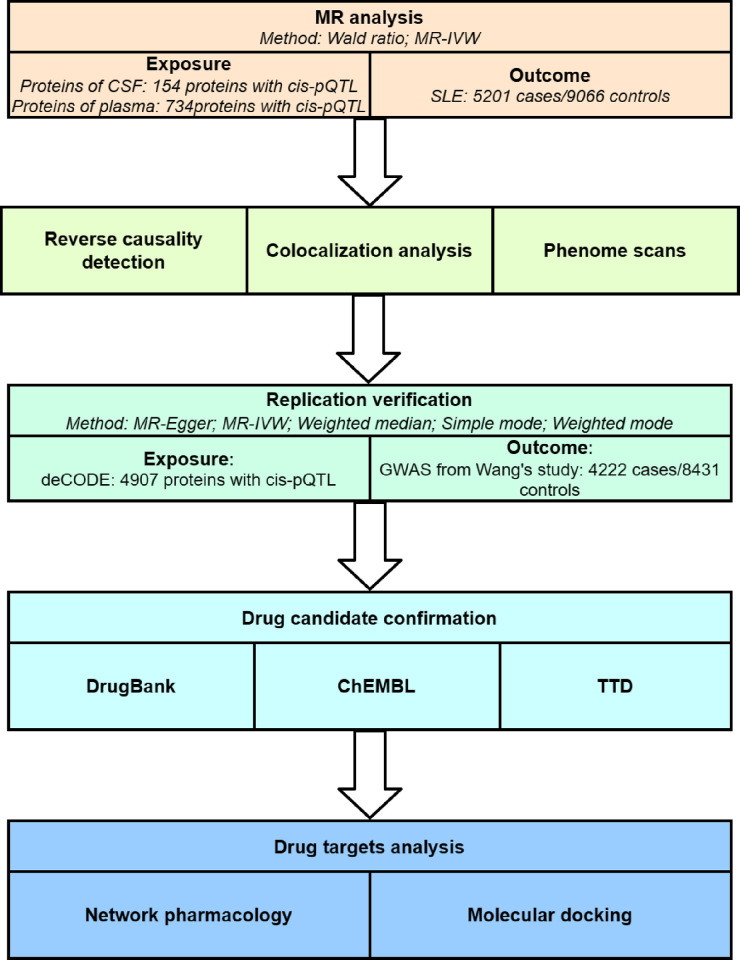
Although there are more and more studies on the treatments of SLE, exploring potential drug targets for SLE through Mendelian randomization in plasma proteins and CSF proteins is still absent. Therefore, the primary objective of this study is to identify potential therapeutic targets for SLE. First, we identify potential causal plasma and CSF proteins for SLE using MR analysis based on GWAS summary data from plasma and CSF proteins. Second, bidirectional MR analysis, colocalization analysis, and phenotype scanning were performed to further validate primary findings. Third, to further ensure the reliability of potential causal proteins, we conducted a replication verification using external data. Final, we find different drugs based on novel drug targets and discuss the druggability (Fig 1). Finally, we search for different drugs based on new drug targets and discuss their drug availability through network pharmacology and molecular docking methods.
Fig 1. Study design flowchart.
2. Method
2.1. SLE and proteins of CSF and plasma data source
In this study, CSF pQTL was selected from Yang’s study, which recruited 713 proteins from Washington University School of Medicine in St. Louis [13]. Plasma pQTL was obtained from Cheng’s study containing 1699 proteins from summary of 5 GWAS [5]. 4907 proteins and 18084 pQTL from the deCODE database which recruited 35,559 Icelanders from 24 August 2000 until 11 January 2019, were used for replication verification [14]. SLE was selected from James’ study whose participants were mostly from southern Europe (ncase = 5201; ncontrol = 9066) for MR analysis [15]. For replication verification, summary statistics were from Wang’s study whose participants came from the same continentas above (ncase = 4222, ncontrol = 8431) [16]. If the effect allele frequency was missing, the matched human genome build was used to fill in the missing.
2.2. Mendelian randomization analysis
Mendelian randomization analysis is an observational study way. For the selection criteria of IV: (1) SNP from cis-pQTL. (2) Significantly correlated with proteins (P<5×10−8). To avoid bias introduced by weak instruments of screening [17], we used F-statistic values to calculate the strength of IV. F > 10 were considered to be strongly correlated, satisfying the MR assumption a. (3) Independence (LD clumping r2 < 0.001), (4) Exclusion of pQTL within the major histocompatibility complex (MHC) region (chr6, from 26 Mb to 34 Mb).
