Summary
Neutral lipids affect the immunosuppressive function of myeloid-derived suppressor cells (MDSCs). Here, we present a protocol for measuring neutral lipids in MDSCs using BODIPY from mouse mammary tumor derived from triple-negative breast cancer cells, 4T1, which is applicable to other mammary tumors of interest. We describe steps for 4T1 cell culture, single-cell isolation from tumors, staining of cells with antibodies and BODIPY, and flow cytometry. Furthermore, we introduce alternative protocols with MDSC sorting to overcome risk of cell death by BODIPY.
For complete details on the use and execution of this protocol, please refer to Kim et al.1
Subject areas: Cell-based Assays, Flow Cytometry, Cancer, Model Organisms, Antibody
Graphical abstract
Highlights
-
•
Establish the 4T1 syngeneic and orthotopic mouse model
-
•
Isolate single cells from mammary tumors followed by cell sorting for MDSCs
-
•
Analyze the lipid contents in cells of interest, including MDSCs
-
•
Alternative methods to improve viability of BODIPY+ MDSCs
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Neutral lipids affect the immunosuppressive function of myeloid-derived suppressor cells (MDSCs). Here, we present a protocol for measuring neutral lipids in MDSCs using BODIPY from mouse mammary tumor derived from triple-negative breast cancer, 4T1, which is applicable to other mammary tumors of interest. We describe steps for 4T1 cell culture, single-cell isolation from tumors, staining of cells with antibodies and BODIPY, and flow cytometry. Furthermore, we introduce alternative protocols with MDSC sorting to overcome risk of cell death by BODIPY.
Before you begin
Myeloid-derived suppressor cells (MDSCs) promote the progression and metastasis of tumors by suppressing cytotoxic T cells and facilitating the expansion of regulatory T cells.2 Fatty acids are stored in the form of neutral lipids, which is important for energy homeostasis.3 Accumulation of lipids can regulate immunosuppressive activity by MDSCs.4 To detect neutral lipids in tumor-infiltrating MDSCs, triple-negative breast cancer (TNBC) 4T1 mammary tumor from mammary fat pads injection were used. BALB/c mice were euthanized and harvested tumors were used for protocols. The protocol below describes the specific steps for using 4T1 tumors. We used antibodies to MDSCs with flow cytometry. Alternatively, we also introduced sorting MDSCs and directly stained sorted MDSCs with BODIPY.
Institutional permissions
Animal procedures were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) of the University of Miami. The permissions from the relevant institutions should be acquired to perform the protocol.
Mammary fat pad injection
Timing: 2.5 h
-
1.Preparation of 4T1 cells
-
a.Culture 4T1 cells in 4T1 Culture media.Note: Split 4T1 cells every 2–3 days when confluency reached 80–90%.Note: Use cells before passage 20 for injection.Note: 4T1 Culture media can be stored at 4°C for up to 1 month.
-
b.Wash 4T1 cells in 10 cm dish with 5 mL of PBS.
-
i.Detach the washed cells with 1 mL of trypsin-EDTA for 5 min in 37°C CO2 incubator.
-
ii.Spin down the detached cells by centrifugation at 500 g, 25°C for 3 min.
-
i.
-
c.Prepare 200K 4T1 cells/per mouse in PBS.Note: 50K cells in 10 μL of Matrigel/PBS (1:1) will be injected to left and right mammary fat pads, respectively.Note: Considering loss during procedure, prepare 200K cells/per mouse. (For example, prepare 1 M (106) cells for 5 mice, contralateral mammary fat pad injection).
-
d.Mix the cells in PBS with Matrigel at 1:1 ratio.Note: Injection volume per mammary fat pads will be 10 μL of Matrigel/PBS (1:1). (For example, mix 1 M resuspended cells in 100 μL of PBS and 100 μL of Matrigel for 5 mice).Note: Final concentration and volume for injection will be 5K cells/μL and 10 μL with 50K cells in Matrigel/PBS (1:1).
-
e.Add 1/100 volume of trypan blue to visualize the cells. (For example, add 2 μL trypan blue for 200 μL of Matrigel/PBS (1:1)).Note: Trypan blue is used to stain the Matrigel/PBS (1:1). It will help to check the injected cells are staying in the mammary fat pads without leaking out. Make sure to use sterile trypan blue.
-
f.Keep on ice. Directly proceed to next steps.Note: Cells can be kept on the ice for 2 h.
-
a.
-
2.Anesthesia and Analgesia
-
a.Prepare 30 μL of ketamine/xylazine combination per mouse.Note: Combination contains 66.6 mg/mL Ketamine and 6.6 mg/mL Xylazine. (For example, mix 200 μL of 100 mg/mL ketamine and 100 μL of 20 mg/mL xylazine to make 300 μL).Note: If dilution is needed, sterile 0.9% NaCl saline can be used.
-
b.Prepare 5 mg/mL meloxicam.Note: If dilution is needed, sterile 0.9% NaCl saline can be used.
-
c.Inject 30 μL of ketamine/xylazine combination intraperitoneally per 20 g mouse with Insulin syringe (28G, ½”).
