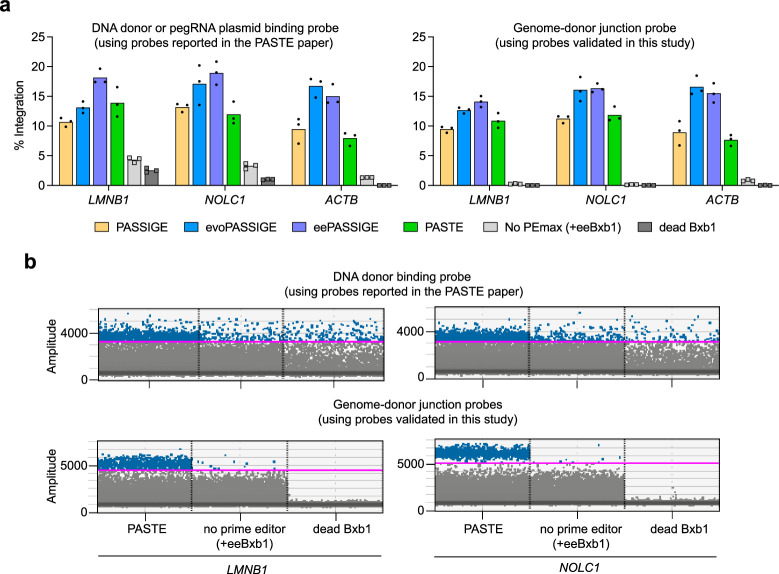
Extended Data Fig. 10. Performance of ddPCR probes at the LMNB1, NOLC1, and ACTB sites.
a, Integration efficiencies for PASSIGE, evoPASSIGE, eePASSIGE, and PASTE at the top three most common sites used to characterize PASTE. Initial PASTE publication uses ddPCR probes that bind either to the DNA donor or pegRNA plasmid26 whereas the initial twin prime editing24 publication and this work primarily employ probes that bind to the genome–donor junction. Bars reflect the mean of three independent replicates and dots show individual replicate values. b, ddPCR plots for PASTE, no PEmax (+eeBxb1) control, and dead Bxb1 control when using different ddPCR probes. The magenta line shows the threshold that was set to assess integration efficiencies in a. Details on threshold calculations are provided in Supplementary Note 1. In a and b, probes used in the original PASTE paper are compared side-by-side with probes used in this study. In the original PASTE report, probes bind to either the DNA donor plasmid (LMNB1, and NOLC1) or the Bxb1 attB sequence, which is also present in the pegRNA plasmid (ACTB). In this study, we primarily used probes that bind to the Bxb1 attL or attR sites, which are only present in cells after recombination. When using the DNA donor plasmid-binding probe reported in the PASTE paper, high background was observed in the negative controls at LMNB1, and NOLC1: no PEmax (+eeBxb1) and dead Bxb1 both showed false positive signals. In contrast, minimal or no background was observed at these sites when using an attL- binding probe. For PASSIGE and PASTE experiments, all components were delivered using single transfection, and a 4.5- kB donor DNA plasmid reported in the PASTE paper was used. The primer pair used for ddPCR was that reported in the PASTE paper for all reactions. All experiments were performed in HEK293T cells. The genome–donor junction probe was used in all experiments except for a few off-target sites, as detailed in Methods.