According to the above selection criteria, we obtained 154 cis-pQTL of 154 CSF proteins and 738 cis-pQTL of 734 plasma proteins. We used "TwoSampleMR" (https://github.com/MRCIEU/TwoSampleMR) to perform two-sample MR analysis. Proteins were used as exposure, and SLE as outcome. If the proteins in MR analysis have only one SNP, the "Wald ratio" method will be used. Meanwhile, "Inverse variance weighted MR (MR-IVW)" was used for proteins with multiple SNPs. In addition, for screened SNPs that meet the above criteria, heterogeneity and pleiotropy tests were performed further. Results were adjusted using Bonferroni correction, and P<0.05/888 (5.63×10−5) were selected for subsequent analysis.
2.3. Reverse causality detection
Steiger filtering was employed to analyze the directionality between proteins and SLE, ensuring the credibility of the results and identifying reverse associations. Besides, we exchanged the exposure and outcome data to perform a secondary analysis in order to more accurately identify reverse causality: SLE was used as the exposure, and data from the original protein literature was obtained as the outcome.
What counts is that IVs were screened using the criteria described above. MR analysis was performed using MR-IVW, MR-Egger, weighted median, simple mode, and weighted mode. Statistical significance was achieved when P<0.05.
2.4. Colocalization analysis
Bayesian colocalization analysis can assess whether proteins and diseases are caused by the same pQTL, eliminating LD bias in MR analysis. We used the “coloc” package (https://github.com/chr1swallace/coloc) for colocalization analysis. Colocalization analysis contains five hypotheses: posterior probability (PP0), functional association only (PP1), GWAS association only (PP2), independent functional/GWAS associations (PP3), or colocalized functional/GWAS associations (PP4) [18]. By obtaining all pQTL data from the original article. We performed colocalization analysis of all SNPs within 1MB of the cis-pQTL obtained from the MR analysis. Finally, we use the result of the coloc.abf algorithm, PPH4 > 0.8, as a basis for co-localization in this study.
2.5. Phenotype scanning
We also further performed "phenome scans" to exclude confounding factors from the results. PhenoScanner V2 was used to search for associations of the IVs with other traits by searching previous GWAS [19]. As described above, IVs that did not satisfy MR assumptions b and c were excluded. Exclusion criteria: significant genome-wide association (p<5×10−8), IV associated with known risk factors.
2.6. Replication verification
To better verify the authenticity of the primary findings, data from different databases were used to repeat the experiments. Due to less CSF pQTL data, the above data were still used. Plasma proteins from the deCODE database were used as exposure. SLE from Wang’s study was used as the outcome [16]. The screen of IVs and MR analyses were consistent with the above. Because of just validation of preliminarily identified proteins, the range of P values was appropriately relaxed to P<0.05 to be considered significant.
2.7. Plasma-CSF proteins correlation
While both plasma and CSF proteins undergo metabolic circulation within the body, they possess distinct differences owing to the presence of the blood-brain barrier. To assess the discrepancy between proteins pQTL in plasma and CSF, we conducted Spearman correlation analysis to investigate the shared pQTL correlation between plasma and CSF. In addition, we set different p-value thresholds to explore whether the correlation changes with increasing significance levels.
2.8. Drug candidate confirmation
The proteins identified by the above screen series proved causally related to SLE. To explore the interactions between those SLE-associated proteins and the targets for medications already on the market, we searched drug databasea to find medications targeting the identified potential causal proteins including DrugBank (https://go.drugbank.com/), ChEMBL (https://www.ebi.ac.uk/chembl/), and Therapeutic Target Database (TTD) (https://idrblab.net/ttd/) [20].
2.9. Network pharmacology analysis
Active components of Voclosporin and Cyclosporine were searched by the Drugbank database. We obtained disease-related targets of SLE by employing the keyword "Systemic Lupus Erythematosus" in the Genecards (https://www.genecards.org/). Then the collected drug targets and SLE targets were calculated to receive overlapping targets in Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/). This study imported the identified shared targets into the STRING Database (https://string-db.org/) whose confidence was set to medium (0.400) to ascertain hub drug and disease targets, subsequently visualizing the results in the form of a PPI network. We performed the MCC algorithm to calculate the hub targes of drug candidates against SLE in the Cytohubba plugin of CytoScape, and constructed the related protein target network.
2.10. Molecular docking
Drug molecule structures of candidate drugs were downloaded from the ChemSpider database (https://www.chemspider.com/Default.aspx), and the target protein from the UniProt Database. Next, we dehydrated and then removed the active center ligands for the drug structure in PyMOL 2.6.0 software, and carried out hydrogenation and charge calculation for the target proteins in Autodock Tools 1.5.7 software. The molecular docking results of the key targets of SLE and the corresponding active components were visualized by Pymol.