-
d.After anesthesia takes effect in mice, inject 10 μL of meloxicam subcutaneously per 20 g mouse with Insulin syringe (28G, ½”).
-
a.
-
3.Surgery
-
a.Open the middle of abdomen skin in sagittal plain with autoclaved scissors and forceps.Note: Surgery should be performed in clean biosafety cabinet in animal room.
-
b.Detach the skin from abdominal walls to exposure right mammary fat pad.Note: Make sure not to damage the peritoneal cavity.Note: Bleeding may occur. Stop the bleeding with pressure or cauterization.
-
c.Wash the autoclaved 10 μL Hamilton syringe with 70% ethanol, autoclaved water, and PBS sequentially.
-
d.Take the 10 μL of 4T1 cells and inject into mammary fat pad (Figure 1).Note: 10 μL is recommended injection volume, but it can be optimized for the purpose of experiments.
 CRITICAL: Make sure needle is inside the mammary fad pad. Trypan blue staining will help to track the injected tumor cells. Check the tumor cells is staying inside. If tumor cells are leaking out, thoroughly clean with alcohol swab and try again.
CRITICAL: Make sure needle is inside the mammary fad pad. Trypan blue staining will help to track the injected tumor cells. Check the tumor cells is staying inside. If tumor cells are leaking out, thoroughly clean with alcohol swab and try again. -
e.Turn upside down and repeat with left mammary fat pad.
-
f.Close the wound with wound closure autoclips (Kent Scientific) or thread with needle.
-
g.Keep mice warm and monitor the mice until recovery from anesthesia.
 CRITICAL: Most deaths during surgery are due to failure to recover from anesthesia. Hypothermia is a major cause of death, so proper heating is necessary, and low-temperature burns caused by excessive heating should also be avoided.
CRITICAL: Most deaths during surgery are due to failure to recover from anesthesia. Hypothermia is a major cause of death, so proper heating is necessary, and low-temperature burns caused by excessive heating should also be avoided. -
h.Clean the Hamilton syringes with 70% alcohol and also autoclave it for next time.
-
a.
Figure 1.
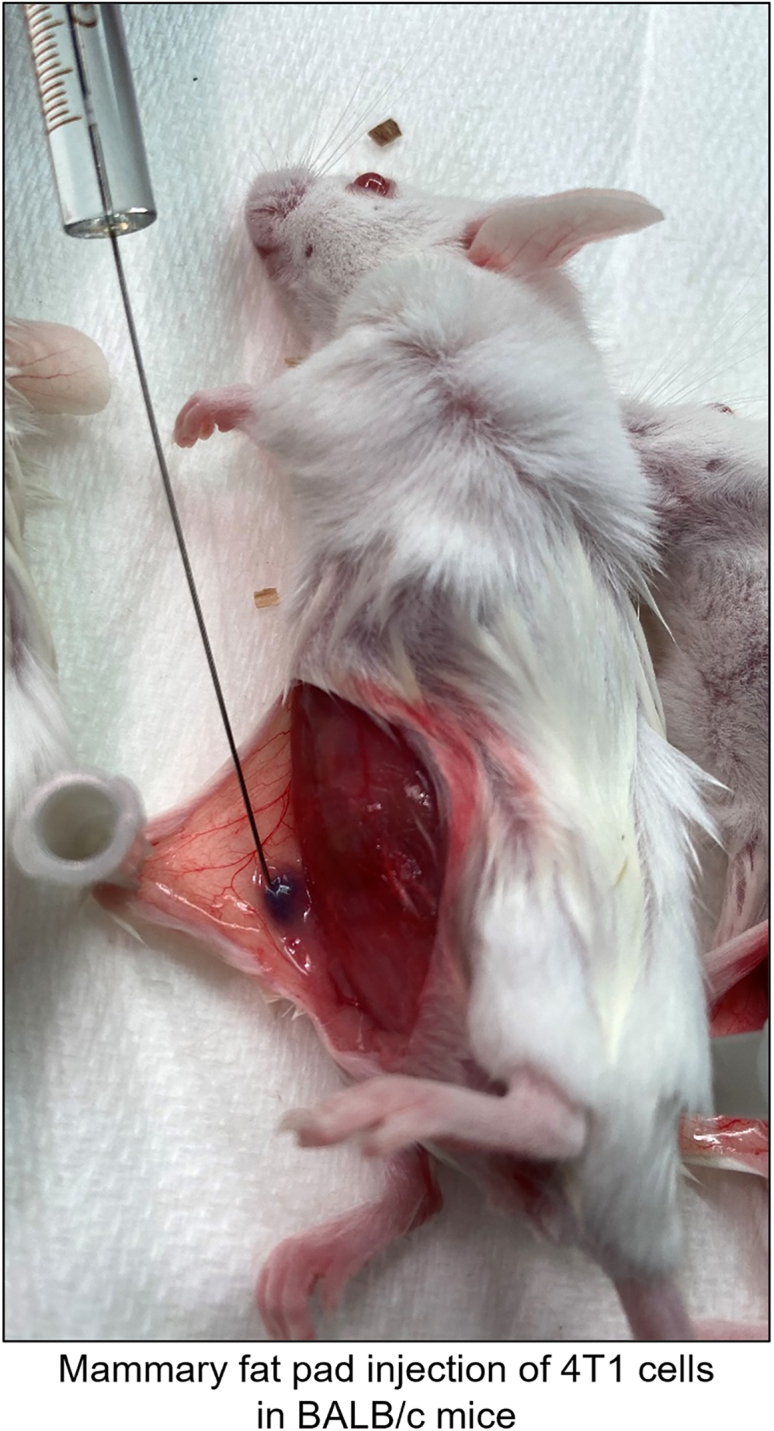
Injection of 4T1 TNBC cells into mammary fat pads of BALB/c mice
After anesthesia, skin adjacent to 4th mammary fat pads was opened in sagittal plain in the clean biosafety cabinet in DVR facility and 4T1 cells were injected into the right 4th mammary fat pad. 4T1 cells were in PBS with Matrigel at 1:1 ratio. Final concentration and volume for injection will be 5K cells/μL and 10 μL with 50K cells in Matrigel/PBS (1:1). Trypan blue (1:100 ratio) staining helped to track the inoculated cells.
