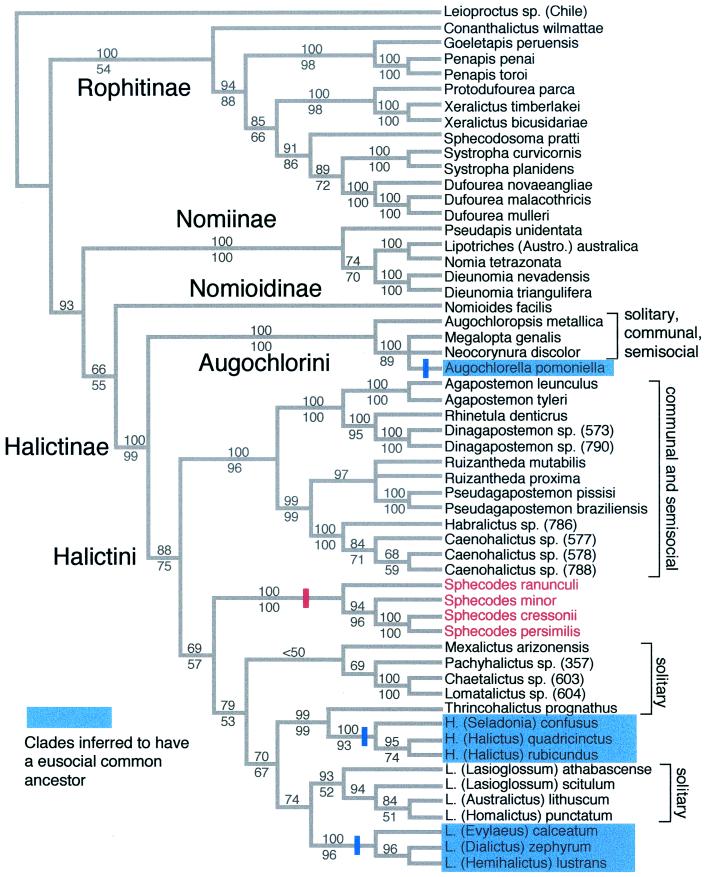
Figure 1.
Phylogeny of the halictid subfamilies, tribes, and genera. Strict consensus of six trees based on equal weights parsimony analysis of the entire data set (spanning positions 145-1271 in the coding region of the Apis mellifera F2 copy). The data set includes three exons and two introns. Two regions within the introns were excluded because they could not be aligned unambiguously. Gaps coded as a fifth state or according to the methods described in ref. 23 yielded the same six trees. Bootstrap values above the nodes indicate bootstrap support based on the exons + introns data set. Bootstrap values below the nodes indicate support based on an analysis of exons only. For the exons + introns analysis the data set included 1,541 total aligned sites (619 parsimony-informative sites), the trees were 3,388 steps in length, and the consistency index (excluding uninformative sites) was 0.3831. For the exons-only analysis there were 1,127 aligned sites (327 parsimony-informative sites), the trees were 1,648 steps in length, and the consistency index (excluding uninformative sites) was 0.3195. The tree was rooted by using a colletid bee (Leioproctus sp.). Maximum-likelihood analyses using the K2P model with rate heterogeneity accounted for by site-specific rates yielded the same overall tree topology irrespective of whether exons or exons + introns were analyzed. Clades inferred to have a eusocial common ancestor (see text) are shown in blue. Cleptoparasitic taxa (Sphecodes) are shown in red. Three origins of eusociality (indicated by blue bars) are indicated on the cladogram.