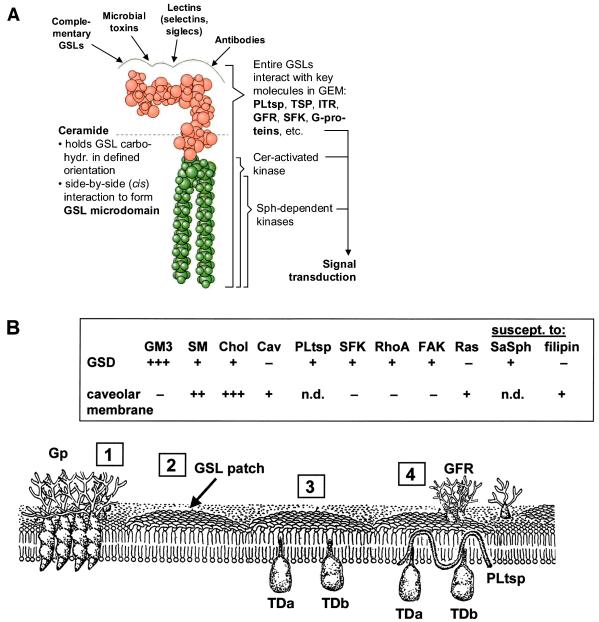
Figure 1.
GSL conformation and organization in membrane. (A) Minimum-energy model of GSL, Gb5 taken as an example, showing that the axis of carbohydrate moiety is perpendicular to the axis of ceramide (133, 134). The surface profile of carbohydrate provides binding sites for Abs, lectins, microbial toxins, and complementary GSLs. (Left) Role of ceramide (Cer) to form microdomain. (Right) Interaction of entire GSLs with key molecules (TSP, tetraspanin; ITR, integrin receptor; GFR, growth factor receptor; SFK, Src family kinase). These molecules, together with Cer or sphingosine (Sph) released from sphingomyelin, activate or modulate their respective kinases and are involved in various ways in control of signal transduction. (B) GSLs are clustered (“GSL patch”) and inserted via ceramide into the outer leaflet of the membrane, without (“2”) or with (“3”) signal transducers (TDa, TDb). GSL clusters organized with signal transducers, PL tetraspanin (PLtsp), and growth factor receptor (GFR) are shown in “4.” Glycoprotein (Gp) clusters (“1”) in many cases may be separated from GSL patches. (Inset) Contrasting properties of glycosignaling domain (GSD), the domain enriched in GSL, TD, and PL, separable from caveolar membrane. Data from refs. 20 and 57 and from K. Handa, D. A. Withers, and S.H. (unpublished data). Cav, caveolin; SaSph, sialyl 2 → 1 sphingosine.