Monitoring and harvest
Timing: 3–4 weeks
-
4.Monitoring
-
a.Check the clinical symptoms including weight loss, lethargy, ulceration, hunched back, and etc. twice a week.
-
b.Measure the tumor size every 3 days.
-
a.
Note: Using a caliper, measure the length (L) and width (W) of tumor.
Note: Tumor size can be calculated as (L)X(W)2 X(Pi)/6.
-
5.Harvest
-
a.Prepare harvest when tumor reaches desirable volume (approximately 1 × 1 cm or humane endpoint).Note: For example, 2 cm of length can be suggested as humane endpoint.
-
b.Euthanize the mice with CO2 followed by cervical dislocation, which is a second method to confirm euthanasia.
-
c.Open the abdomen with autoclaved scissors and forceps to evaluate the gross tumor morphology (Figure 2).
-
d.Resect the tumors and keep in cold PBS in a dish (Figure 3).
-
e.Chop the tumor and make chunks (approximately 1 mm3) on sterile 60 mm petri dish.
-
i.Grab a big tumor with forceps and cut with scissors.
-
ii.Then, hold a sterile scalpel in each hand and chop the tumor by crossing the blades (Figure 3).Note: Tumor chunks can be saved in Tumor Freezing Media in Mr. frosty. Freeze chunks slowly until −80°C and transfer to the liquid nitrogen tank subsequently.Note: Tumor Freezing Media should be made fresh.
-
i.
-
a.
Figure 2.

Harvesting 4T1 TNBC tumor after euthanasia of mice
BALB/c mice were euthanized by cervical dislocation and confirmed by completely disconnecting the cervical spine. Skin adjacent to 4th mammary gland was cut in sagittal plain with sterile forceps and scissors. 4T1 tumors were harvested by blunt dissection with sterile forceps and scissors in the clean biosafety cabinet in the necropsy room. Image showed tumor in 4th mammary gland position (yellow line). Scale bar, 1 cm.
Figure 3.
Isolation of mammary tumors into single cells
Tumors are minced and digested with Mammary gland digestion media (MGDM) for 45–50 min in 37°C water bath. After spinning down at 500 g, 4°C for 3 min, tumors are incubated with 0.25% trypsin-EDTA, Dispase/DNase I solution, and 0.64% NH4Cl sequentially in 37°C water bath. Between each digestion, flow buffer with serum was added to stop the digestion and spun down to remove supernatant.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE/Cy7 anti-CD45 antibody (1:50) | BD Biosciences | Cat#552848, RRID:AB_394489 |
| PerCP/Cy5.5 anti-CD11b antibody (1:50) | BD Biosciences | Cat#550993, RRID:AB_394002 |
| APC/Cy7 anti-GR1 antibody (1:50) | BD Biosciences | Cat#557661, RRID:AB_396775 |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM | Thermo Fisher Scientific | Cat#11960-051 |
| PBS | Thermo Fisher Scientific | Cat#10010-023 |
| Fetal bovine serum/FBS | GeminiBio | Cat#900-108 |
| Penicillin/streptomycin | Thermo Fisher Scientific | Cat#15140-122 |
| Insulin | Sigma-Aldrich | Cat#I9278 |
| Collagenase from Clostridium histolyticum | Sigma-Aldrich | Cat#C2674 |
| Hyaluronidase | Sigma-Aldrich | Cat#H3884 |
| DMEM/F-12 | Thermo Fisher Scientific | Cat#11330-032 |
| Gentamicin | Thermo Fisher Scientific | Cat#15750-060 |
| Cholera toxin from Vibrio cholerae | Sigma-Aldrich | Cat#C8052 |
| Hydrocortisone | Sigma-Aldrich | Cat#H0888 |
| Trypsin-EDTA | Thermo Fisher Scientific | Cat#25200-056 |
| Dispase | Thermo Fisher Scientific | Cat#100-10-023 |
| DNase I | Sigma-Aldrich | Cat#D5025 |
| Fetal calf serum/FCS | GeminiBio | Cat#100-504 |
| Ammonium chloride | Calbiochem | 168320 |
| Recombinant human epidermal growth factor/EGF | Novoprotein | Cat#C029 |
| DAPI | Sigma-Aldrich | Cat#D9542 |
| BODIPY | Thermo Fisher Scientific | Cat#D3922 |
| UltraComp eBeads Plus compensation beads | Thermo Fisher Scientific | Cat#01-3333-42 |
| Dimethyl sulfoxide/DMSO | MP Biomedicals | Cat#219605590 |
| Trypan blue | Sigma-Aldrich | Cat#T8154-100ML |
| Experimental models: Cell lines | ||
| 4T1/LIG murine TNBC cells | Laboratory of Dr. Yibin Kang | N/A |
| Experimental models: Organisms/strains | ||
| BALB/c Mus musculus, 4–5 weeks old, female | The Jackson Laboratory | Cat#000651 RRID:IMSR_JAX:000651 |
Materials and equipment
4T1 Culture Media
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM | N/A | 1,000 mL |
| Insulin (10 mg/mL) | 10 μg/mL | 1 mL |
| FBS | 10% | 100 mL |
| Penicillin/Streptomycin (10,000 U/mL) | 100 U/mL | 10 mL |
| Total | N/A | 1,111 mL |
Store at 4°C for up to 1 month.