3. Results
3.1. MR analysis
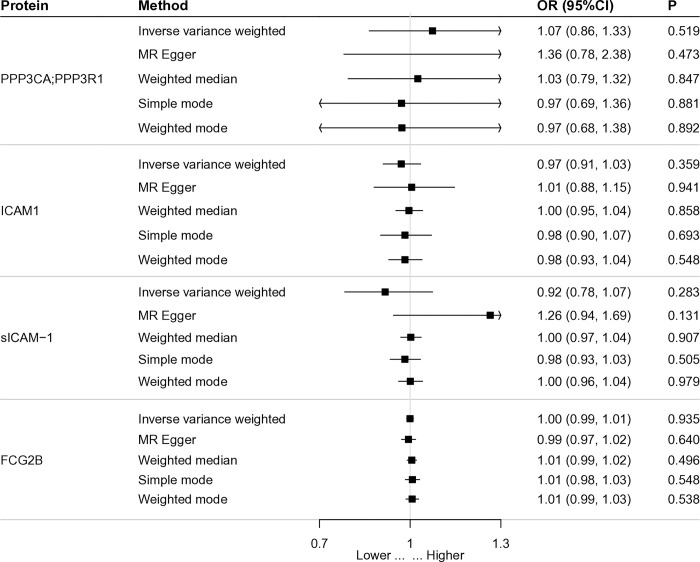
The causal relationship of CSF and plasma proteins on SLE were assessed using two-sample MR analysis. At Bonferroni correction (p<0.05/888), the result revealed that four potential causal proteins were finally obtained for SLE (Table 1 and S2 Fig). Intercellular adhesion molecule 1 (sICAM-1)(P = 4.62×10−5, OR = 0.49(0.35, 0.69)), Low affinity immunoglobulin gamma Fc region receptor II-b (FCG2B) (P = 7.63×10−11, OR = 0.57(0.48, 0.67)), and Protein phosphatase 3 catalytic subunit alpha (PPP3CA); Calcineurin subunit B type 1 (PPP3R1)(P = 5.47×10−7, OR = 0.66(0.57, 0.78)), Intercellular adhesion molecule 1 (ICAM1)(P = 4.62×10−5, OR = 0.90(0.86, 0.95)). Although sICAM-1 in CSF and ICAM-1 in plasma are same protein, they distribute in different microenvironments. Subsequent analyses will also be treated separately to distinguish whether this protein differs in CSF and plasma. By checking the original literature, PPP3CA; PPP3R1 corresponds to Calcineurin, so it is placed in one group. Increasing these four proteins will decrease the risk of SLE. No heterogeneity and pleiotropy were detected in the preliminary analyses.
Table 1. MR results for plasma and CSF proteins after Bonferroni correction.
| Tissue | Protein | SNP | Outcome | Method | Beta | SE | P value | PVE | F | Steiger pval | OR(95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma | PPP3CA; PPP3R1 | rs17266357 | SLE | Wald ratio | -0.41 | 0.08 | 5.47E-07 | 6.29% | 66.9 | 6.15E-11 | 0.66(0.57, 0.78) |
| Plasma | ICAM1 | rs5498 | SLE | Wald ratio | -0.1 | 0.03 | 4.62E-05 | 70.18% | 7769.9 | 0 | 0.90 (0.86, 0.95) |
| CSF | sICAM-1 | rs5498 | SLE | Wald ratio | -0.72 | 0.18 | 4.62E-05 | 57.39% | 1124.84 | 1.04E-158 | 0.49 (0.35, 0.69) |
| CSF | FCG2B | rs4657041 | SLE | Wald ratio | -0.57 | 0.09 | 7.63E-11 | 47.57% | 757.73 | 9.53E-110 | 0.57 (0.48, 0.67) |
• PVE = proportion of variance explained.
• F-value assesses the bias of instrumental variables; when F > 10, weak instrumental variables do not exist.
3.2. Reverse causality detection and Replication verification
Steiger filtering obtained results confirming the unidirectionality of causality. When exposure and outcome were interchanged, reverse MR analysis showed that SLE was no causality for the four proteins, which further ensured directionality (Fig 2). Using data from different databases and the same analytical method, the result display that PPP3CA;PPP3R1 (P = 3.11×10−7, OR = 0.61), and FCG2B (P = 3.91×10−5, OR = 0.68) were re-validated a causal effect for SLE (S1 Table). Meanwhile, increasing these two proteins reduced the risk of SLE, consistent with the results of previous analyses.
Fig 2. Forest map of reverse causality detection about significant protein.
3.3. Colocalization analysis and phenotype scanning
We subsequently performed the co-localization analysis of the four proteins obtained from MR analysis. The proteins were tested by the coloc.abf algorithm to identify if they shared a common causal variant with SLE. The results strongly suggest that PPP3CA;PPP3R1 (PH4.coloc.abf = 0.872), FCG2B (PH4.coloc.abf = 0.985), and SLE share the same variants (S2 Table).
