Extended Data Fig. 2 |. Classification of IS605-family elements encoded by G. stearothermophilus strain DSM 458.
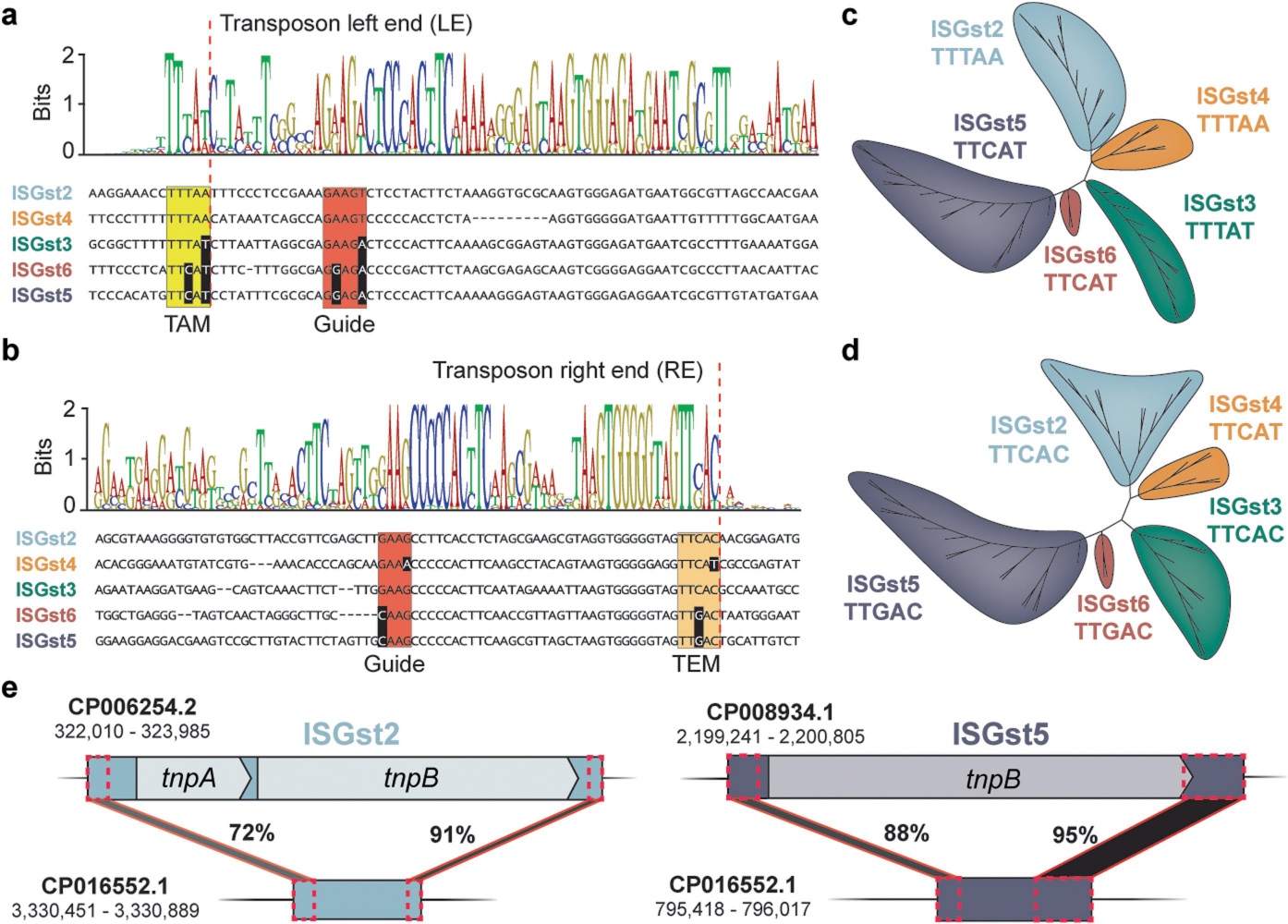
a, DNA multiple sequence alignment of transposon left ends (LE) for IS200/IS605-family elements from G. stearothermophilus. The weblogo (top) is built from 47 unique elements (Supplementary Figs. 2 & 3), and one representative sequence from each family is shown below, with the TAM highlighted in yellow and DNA guide sequences highlighted in red. Nucleotides highlighted in black exhibit covarying mutations, relative to ISGst2. TAM, transposon-adjacent motif; the dotted red line indicates the transposon boundary. b, DNA multiple sequence alignment of transposon right ends (RE) for IS200/IS605-family elements from G. stearothermophilus, shown as in a. TEM, transposon-encoded motif is shown in orange. c, Phylogenetic tree of ISGst elements based on the transposon left end. Each colored clade encodes an associated TnpB/IscB protein homolog and is flanked by the indicated TAM sequence. d, Phylogenetic tree of ISGst elements based on the transposon right end, shown as in b but with TEM sequence in lieu of TAM. e, Schematic of PATEs (palindrome-associated transposable elements) related to ISGst2 and ISGst5, which contain similar transposon ends but no protein-coding genes. The percent sequence identity between shaded regions (black) is shown, as are the genomic accession IDs and coordinates.