Mammary Epithelial Cell Growth Media (MEGM)
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/F-12 | N/A | 500 mL |
| Insulin (10 mg/mL) | 5 μg/mL | 250 μL |
| Hydrocortisone (1 mg/mL) | 500 ng/mL | 250 μL |
| EGF (100 μg/mL) | 10 ng/mL | 50 μL |
| Cholera toxin (200 μg/mL) | 20 ng/mL | 50 μL |
| FBS | 5% | 25 mL |
| Gentamicin (50 mg/mL) | 50 μg/mL | 500 μL |
| Total | N/A | 526.1 mL |
Filter with 0.2 μm bottle top filtration device. Store at 4°C for up to 1 month.
Tumor Freezing Media
| Reagent | Final concentration | Amount |
|---|---|---|
| MEGM | N/A | 25 mL |
| DMSO | 10% | 5 mL |
| FBS | 40% | 20 mL |
| Total | N/A | 50 mL |
Make fresh.
Mammary Gland Dissociation Media (MGDM)
| Reagent | Final concentration | Amount |
|---|---|---|
| MEGM | N/A | 50 mL |
| Collagenase | 300 U/mL | depending on U/g |
| Hyaluronidase | 100 U/mL | depending on U/g |
| Total | N/A | 50 mL |
Make fresh. Sterilize using a 0.45 μm syringe filter.
Dispase/DNase I solution
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | N/A | 40 mL |
| Dispase | 2.5 U/mL | depending on U/g |
| DNase I | 200 U/mL | depending on U/g |
| Total | N/A | 40 mL |
Sterilize using a 0.45 μm syringe filter. Store at 4°C for up to 2 weeks.
0.64% NH4Cl
| Reagent | Final concentration | Amount |
|---|---|---|
| Milli-Q water | N/A | 1 L |
| NH4Cl | 0.64% | 6.4 g |
| Total | N/A | 1 L |
Autoclave. Store at 4°C for up to 3 months.
Flow buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | N/A | 500 mL |
| FCS | 5% | 25 mL |
| Gentamicin (50 mg/mL) | 50 μg/mL | 500 μL |
| Total | N/A | 525.5 mL |
Filter with 0.2 μm bottle top filtration device. Store at 4°C for up to 1 month.
Flow buffer with DAPI
| Reagent | Final concentration | Amount |
|---|---|---|
| Flow buffer | N/A | 50 mL |
| DAPI (1 mg/mL) | 333 ng/mL | 16.7 μL |
| Total | N/A | 50 mL |
Store at 4°C for up to 1 month.
BODIPY stock
| Reagent | Final concentration | Amount |
|---|---|---|
| DMSO | N/A | 1.63 mL |
| BODIPY | 5 mM | 10 mg |
| Total | N/A | 1.63 mL |
Aliquot and store at −20°C up to 6 months.
Step-by-step method details
Single cell isolation with 4T1 tumor
Timing: 2 h
This step makes the single cell suspension from 4T1 tumor enabling the staining with antibodies and BODIPY (Figure 3).
Note: Depending on tumor type, incubation time and concentration of digesting reagents may need to be optimized.
-
1.Tissue digestion.
-
a.Place the 1 mm3 each 4–5 tumor chunks in 60 mm dish.
-
b.Mince the chunks with 2 scalpels.
-
i.Hold a sterile scalpel in each hand and mince the tumor by crossing the blades.
-
i.
-
c.Take 20 mL of MGDM with 25 mL pipette.Note: MEGM can be stored at 4°C for up to 1 month.Note: MGDM should be made fresh.
-
d.Collect minced tumor with 25 mL pipette and transfer to 50 mL conical tube.