Finally, confounders were removed by phenotypic scanning (S3 Table). FCG2B (rs4657041) was found to be associated with systemic lupus erythematosus, inflammatory bowel disease, ulcerative colitis, and low affinity immunoglobulin gamma fc region receptor II-a/b. Because FCG2B (rs4657041) was directly associated with SLE, it did not meet MR assumption a. So it was not included in the analysis of drug-targeted proteins. sICAM-1 (rs5498) and ICAM-1 (rs5498) were associated with White blood cell count, soluble intercellular adhesion molecule 1 ICAM 1, Intercellular adhesion molecule 1, etc. PPP3CA;PPP3R1 (rs17266357) correlated with mean platelet volume, calcineurin.
3.4. Correlation and drug targets analysis
There was a non-significant negative correlation between the CSF and plasma proteins MR results (Spearman correlation coefficient = −0.035). When restricting the number of proteins included in the analysis with different P-value thresholds, a negative correlation was still present and remained insignificant (S3 Fig). Through a series of analyses, it was finally shown that PPP3CA;PPP3R1 are genetically linked to SLE. Look for the drug records of the proteins in the database. Three PPP3R1 target proteins were looked up in the DrugBank, including Calcineurin subunit B type 1, Calcineurin subunit B type 2, and Calcium and integrin-binding protein 1. For PPP3R1, five drugs have been approved, and two drugs have explicit pharmacological action (Table 2). Interestingly, Voclosporin is a potential drug target for both causal proteins, which has clear pharmacological action and has been approved on the market.
Table 2. Drug targets analysis.
| Protein | Targe | Drug name | Source | Drug group | Pharmacological action | Actions |
|---|---|---|---|---|---|---|
| PPP3R1 | Calcineurin subunit B type 1 | Voclosporin | DrugBank | approved, investigational | yes | inhibitor |
| Calcineurin subunit B type 2 | Cyclosporine | DrugBank | approved,investigational | yes | inhibitor | |
| Voclosporin | DrugBank | approved, investigational | yes | inhibitor | ||
| PPP3CA | - | Voclosporin | ChEMBL; TTD | approved, investigational | yes | inhibitor |
3.5. Network pharmacology analysis
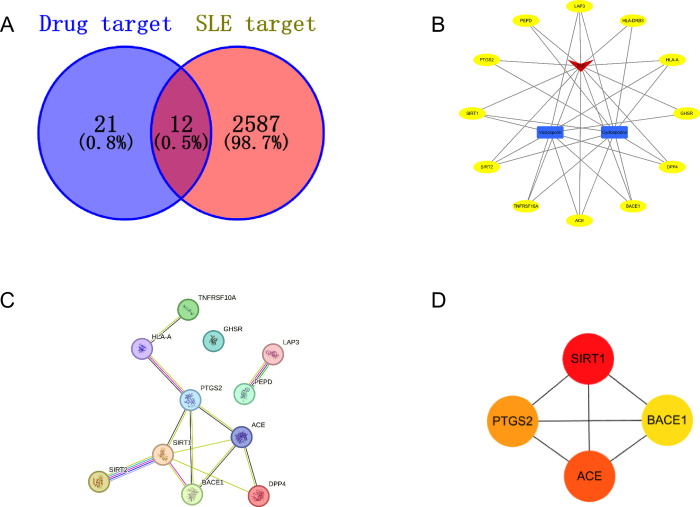
33 drug targets were screened in Drugbank and 2599 targets of SLE in the Genecards Database. The Venn plots show that SLE targets and Voclosporin and Cyclosporine targets have shared 12 common ones (Fig 3A). Next, we draw targets of drug candidates into a drug disease-target network diagram to show intuitively this relationship (Fig 3B). The PPI network by feeding 12 overlapping targets into the STRING database revealed 11 nodes,12 edges, and 2.18 average node degree (Fig 3C). Then we imported the PPI network data from the STRING platform into Cytoscape to calculate and select the top 4 core targets with the Cytohubba plug-in (Fig 3D). The results of algorithm reveal that SIRT1, ACE, PTGS2 and BACE1 may play important roles in the treatment of SLE.
Fig 3. Network pharmacology analysis.
(A) Drug-disease target Venn diagram; (B) Drug-Disease-Target network. The red color indicates SLE; The blue color indicates the drugs; the yellow color indicates the targets; (C) PPI network of shared targets of Voclosporin and Cyclosporine in treatment of SLE; (D) Core targets of overlapping targets.
3.6. Molecular docking analysis
The key targets SIRT1 (PDB: 4I5I), ACE (PDB: 1O86), and BACE1 (PDB: 1TQF) with high degree values in the PPI network were selected for molecular docking verification with Voclosporin and Cyclosporine. The results of Molecular docking analysis exhibited that bioactive compounds of Voclosporin and Cyclosporine could be bound into the docking pocket, and demonstrated strong docking activity (Table 3; Fig 4). Cyclosporine bound to ACE by forming hydrogen bonds with TYR-213 (length: 3.4 Å), ASN-105 (2.8 Å), ASN-105 (2.7 Å) and LYS-113 (2.2 Å). Voclosporin bound to SIRT1 by forming hydrogen bonds with LYS-499 (2.9 Å), CYS-501 (2.1 Å) and LYS-499 (3.3 Å).
