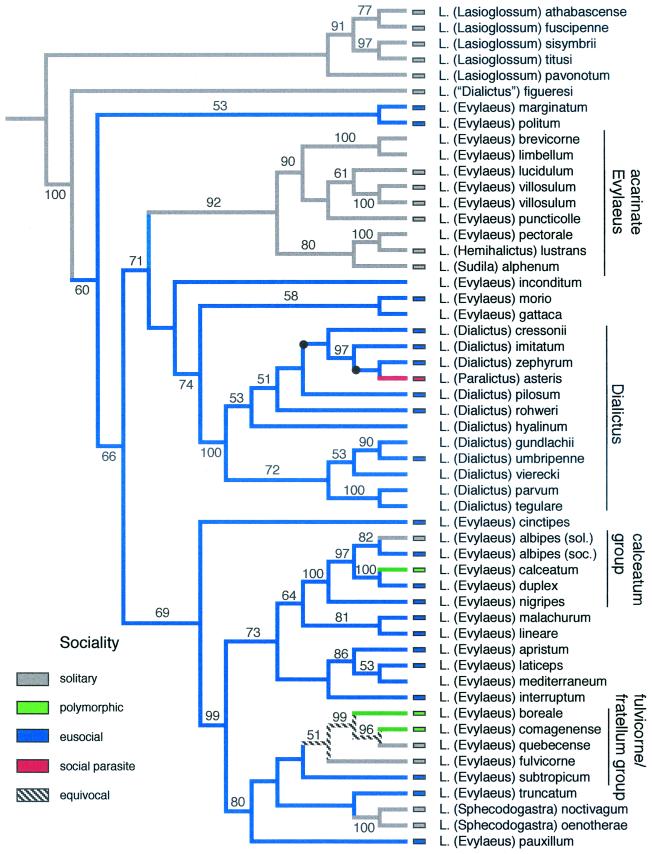
Figure 3.
Phylogeny of the eusocial clade of Lasioglossum. One of six trees based on equal weights parsimony analysis of the combined cytochrome oxidase I (COI) (24) and elongation factor-1α (25) data set. (Nodes that collapse in the strict consensus are marked with a black dot and do not alter the character mapping.) Third position sites in COI were excluded (downweighting yielded the same overall tree topologies) because this data partition showed a highly skewed base composition (91% A/T), significant base compositional heterogeneity among taxa (P ≤ 0.001), and an 8-fold higher rate of substitution than any of the other six data partitions. The total data set included 2,734 aligned nucleotide sites (2,321 with COI nucleotide 3 excluded; 394 parsimony informative sites). Trees were 1,765 steps in length and the consistency index was 0.3669. The tree was rooted by using five species of Lasioglossum s.s. The tree recovers several of the recognized subgenera of Lasioglossum (including Sphecodogastra and Dialictus) as well as many of the species groups of Evylaeus. Social behavior and parasitism (obtained from published studies listed in ref. 37) were mapped onto the tree by using macclade version 3.07 (27). Eusociality arose once in the common ancestor of the ingroup species excluding L. (“Dialictus”) figueresi. Reversals to either solitary nesting or social polymorphism (eusocial and solitary populations of the same species) occurred as many as five times. Social parasitism (in the subgenus Paralictus) is inferred to have arisen once. Methods are outlined in the text.