-
e.Keep the tube on ice and repeat with next samples.
 CRITICAL: Keeping tube at 25°C will affect digestion and cell viability.
CRITICAL: Keeping tube at 25°C will affect digestion and cell viability. -
f.Incubate tubes in 37°C water bath for 45–50 min and shake it with hands every 10 min.
 CRITICAL: Over incubation with MGDM will affect viability.
CRITICAL: Over incubation with MGDM will affect viability. -
g.Centrifuge at 500 g, 4°C for 3 min.
-
a.
-
2.Single cell isolation
-
a.Remove supernatant and add 1 mL 0.25% trypsin-EDTA.
-
b.Incubate tubes in 37°C water bath for 90 s.
 CRITICAL: Over incubation with trypsin is detrimental to cells.
CRITICAL: Over incubation with trypsin is detrimental to cells. -
c.Add 5 mL of flow buffer and centrifuge at 500 g, 4°C for 3 min.Note: Flow buffer can be stored at 4°C for up to 1 month.
-
d.Remove supernatant and add 5 mL Dispase/DNase I solution.Note: Dispase/DNase I solution can be stored at 4°C for up to 2 weeks.
-
e.Incubate tubes in 37°C water bath for 5 min.
-
f.Add 5 mL of flow buffer and centrifuge at 500 g, 4°C for 3 min.
-
g.Remove supernatant and add 5 mL 0.64% NH4Cl.Note: 0.64% NH4Cl can be stored at 4°C for up to 3 months.
-
h.Incubate tubes in 37°C water bath for 5 min.
-
i.Add 5 mL of flow buffer and centrifuge at 500 g, 4°C for 3 min.
-
j.Remove supernatant and add 1 mL of flow buffer.
-
k.Pipette 10 times with 1000P pipette and filter with 40 μm cell strainer
-
l.Count cells on a hemocytometer with Trypan blue.
-
a.
Staining cells with antibodies and BODIPY
Timing: 1.5 h
Antibodies to stain MDSC will be used first. Labeled cells will be stained with BODIPY to measure neutral lipid contents in MDSC populations.
-
3.Preparing control for compensation
-
a.For unstained control, take small portion (∼500K cells) from sample and resuspend unstained cells in 500 μL flow buffer. Keep on the ice.
-
b.For DAPI control, take small portion (∼500K cells) from sample and resuspend unstained cells in 500 μL flow buffer with DAPI. Block the light and keep on the ice.Note: Flow buffer with DAPI can be stored at 4°C for up to 1 month.
-
c.For BODIPY control, take small portion (∼500K cells) from sample and stain them with BODIPY as shown below and resuspend in 500 μL flow buffer. Keep on the ice.
-
d.Vortex UltraComp eBeads Plus Compensation Beads well before using.
-
e.Aliquot 1 drop of beads for 3 color controls.
-
f.Add 1 μL of PE/Cy7 Anti-CD45 Antibody, 1 μL of PerCP/Cy5.5 Anti-CD11b Antibody, and 1 μL of APC/Cy7 Anti-GR1 Antibody in each color controls.
-
g.Mix well and incubate tubes at 25°C for 30 min in the dark.
-
h.Add 1 mL flow buffer and centrifuge at 500 g, 4°C for 3 min.
-
i.Remove supernatant and resuspend 500 μL flow buffer. Block the light and keep on the ice.
-
a.
-
4.Antibody staining.
-
a.Prepare 50 μL of cocktail per ∼500K cells with PE/Cy7 Anti-CD45 Antibody, PerCP/Cy5.5 Anti-CD11b Antibody, and APC/Cy7 Anti-GR1 Antibody (1:50 dilution for each antibody in flow buffer).
-
b.Centrifuge the filtered samples at 500 g, 4°C for 3 min.
-
c.Remove the supernatant and add 50 μL cocktail of antibody (1:50) per ∼500K cells for panel.Note: Required volume of cocktail may vary depending on the cell number.
-
d.Mix well and incubate tubes at 25°C for 30 min in the dark.
-
e.Add 1 mL flow buffer and centrifuge at 500 g, 4°C for 3 min.
-
a.
-
5.BODIPY staining.
-
a.Dilute 5 mM BODIPY stock with sterile Milli-Q water at 1:2500 ratio to make 2 μM BODIPY working solution.Note: BODIPY stock can be stored at −20°C for up to 6 months.
-
b.Add 200 μL BODIPY working solution per ∼500K cells in the cell pellet.
-
c.Incubate the tube in 37°C for 30 min.
 CRITICAL: Over incubation will affect the viability of the cells.
CRITICAL: Over incubation will affect the viability of the cells. -
d.After 30 min, add 1 mL flow buffer per ∼500K cells to the cells.
-
e.Centrifuge at 500 g, 4°C for 3 min and discard supernatant.
-
f.Resuspend cells with 500 μL of flow buffer with DAPI.
-
a.
Sorting MDSCs and BODIPY staining
Timing: 2.5 h
BODIPY staining in 37°C might be detrimental to cells of interest. Alternatively, MDSCs can be sorted with BD Aria Fusion first and stained with BODIPY to improve viability (Figure 4). Alternative flow cytometric machine can be applied.