Table 3. Docking simulation for active molecular and targets of SLE.
| Molecular name | Targets | PBD ID | Residue involved in H bonding | Docking score (kcal/mol) |
|---|---|---|---|---|
| Cyclosporine | ACE | 1O86 | TYR-213; ASN-105; LYS-113 | -2.59 |
| Voclosporin | SIRT1 | 4I5I | LYS-499; CYS-501 | -1.97 |
| Cyclosporine | BACE1 | 1TQF | GLU-364; HIS-362; HIS-362 | -0.19 |
| Voclosporin | ACE | 1O86 | SER-594; GLU-535 (1.7 Å) | -0.17 |
Fig 4. Molecular docking diagrams of SLE related targets with main compounds of Voclosporin and Cyclosporine.
4. Discussion
In this study, we used CSF and plasma proteins for MR analysis and co-localization analysis to find proteins causally associated with SLE. We used cis-pQTL as IV because it directly affects transcription and translation, and reduces horizontal pleiotropy [21]. Preliminary analyses of the proteins provided four causal proteins. At the same time, we found one identical protein, ICAM1, in CSF and plasma. Because the results of MR analysis may be affected by reverse causality, we corroborated each other by Steiger filtering and reverse causality detection to minimize the error. The results showed that the proteins analyzed by MR did not have reverse causality. Bayesian co-localization analysis was performed with a threshold of 0.8 to exclude the bias introduced by LD. In the co-localization analysis, although ICAM1 was a failure, it could not be ignored in explaining the mechanism of the disease. When searching for relevant phenotypes for instrumental variables, FCG2B was considered a confounder in the MR analysis because it was directly associated with SLE. Finally, we found that PPP3CA and PPP3R1 are causal proteins of the disease that can be potential drug targets. For PPP3CA and PPP3R1, the target drug is Voclosporin. It has been clearly available for the treatment of lupus nephritis. Then, the therapeutic effects of Voclosporin and Cyclosporine on SLE were evaluated by network pharmacology and molecular docking methods.
ICAM-1, a cell surface glycoprotein and an adhesion receptor, is involved in antigen presentation, immune cell initiation, leukocyte transport, and effector functions [22]. In addition, it is involved in regulating endothelial and epithelial barriers and influences tumor development and metastasis [23]. ICAM-1 has been implicated in a variety of diseases. FCG2B is engaged in a variety of effects and regulation, including phagocytosis of immune complexes and regulation of B cell antibodies. Kyogoku suggests that FCG2B is implicated in SLE [24].
By searching the original literature [25], PPP3CA; PPP3R1 targets Calcineurin (CaN). CaN, primarily located in the cytosol, is a serine/threonine phosphatase needing calcium and calmodulin (CaM) [26]. CaN and CaM bind calcium, and then the two bind to each other to form an active phosphatase. This way couples calcium signaling to dephosphorylation of proteins [27]. CaN activates the nuclear factor family of T cells, regulates a number of receptors and channels, and regulates mitochondria-associated proteins and microtubule-associated proteins [26]. The best known of these is that CaN activates T cells and drives the immune system. Therefore, calcineurin inhibitors (FK506 (tacrolimus), and cyclosporin A) are used as immunosuppressants including organ transplantation [28]. Calcineurin inhibitors have been used to treat autoimmune diseases with some adverse effects, possibly due to some effects in non-immune tissues [28]. Therefore, it is crucial to clarify the molecular biological mechanism of CaN as well as the substrates for the reaction. PPP3CA is calmodulin-stimulated protein phosphatase [29]. Several studies have shown that variants in PPP3CA lead to epilepsy [30, 31]. Maria showed that PPP3CA is associated with SLE disease subtypes [32]. PPP3R1 is a regulatory subunit of calcineurin. It is thought to be associated with Alzheimer’s disease [33, 34].
