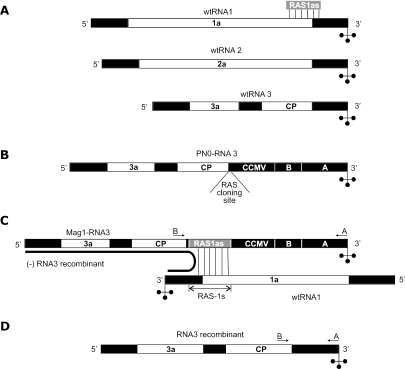
Figure 1.
The BMV-based recombination system. White, black and gray boxes represent coding, noncoding and recombinationally active sequences, respectively. The location of the primers (A and B) used for specific RT–PCR amplification of the 3′-portion of BMV RNA3 (parental or recombinant) is indicated by arrows. (A) BMV genome. The BMV genome consists of three RNA segments: RNA1, RNA2 and RNA3. All three BMV RNAs share an almost identical 3′-noncoding region with a tRNA-like structure at the very end. (B) Recombination vector. The PN0-RNA3 vector is a wtRNA3 derivative with a modified 3′-noncoding end [(for details see ref. (33)]. The latter includes (i) the RAS cloning site, (ii) a 197 nt sequence derived from the 3′-noncoding region of cowpea chlorotic mottle virus RNA3 (marked as CCMV), (iii) the sequence of wtRNA3 between nt 7 and 200 (counting from the 3′ end—marked as the region B) and (iv) the last 236 nt from the 3′ end of BMV wtRNA1 (marked as A). (C) Non-homologous recombination. Non-homologous recombination was observed if an ∼140 nt sequence, called RAS1as (shown in A), complementary to wtRNA1 between positions 2856 and 2992 was inserted into the PN0-RNA3 vector. The presence of the RAS1as sequence in RNA3 derivative (called Mag1-RNA3) allows local RNA1-RNA3 hybridization that mediates frequent non-homologous crossovers. It is thought that polymerase starts nascent strand synthesis on the 3′ end of RNA1 and then switches to RNA3 within a local double-stranded region. (D) Non-homologous recombinant. Non-homologous recombination repairs Mag1-RNA3 by replacing its modified 3′ end with the 3′-noncoding fragment coming from RNA1.