-
6.Preparing control for compensation.
-
a.For unstained control, take small portion (∼500K cells) from sample and resuspend unstained cells in 500 μL flow buffer. Keep on the ice.
-
b.For DAPI control, take small portion from sample (∼500K cells) and resuspend unstained cells in 500 μL flow buffer with DAPI. Block the light and keep on the ice.
-
c.For BODIPY control, take small portion (∼500K cells) from sample and stain them with BODIPY as shown below and resuspend in 500 μL flow buffer. Keep on the ice.Note: BODIPY control will be used for analysis of BODIPY+ MDSCs.Note: DAPI control can be kept after sorting and reused in analysis of BODIPY+ MDSCs.
-
d.Vortex UltraComp eBeads Plus Compensation Beads well before using.
-
e.Aliquot 1 drop of beads for 3 color controls.
-
f.Add 1 μL of PE/Cy7 Anti-CD45 Antibody, 1 μL of PerCP/Cy5.5 Anti-CD11b Antibody, and 1 μL of APC/Cy7 Anti-GR1 Antibody in each color controls.
-
g.Mix well and incubate tubes at 25°C for 30 min in the dark.
-
h.Add 1 mL flow buffer and centrifuge at 500 g, 4°C for 3 min.
-
i.Remove supernatant and resuspend 500 μL flow buffer. Block the light and keep on the ice.
-
a.
-
7.Antibody staining.
-
a.Prepare 50 μL of cocktail per ∼500K cells with PE/Cy7 Anti-CD45 Antibody, PerCP/Cy5.5 Anti-CD11b Antibody, and APC/Cy7 Anti-GR1 Antibody (1:50 dilution for each antibody in flow buffer).
-
b.Centrifuge the filtered samples at 500 g, 4°C for 3 min.
-
c.Remove the supernatant and add 50 μL cocktail of antibody (1:50) per ∼500K cells for panel.Note: Required volume of cocktail may vary depending on the cell number.
-
d.Mix well and incubate tubes at 25°C for 30 min in the dark.
-
e.Add 1 mL flow buffer and centrifuge at 500 g, 4°C for 3 min.
-
a.
-
8.MDSCs sorting.
-
a.Turn on the BD Aria Fusion and start BD FACSDiva Software. Create new experiment and add PE/Cy7 (CD45), PerCP/Cy5.5 (CD11b), APC/Cy7 (GR1), and DAPI panels.
-
b.Perform compensation and calculate compensation.
-
c.Draw the gate as shown in Figure 5.Note: P2 gate (95.9%) comes from P1 gate (37.9%). Live gate (47.6%) comes from P2 gate. CD45+ gate (7.32%) comes from Live gate. MDSC gate (14.1%) comes from CD45+ gate.Note: CD45 was used for immune cell marker. CD11b was used for myeloid cell marker and GR1 was used for granulocytes and myeloid differentiation marker.Note: Percentage of each population may vary according to samples.
-
d.Sort MDSCs.Note: 100 μm sorter nozzles will be helpful to reduce sort pressure.
-
a.
-
9.BODIPY staining.
-
a.Dilute 5 mM BODIPY stock with sterile Milli-Q water at 1:2500 ratio to make 2 μM BODIPY working solution.
-
b.Spin down the sorted MDSCs at 500 g, 4°C for 3 min and discard supernatant.
-
c.Add 200 μL BODIPY working solution per ∼500K cells in the cell pellet.
-
d.Incubate the tube in 37°C for 30 min.
 CRITICAL: Over incubation will affect the viability of the cells.
CRITICAL: Over incubation will affect the viability of the cells. -
e.After 30 min, add 1 mL flow buffer per ∼500K cells to the cells.
-
f.Centrifuge at 500 g, 4°C for 3 min and discard supernatant.
-
g.Resuspend cells with 500 μL of flow buffer with DAPI.
-
a.
Figure 4.
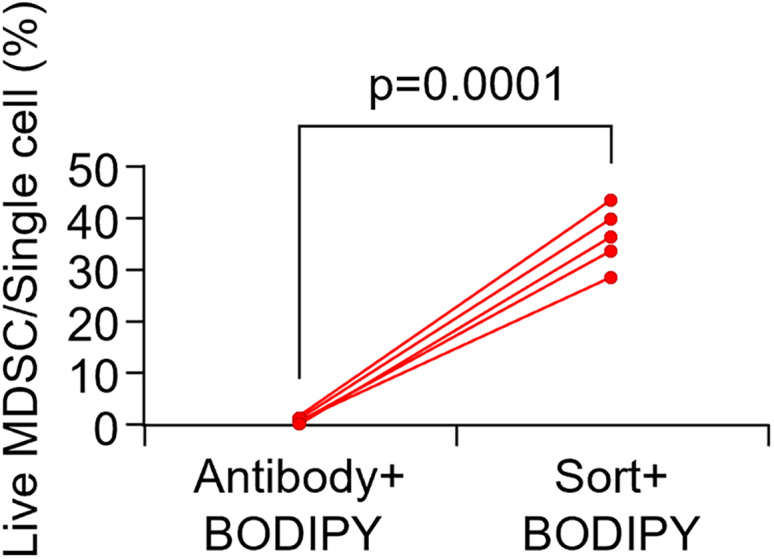
Sorting and BODIPY staining improved viability of MDSCs
After single cell isolation from tumor, single cell suspension was divided and subjected different steps to compare viability of MDSCs. The half of the single cell suspension was stained with antibodies and BODIPY. The other half of the single cell suspension was sorted with BD Aria Fusion for MDSCs followed by staining with BODIPY. Single cell suspension was analyzed with BD Fortessa and ratio of live MDSCs to single cell population (P2 gate) were compared. Sorted MDSCs showed improved viability compared to unsorted MDSCs together with BODIPY, n = 5 tumors/group. Data are presented as each sample connected with corresponding counterparts. p values were calculated using two-tailed paired Student’s t-test.