Angiotensin-converting enzyme (ACE) plays a key role in the regulation of blood pressure by converting angiotensin I to the potent vasoconstrictor angiotensin II [35]. At present, the relationship between ACE and SLE has been studied, especially the polymorphism of the ACE gene. The DD genotype (associated with higher ACE activity levels) was found to be more frequent in patients with SLE and associated with visceral damage in this population [36]. This suggests that the ACE gene I/D polymorphism may affect the pathological process of SLE and may be involved through angiotensin II-mediated inflammatory and immunomodulatory mechanisms. Silent information regulator 2 related enzyme 1 (SIRT1) is a NAD+ -dependent histone deacetylase with regulatory effects on a variety of physiological processes including immune responses, which may be related to the development of SLE [37]. Related studies have shown that the long non-coding RNA MALAT-1 plays a role in the pathogenesis of SLE by regulating SIRT1 expression, suggesting a possible relationship between SIRT1 and the development of SLE [38]. BACE1, β-secretase 1, plays A key role in the production of β-amyloid peptide (Aβ) by cleaving amyloid precursor protein (APP). It is best known for its function in promoting the accumulation of Aβ peptides in the pathogenesis of Alzheimer’s disease [39]. However, the activity of BACE1 is not limited to the brain, it is identified in various cell types and implicated in a variety of physiological and pathological processes, including metabolic diseases and obesity [40]. Although the direct relationship between BACE1 and SLE has not been extensively studied, the broad effects of BACE1 on immune function and cellular signaling pathways imply a potential intersection with autoimmune diseases such as SLE.
Although this study did not find SLE-associated proteins from those analyzed CSF proteins, it was because the available pQTL data in the GWAS data of CSF proteins were limited. For example, PPP3CA;PPP3R1 did not exist in this CSF GWAS data, but the utility of PPP3CA;PPP3R1 in SLE in CSF cannot be ruled out. Neuropsychiatric SLE (NPSLE) means that the lesions involve the nervous system, which may affect 12% ~ 95% of patients with SLE. Due to the existence of the blood-brain barrier, research on PPP3CA;PPP3R1 protein for NPSLE has not been carried out and is relatively scarce. For future research, we will focus on the dual role of PPP3CA;PPP3R1 in CSF and plasma and explore the impact of the blood-brain barrier on target drug. We hope future research can conduct more basic and animal experiments.
The strength of our study lies in the use of MR analysis and co-localization analysis, which eliminates the effects of confounding factors and reverses causation as much as possible. Using cis-pQTL as an instrumental variable ensured robustness between SNP and proteins. Demonstrate the reproducibility of the results by replication validation. Avoid the chance of results due to differences between data. In addition, using large samples of GWAS data, there is enough statistical strength to prove the relationship between proteins and diseases. AND network pharmacology and molecular docking technology were used to evaluate the efficacy of target drugs in the treatment of SLE. Nonetheless, the study has some limitations. While cis-pQTL is an ideal tool for analyzing drug targets, this would also ignore some of the roles of trans-pQTL in disease. Since the population is limited to Europe, the results may be biased relative to other regions. In addition, information related to network pharmacology is obtained through online databases, and the database information still needs to be further improved.
5. Conclusions
By using MR analysis, we identified proteins that have a causal relationship with SLE. These proteins are essential in explaining the disease’s mechanisms and molecular pathways. Finally, we discuss the role of PPP3CA, and PPP3R1 in SLE and potential drug use. Based on data mining network pharmacology, and molecular docking method validation, it provides new evidence for potential drug as an SLE treatment, and warrants further clinical investigation.
Supporting information
(TIF)
(TIF)
(TIF)
(CSV)
(CSV)
(CSV)
Abbreviations
- SLE
Systemic lupus erythematosus
- MR
Mendelian randomization
- pQTL
protein quantitative trait loci
- GWAS
published genome-wide association studies
- CSF
cerebrospinal fluid
- PPI
Protein-protein interaction
- LD
linkage disequilibrium
- NSAIDs
nonsteroidal anti-inflammatory drugs
- sICAM-1
Intercellular adhesion molecule 1
- FCG2B
Fc region receptor II-b
- PPP3CA
Protein phosphatase 3 catalytic subunit alpha
- PPP3R1
Calcineurin subunit B type 1
- ACE
Angiotensin-converting enzyme
- SIRT1
Silent information regulator 2 related enzyme 1
- APP
amyloid precursor protein
Data Availability
Protein pQTL data from original literature(5, 13, 14). SLE data from the IEU Open GWAS (https://gwas.mrcieu.ac.uk/; Accession numbers: ebi-a-GCST003156 and ebi-a-GCST90011866).