Figure 5.
Gating strategy for MDSC sorting post mammary tumor cell digestion
Stained single cell suspension was analyzed with BD Fortessa. P1 population was gated with forward scatter-area (FSC-A) and side scatter-area (SSC-A) to exclude debris. P2 populations was selected with SSC-high (SSC-H) and SSC-A to select single cell populations. P2 populations were subjected with DAPI for live DAPI- cells. CD45+ immune cells populations were gated with CD45. CD11b and GR1 were used to gate double positive MDSC populations.
Flow cytometry to measure BODIPY+ MDSCs
Timing: 1 h
Stained cells will be analyzed with BD Fortessa. Alternative flow cytometric machine can be applied.
-
10.Setup compensation and gate.
-
a.Create new experiment and add PE/Cy7 (CD45), PerCP/Cy5.5 (CD11b), APC/Cy7 (GR1), DAPI, and FITC (BODIPY) panels.
-
b.Perform compensation and calculate compensation.
-
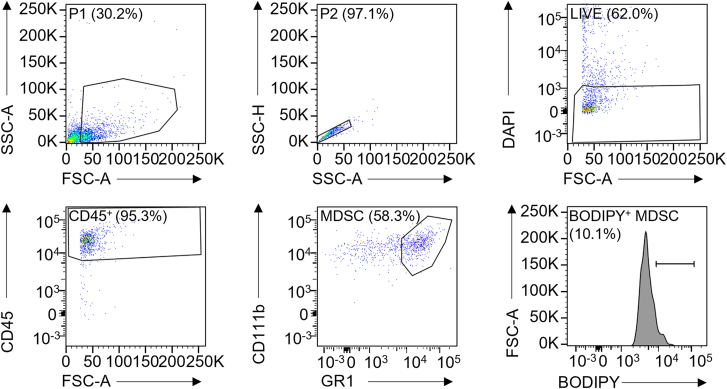
c.Draw the gate as shown in Figure 6.Note: P2 gate (97.1%) comes from P1 gate (30.2%). Live gate (62.0%) comes from P2 gate. CD45+ gate (95.3%) comes from Live gate. MDSC gate (58.3%) comes from CD45+ gate. BODIPY+ MDSC gate (10.1%) comes from MDSC gate.Note: CD45 was used for immune cell marker. CD11b was used for myeloid cell marker and GR1 was used for granulocytes and myeloid differentiation marker.
-
d.Collect all sample and analyze BODIPY+ MDSC.
-
a.
Figure 6.
Gating strategy for BODIPY staining in MDSCs in flow cytometry
Stained single cell suspension was analyzed with BD Fortessa. P1 population was gated with forward scatter-area (FSC-A) and side scatter-area (SSC-A) to exclude debris. P2 populations was selected with SSC-high (SSC-H) and SSC-A to select single cell populations. P2 populations were subjected with DAPI for live DAPI- cells. CD45+ immune cells populations were gated with CD45. CD11b and GR1 were used to gate double positive MDSC populations. BODIPY+ MDSCs were gated with FITC channel for BODIPY.
Expected outcomes
Herein, we describe protocol to measure lipid metabolism of MDSCs. This protocol also can be used for human or mouse cells with specific antibodies and flow cytometry. As shown in Figure 6, the proportion of BODIPY+ MDSCs can be analyzed which have higher neutral lipid contents. Neutral lipids are utilized as a storage of fatty acids,3 and lipid accumulation is related to the suppressive activity of MDSCs.4 Research targeting lipid metabolism of MDSCs has been proposed, and BODIPY staining will be a useful technique for evaluating the efficacy of targeting lipid accumulation of MDSCs.
Limitations
The protocol described here can detect neutral lipid in cells of interest. However, this protocol is limited in that component of neutral lipids such as cholesteryl esters, triacylglycerol, and free fatty acids cannot be distinguished. Commercial assay kit will be one option to further examine the neutral lipid components. The advantage of BODIPY is that BODIPY is more specific for cellular lipid droplets than Nile Red.5 However, BODIPY is using FITC channel which is not compatible with tissue expressing GFP. We suggest alternative dyes like Nile Red, HCS LipidTOX will be good option to detect neutral lipid in GFP expressing tissues.