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Kiriakidou M, Ching CL. Systemic Lupus Erythematosus. Annals of internal medicine. 2020;172(11):Itc81–itc96. doi: 10.7326/AITC202006020 [DOI] [PubMed] [Google Scholar]
- 2.Rees F, Doherty M, Grainge MJ, Lanyon P, Zhang W. The worldwide incidence and prevalence of systemic lupus erythematosus: a systematic review of epidemiological studies. Rheumatology (Oxford, England). 2017;56(11):1945–61. doi: 10.1093/rheumatology/kex260 [DOI] [PubMed] [Google Scholar]
- 3.Huang X, Zhang Q, Zhang H, Lu Q. A Contemporary Update on the Diagnosis of Systemic Lupus Erythematosus. Clinical reviews in allergy & immunology. 2022;63(3):311–29. doi: 10.1007/s12016-021-08917-7 [DOI] [PubMed] [Google Scholar]
- 4.Allen ME, Rus V, Szeto GL. Leveraging Heterogeneity in Systemic Lupus Erythematosus for New Therapies. Trends in molecular medicine. 2021;27(2):152–71. doi: 10.1016/j.molmed.2020.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng J, Haberland V, Baird D, Walker V, Haycock PC, Hurle MR, et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nature genetics. 2020;52(10):1122–31. doi: 10.1038/s41588-020-0682-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansson O. Biomarkers for neurodegenerative diseases. Nature medicine. 2021;27(6):954–63. doi: 10.1038/s41591-021-01382-x [DOI] [PubMed] [Google Scholar]
- 7.Swerdlow DI, Kuchenbaecker KB, Shah S, Sofat R, Holmes MV, White J, et al. Selecting instruments for Mendelian randomization in the wake of genome-wide association studies. International journal of epidemiology. 2016;45(5):1600–16. doi: 10.1093/ije/dyw088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ (Clinical research ed). 2018;362:k601. doi: 10.1136/bmj.k601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. European journal of epidemiology. 2015;30(7):543–52. doi: 10.1007/s10654-015-0011-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emdin CA, Khera AV, Kathiresan S. Mendelian Randomization. Jama. 2017;318(19):1925–6. [DOI] [PubMed] [Google Scholar]
- 11.Reay WR, Cairns MJ. Advancing the use of genome-wide association studies for drug repurposing. Nature reviews Genetics. 2021;22(10):658–71. doi: 10.1038/s41576-021-00387-z [DOI] [PubMed] [Google Scholar]
- 12.Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, et al. The support of human genetic evidence for approved drug indications. Nature genetics. 2015;47(8):856–60. doi: 10.1038/ng.3314 [DOI] [PubMed] [Google Scholar]
- 13.Yang C, Farias FHG, Ibanez L, Suhy A, Sadler B, Fernandez MV, et al. Genomic atlas of the proteome from brain, CSF and plasma prioritizes proteins implicated in neurological disorders. Nature neuroscience. 2021;24(9):1302–12. doi: 10.1038/s41593-021-00886-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferkingstad E, Sulem P, Atlason BA, Sveinbjornsson G, Magnusson MI, Styrmisdottir EL, et al. Large-scale integration of the plasma proteome with genetics and disease. Nature genetics. 2021;53(12):1712–21. doi: 10.1038/s41588-021-00978-w [DOI] [PubMed] [Google Scholar]
- 15.Bentham J, Morris DL, Graham DSC, Pinder CL, Tombleson P, Behrens TW, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nature genetics. 2015;47(12):1457–64. doi: 10.1038/ng.3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YF, Zhang Y, Lin Z, Zhang H, Wang TY, Cao Y, et al. Identification of 38 novel loci for systemic lupus erythematosus and genetic heterogeneity between ancestral groups. Nature communications. 2021;12(1):772. doi: 10.1038/s41467-021-21049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. International journal of epidemiology. 2011;40(3):755–64. doi: 10.1093/ije/dyr036 [DOI] [PubMed] [Google Scholar]
- 18.Cullell N, Gallego-Fábrega C, Cárcel-Márquez J, Muiño E, Llucià-Carol L, Lledós M, et al. ICA1L Is Associated with Small Vessel Disease: A Proteome-Wide Association Study in Small Vessel Stroke and Intracerebral Haemorrhage. International journal of molecular sciences. 2022;23(6). doi: 10.3390/ijms23063161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics (Oxford, England). 2019;35(22):4851–3. doi: 10.1093/bioinformatics/btz469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y, Zhang Y, Lian X, Li F, Wang C, Zhu F, et al. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic acids research. 2022;50(D1):D1398–d407. doi: 10.1093/nar/gkab953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montgomery SB, Dermitzakis ET. From expression QTLs to personalized transcriptomics. Nature reviews Genetics. 2011;12(4):277–82. doi: 10.1038/nrg2969 [DOI] [PubMed] [Google Scholar]
- 22.Martin TM, Burke SJ, Wasserfall CH, Collier JJ. Islet beta-cells and intercellular adhesion molecule-1 (ICAM-1): Integrating immune responses that influence autoimmunity and graft rejection. Autoimmunity reviews. 2023;22(10):103414. doi: 10.1016/j.autrev.2023.103414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bui TM, Wiesolek HL, Sumagin R. ICAM-1: A master regulator of cellular responses in inflammation, injury resolution, and tumorigenesis. Journal of leukocyte biology. 2020;108(3):787–99. doi: 10.1002/JLB.2MR0220-549R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyogoku C, Dijstelbloem HM, Tsuchiya N, Hatta Y, Kato H, Yamaguchi A, et al. Fcgamma receptor gene polymorphisms in Japanese patients with systemic lupus erythematosus: contribution of FCGR2B to genetic susceptibility. Arthritis and rheumatism. 2002;46(5):1242–54. doi: 10.1002/art.10257 [DOI] [PubMed] [Google Scholar]
- 25.Suhre K, Arnold M, Bhagwat AM, Cotton RJ, Engelke R, Raffler J, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nature communications. 2017;8:14357. doi: 10.1038/ncomms14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creamer TP. Calcineurin. Cell communication and signaling: CCS. 2020;18(1):137. doi: 10.1186/s12964-020-00636-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swulius MT, Waxham MN. Ca(2+)/calmodulin-dependent protein kinases. Cellular and molecular life sciences: CMLS. 2008;65(17):2637–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy J, Cyert MS. Identifying New Substrates and Functions for an Old Enzyme: Calcineurin. Cold Spring Harbor perspectives in biology. 2020;12(3). doi: 10.1101/cshperspect.a035436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rydzanicz M, Wachowska M, Cook EC, Lisowski P, Kuźniewska B, Szymańska K, et al. Novel calcineurin A (PPP3CA) variant associated with epilepsy, constitutive enzyme activation and downregulation of protein expression. European journal of human genetics: EJHG. 2019;27(1):61–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panneerselvam S, Wang J, Zhu W, Dai H, Pappas JG, Rabin R, et al. PPP3CA truncating variants clustered in the regulatory domain cause early-onset refractory epilepsy. Clinical genetics. 2021;100(2):227–33. doi: 10.1111/cge.13979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers CT, Stong N, Mountier EI, Helbig KL, Freytag S, Sullivan JE, et al. De Novo Mutations in PPP3CA Cause Severe Neurodevelopmental Disease with Seizures. American journal of human genetics. 2017;101(4):516–24. doi: 10.1016/j.ajhg.2017.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilbe M, Kozyrev SV, Farias FH, Bremer HD, Hedlund A, Pielberg GR, et al. Multiple Changes of Gene Expression and Function Reveal Genomic and Phenotypic Complexity in SLE-like Disease. PLoS genetics. 2015;11(6):e1005248. doi: 10.1371/journal.pgen.1005248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Z, Bai J, Zhong S, Zhang R, Kang K, Zhang X, et al. Integrative genomic analysis of PPP3R1 in Alzheimer’s disease: a potential biomarker for predictive, preventive, and personalized medical approach. The EPMA journal. 2021;12(4):647–58. doi: 10.1007/s13167-021-00261-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson D, Munger C, Crowley J, Corcoran C, Cruchaga C, Goate AM, et al. Variants in PPP3R1 and MAPT are associated with more rapid functional decline in Alzheimer’s disease: the Cache County Dementia Progression Study. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2014;10(3):366–71. doi: 10.1016/j.jalz.2013.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thorn CF, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for angiotensin-converting enzyme. Pharmacogenetics and genomics. 2010;20(2):143–6. doi: 10.1097/FPC.0b013e3283339bf3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong AM, Li XY, Wang YQ, Yan HX, Xu ZX, Feng Z, et al. Association study of ACE polymorphisms and systemic lupus erythematosus in Northern Chinese Han population. Molecular biology reports. 2012;39(10):9485–91. doi: 10.1007/s11033-012-1813-7 [DOI] [PubMed] [Google Scholar]
- 37.Qiu Y, Zhou X, Liu Y, Tan S, Li Y. The Role of Sirtuin-1 in Immune Response and Systemic Lupus Erythematosus. Frontiers in immunology. 2021;12:632383. doi: 10.3389/fimmu.2021.632383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang H, Liang N, Wang M, Fei Y, Sun J, Li Z, et al. Long noncoding RNA MALAT-1 is a novel inflammatory regulator in human systemic lupus erythematosus. Oncotarget. 2017;8(44):77400–6. doi: 10.18632/oncotarget.20490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meakin PJ, Jalicy SM, Montagut G, Allsop DJP, Cavellini DL, Irvine SW, et al. Bace1-dependent amyloid processing regulates hypothalamic leptin sensitivity in obese mice. Scientific reports. 2018;8(1):55. doi: 10.1038/s41598-017-18388-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor HA, Przemylska L, Clavane EM, Meakin PJ. BACE1: More than just a β-secretase. Obesity reviews: an official journal of the International Association for the Study of Obesity. 2022;23(7):e13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(CSV)
(CSV)
(CSV)
Data Availability Statement
Protein pQTL data from original literature(5, 13, 14). SLE data from the IEU Open GWAS (https://gwas.mrcieu.ac.uk/; Accession numbers: ebi-a-GCST003156 and ebi-a-GCST90011866).