Troubleshooting
Problem 1
Failed establishment of 4T1 tumor (step 1 of before you begin).
Improper 4T1 cell culture can lead to failure of tumor establishment in BALB/c. Mycoplasma infection of 4T1 cells can have unexpected impacts on tumor growth in mice.
Potential solution
It is recommended to screen quarterly using a commercial Mycoplasma detection kit. If Mycoplasma infection occurs, treat anti-Mycoplasma drugs such as Plasmocin according to the manufacturer’s protocol.
Problem 2
Low viability of cells (step 1f and 2b of step-by-step method details).
Inadequate optimization in tissue digestion and single cell isolation may result in low viability of cells. Necrotic tumor tissues also can lead to low viability. BODIPY incubation at 37°C might be detrimental to cells.
Potential solution
We recommend optimization of tissue digestion and single cell isolation steps before experiments. Too long incubation with MGDM and trypsin-EDTA can compromise the cell viability. We recommend testing with multiple incubation time and check the viability. To avoid necrotic tissues, mouse tumor should be harvested in proper time depending on purpose of experiments. You may start to harvest when tumor reaches 1 × 1 cm. Monitor the mice at least twice a week and check if tumors have wound or ulceration. Additionally, depending on the mouse model, necrosis of old tumors may begin in deep area of the tumor even without external contact. Therefore, setting an appropriate endpoint is required. To overcome damage from BODIPY incubation at 37°C, we introduced alternative method that MDSCs can be sorted first and stained with BODIPY.
Problem 3
Sticky pellets during digestion (step 2d and 2e of step-by-step method details).
Incomplete elimination of DNA forms sticky pellets. During aspiration of media from the supernatant, sticky pellets are prone to be removed together loosing big part of cell pellets. Furthermore, sticky pellets make it difficult to obtain single cells.
Potential solution
Check that Dispase/DNase I solution is not expired. If expired, prepare fresh solution. DNase I from different batch may have different U/g. Follow correct U/mL not g/mL.
Problem 4
Suboptimal BODIPY staining (step 5 and 9 of step-by-step method details)
BODIPY staining may need optimization. The multiple concentration of BODIPY working solution and incubation time can be tested to find better conditions for experiment.
Potential solution
We suggested protocol with 2 μM BODIPY working solution with 30 min incubation. If staining is not significant, the concentration BODIPY working solution can be increased up to 10 μM. Furthermore, cells can be incubated for minimum 5 min to minimize to detrimental effects of BODIPY staining.
Problem 5
Gating issues with flow cytometry (step 8 and 10 of step-by-step method details).
Suboptimal staining with antibody may lead to incomplete staining of cells.
Improper compensation can affect readout of the flow cytometry.
Potential solution
Make sure that enough antibodies are used for numbers. The amount of antibodies should be increased for high numbers of cells. Antibodies may need optimization of concentrations. Compensation should be performed by experienced personnel to minimize the interferences by fluorochromes. Forward scatter and side scatter should be optimized for better discrimination of populations.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Rumela Chakrabarti (rxc1335@med.miami.edu).
Technical contact
Technical questions on executing this protocol should be directed to and will be answered by the technical contact, Rumela Chakrabarti (rxc1335@med.miami.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate/analyze datasets.
Acknowledgments
We thank the members of the Flow Cytometry Shared Resource (FCSR: SCR_022501) at Sylvester Comprehensive Cancer Center of University of Miami. This work was supported by a grant from NCI-R01 (R01CA237243) to R.C.
Author contributions
U.K. performed all the experiments. U.K. and R.C. wrote the manuscript. All authors discussed the protocol and commented on the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Kim U., Debnath R., Maiz J.E., Rico J., Sinha S., Blanco M.A., Chakrabarti R. ΔNp63 regulates MDSC survival and metabolism in triple-negative breast cancer. iScience. 2024;27 doi: 10.1016/j.isci.2024.109366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Umansky V., Blattner C., Gebhardt C., Utikal J. The role of myeloid-derived suppressor cells (MDSC) in cancer progression. Vaccines. 2016;4:36. doi: 10.3390/vaccines4040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turkish A.R., Sturley S.L. The genetics of neutral lipid biosynthesis: an evolutionary perspective. Am. J. Physiol. Endocrinol. Metab. 2009;297:E19–E27. doi: 10.1152/ajpendo.90898.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adeshakin A.O., Liu W., Adeshakin F.O., Afolabi L.O., Zhang M., Zhang G., Wang L., Li Z., Lin L., Cao Q., et al. Regulation of ROS in myeloid-derived suppressor cells through targeting fatty acid transport protein 2 enhanced anti-PD-L1 tumor immunotherapy. Cell. Immunol. 2021;362 doi: 10.1016/j.cellimm.2021.104286. [DOI] [PubMed] [Google Scholar]
- 5.Gocze P.M., Freeman D.A. Factors underlying the variability of lipid droplet fluorescence in MA-10 Leydig tumor cells. Cytometry. 1994;17:151–158. doi: 10.1002/cyto.990170207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze datasets.